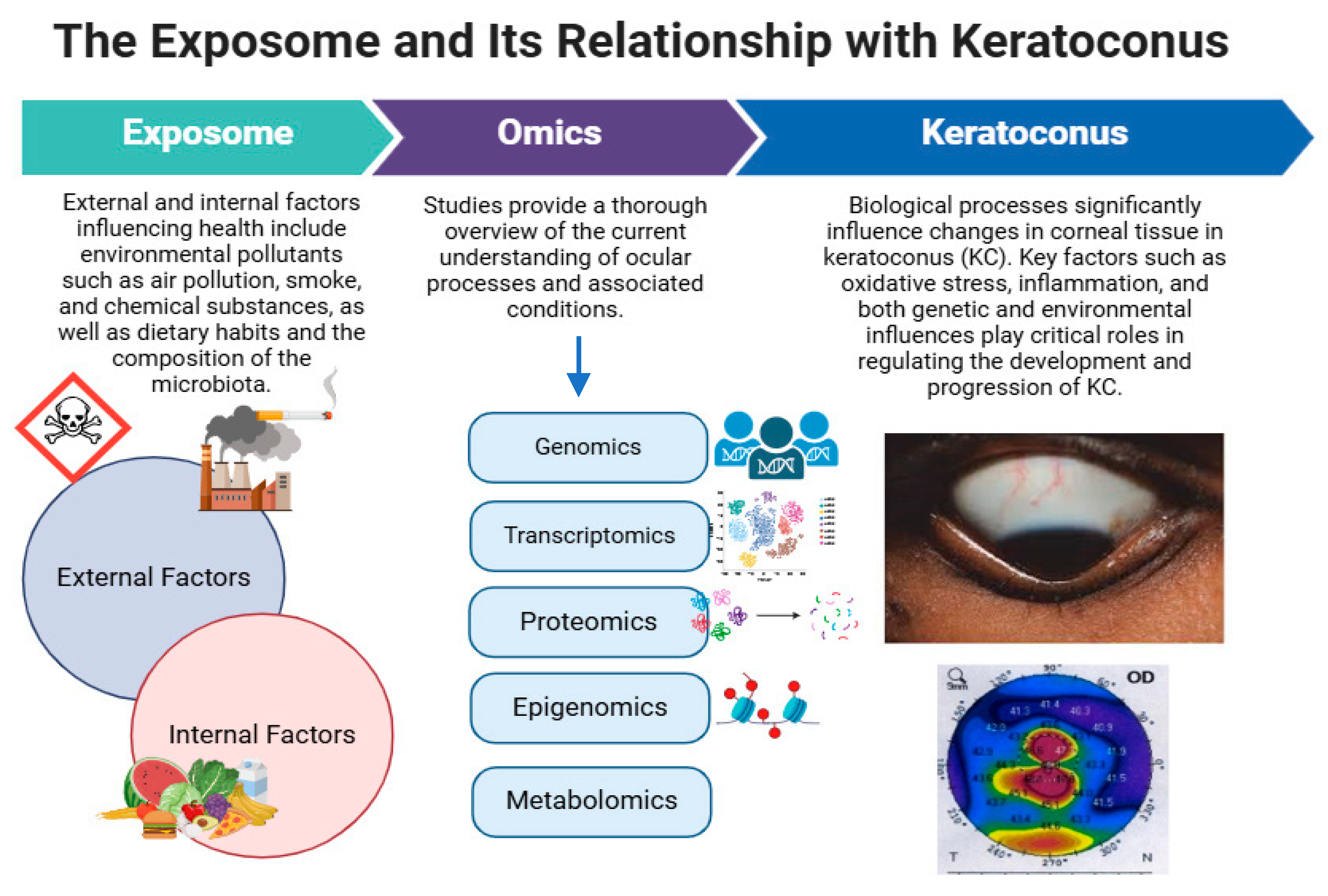
Omics in Keratoconus: From Molecular to Clinical Practice
Abstract
1. Overview of Keratoconus
2. Physiopathology of Keratoconus
2.1. Genetic and Epigenetic Factors
2.2. Inflammation, Oxidative Stress, and Cell Death
3. Multi-Omics in Keratoconus
3.1. Importance of Omics Technologies in Medicine
3.2. Omics Approaches to Understanding Keratoconus at the Molecular Level
3.3. Genomics
| Type of Analysis | Methodology Employed | Population Age (Mean) | Sample | Findings | Reference |
|---|---|---|---|---|---|
| Whole Exome Sequencing | PBMCs | 55.5 ± 0.5 | KC | Eight candidate genes (COL23A1, CD248, ADAMTS16, ADAMTS3, COL4A3, COL18A1, WNT16, and COL6A2). | [18] |
| GWAS | PBMCs | 57 ± 8 | Control/KC | The authors identified several CRF and CCT loci harbored within 1 Mb of mendelian genes associated with rare corneal or connective tissue diseases. These included FECD7, TCF4, SLC4A11, UBIAD1, MPV17, ZNF513, COL5A1, ZNF469, COL8A2, AGBL1, SMAD3, DCN, KERA, and TGFB2. | [87] |
| GWAS | PBMCs | Control/KC | Six loci were previously associated with keratoconus (FOXO1, COL5A1, FNDC3B, ZNF469, LOX, and near PNPLA2), and these were associated with central corneal thickness. In addition, strong associations were found near or within genes that code for fibrillar collagens (types I and V), microfibrillar (VI), and peri-fibrillar (XII) structures, implicating impaired cohesion of the collagen matrix in the pathogenesis of keratoconus. | [84] | |
| GWAS | PBMCs | 56.4 ± 13.0 | Control | STON2 rs2371597 showed a significant association with corneal central thickness. On the other hand, STON2 rs2371597 C allele showed increased CCTs in a Latino population. | [86] |
| GWAS | GitHub repository (last updated in May 2019) | GWAS for central corneal thickness (CCT) and corneal resistance factor (CRF) | [26] |
3.4. Transcriptomics
3.5. Proteomics
3.6. Epigenomics
3.7. Metabolomics
4. Translational and Clinical Implications
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Galvis, V.; Sherwin, T.; Tello, A.; Merayo, J.; Barrera, R.; Acera, A. Keratoconus: An inflammatory disorder? Eye 2015, 29, 843–859. [Google Scholar] [CrossRef] [PubMed]
- Mohammadpour, M.; Heidari, Z.; Hashemi, H. Updates on Managements for Keratoconus. J. Curr. Ophthalmol. 2017, 30, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Valdez-García, J.E.; Sepúlveda, R.; Salazar-Martínez, J.J.; Lozano-Ramírez, J.F. Prevalence of keratoconus in an adolescent population. Rev. Mex. Oftalmol. 2014, 88, 95–98. [Google Scholar] [CrossRef]
- Gomes, J.A.P.; Rodrigues, P.F.; Lamazales, L.L. Keratoconus epidemiology: A review. Saudi J. Ophthalmol. 2022, 36, 3–6. [Google Scholar] [CrossRef]
- Galvis, V.; Tello, A.; Jaramillo, J.A.; Gutierrez, A.J.; Rodriguez, L.; Quintero, M.P. Prevalence of keratoconus patients who consulted with a desire refractive surgery in ophthalmology center reference Bucaramanga, Colombia. Rev. Soc. Colomb. Oftal 2011, 44, 129–134. [Google Scholar]
- Al Qahtani, N.A.; Abahussin, M.O.; Assiri, A.A. Demographic and clinical variations of keratoconus in Saudi population. Saudi J. Ophthalmol. 2022, 36, 42–46. [Google Scholar] [CrossRef]
- Marx-Gross, S.; Fieß, A.; Münzel, T.; Wild, P.S.; Beutel, M.E.; Schmidtmann, I.; Lackner, K.J.; Pfeiffer, N.; Schuster, A.K.-G. Much higher prevalence of keratoconus than announced results of the Gutenberg Health Study (GHS). Graefes Arch. Clin. Exp. Ophthalmol. 2023, 261, 3241–3247. [Google Scholar] [CrossRef]
- Mejia-Salgado, G.; Cifuentes-González, C.; Rojas-Carabali, W.; Zarate-Pinzón, L.; Peña-Pulgar, L.F.; Polania, D.; Cruz-Reyes, D.L.; de-la-Torre, A. Colombian Ocular Diseases Epidemiology Study (CODES): Incidence and sociodemographic characterisation of keratoconus between 2015 and 2020. BMJ Open Ophthalmol. 2023, 8, e001238. [Google Scholar] [CrossRef]
- Lasagni Vitar, R.M.; Bonelli, F.; Rama, P.; Ferrari, G. Nutritional and Metabolic Imbalance in Keratoconus. Nutrients 2022, 14, 913. [Google Scholar] [CrossRef]
- Alves, M.; Asbell, P.; Dogru, M.; Giannaccare, G.; Grau, A.; Gregory, D.; Kim, D.H.; Marini, M.C.; Ngo, W.; Nowinska, A.; et al. TFOS Lifestyle Report: Impact of environmental conditions on the ocular surface. Ocul. Surf. 2023, 29, 1–52. [Google Scholar] [CrossRef]
- Peris-Martínez, C.; Piá-Ludeña, J.V.; Rog-Revert, M.J.; Fernández-López, E.; Domingo, J.C. Antioxidant and Anti-Inflammatory Effects of Oral Supplementation with a Highly-Concentrated Docosahexaenoic Acid (DHA) Triglyceride in Patients with Keratoconus: A Randomized Controlled Preliminary Study. Nutrients 2023, 15, 1300. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Shaag, A.; Millodot, M.; Shneor, E. The Epidemiology and Etiology of Keratoconus. Int. J. Keratoconus Ectatic Corneal Dis. 2012, 1, 7–15. [Google Scholar] [CrossRef]
- Santodomingo-Rubido, J.; Carracedo, G.; Suzaki, A.; Villa-Collar, C.; Vincent, S.J.; Wolffsohn, J.S. Keratoconus: An updated review. Cont. Lens Anterior Eye 2022, 45, 101559. [Google Scholar] [CrossRef]
- Lucas, S.E.M.; Burdon, K.P. Genetic and Environmental Risk Factors for Keratoconus. Annu. Rev. Vis. Sci. 2020, 6, 25–46. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, H.; Shan, M.; Dong, Y.; Zhang, L.; Chen, L.; Wang, Y. Comprehensive Transcriptome Analysis of Patients With Keratoconus Highlights the Regulation of Immune Responses and Inflammatory Processes. Front. Genet. 2022, 13, 782709. [Google Scholar] [CrossRef]
- Christensen, N.J.; Demharter, S.; Machado, M.; Pedersen, L.; Salvatore, M.; Stentoft-Hansen, V.; Iglesias, M.T. Identifying interactions in omics data for clinical biomarker discovery using symbolic regression. Bioinformatics 2022, 38, 3749–3758. [Google Scholar] [CrossRef]
- Hao, X.-D.; Chen, X.-N.; Zhang, Y.-Y.; Chen, P.; Wei, C.; Shi, W.-Y.; Gao, H. Multi-level consistent changes of the ECM pathway identified in a typical keratoconus twin’s family by multi-omics analysis. Orphanet J. Rare Dis. 2020, 15, 227. [Google Scholar] [CrossRef]
- Chen, X.; Liu, C.; Cui, Z.; Huang, Y.; Luo, Q.; Chen, S.; Wang, X.; Hou, X.; Gong, Q.; Li, Y.; et al. Integrative transcriptomics analysis and experimental validation reveal immunomodulatory patterns in keratoconus. Exp. Eye Res. 2023, 230, 109460. [Google Scholar] [CrossRef]
- Kannan, R.; Shetty, R.; Panigrahi, T.; Koh, S.K.; Khamar, P.; Deshpande, V.; Nuijts, R.M.M.A.; Gijs, M.; Nishtala, K.; Zhou, L.; et al. Untargeted Tear Proteomics in a Large South-Asian Cohort Reveals Inflammatory Signaling, ECM Remodeling, and Altered Metabolism in Keratoconus. Investig. Ophthalmol. Vis. Sci. 2025, 66, 60. [Google Scholar] [CrossRef]
- Volatier, T.L.A.; Figueiredo, F.C.; Connon, C.J. Keratoconus at a Molecular Level: A Review. Anat. Rec. 2020, 303, 1680–1688. [Google Scholar] [CrossRef]
- Nowak, D.M.; Gajecka, M. The genetics of keratoconus. Middle East Afr. J. Ophthalmol. 2011, 18, 2–6. [Google Scholar] [CrossRef]
- Lalgudi, V.G.; Shetty, R.; Nischal, K.K.; Ziai, S.; Koaik, M.; Sethu, S. Biochemical and molecular alterations and potential clinical applications of biomarkers in keratoconus. Saudi J. Ophthalmol. 2022, 36, 7–16. [Google Scholar] [CrossRef]
- Shetty, R.; Sathyanarayanamoorthy, A.; Ramachandra, R.A.; Arora, V.; Ghosh, A.; Srivatsa, P.R.; Pahuja, N.; Nuijts, R.M.M.A.; Sinha-Roy, A.; Mohan, R.R.; et al. Attenuation of lysyl oxidase and collagen gene expression in keratoconus patient corneal epithelium corresponds to disease severity. Mol. Vis. 2015, 21, 12–25. [Google Scholar]
- Bisceglia, L.; Ciaschetti, M.; De Bonis, P.; Campo, P.A.P.; Pizzicoli, C.; Scala, C.; Grifa, M.; Ciavarella, P.; Delle Noci, N.; Vaira, F.; et al. VSX1 mutational analysis in a series of Italian patients affected by keratoconus: Detection of a novel mutation. Investig. Ophthalmol. Vis. Sci. 2005, 46, 39–45. [Google Scholar] [CrossRef]
- Jiang, X.; Boutin, T.; Vitart, V. Colocalization of corneal resistance factor GWAS loci with GTEx e/sQTLs highlights plausible candidate causal genes for keratoconus postnatal corneal stroma weakening. Front. Genet. 2023, 14, 1171217. [Google Scholar] [CrossRef]
- Kim, S.-H.; Mok, J.-W.; Kim, H.-S.; Joo, C.K. Association of -31T>C and -511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol. Vis. 2008, 14, 2109–2116. [Google Scholar]
- Wheeler, J.; Hauser, M.A.; Afshari, N.A.; Allingham, R.R.; Liu, Y. The Genetics of Keratoconus: A Review. Reprod. Syst. Sex. Disord. Curr. Res. 2012, S6, 1. [Google Scholar] [CrossRef]
- Tyynismaa, H.; Sistonen, P.; Tuupanen, S.; Tervo, T.; Dammert, A.; Latvala, T.; Alitalo, T. A locus for autosomal dominant keratoconus: Linkage to 16q22.3-q23.1 in Finnish families. Investig. Ophthalmol. Vis. Sci. 2002, 43, 3160–3164. [Google Scholar]
- Brancati, F.; Valente, E.M.; Sarkozy, A.; Fehèr, J.; Castori, M.; Del Duca, P.; Mingarelli, R.; Pizzuti, A.; Dallapiccola, B. A locus for autosomal dominant keratoconus maps to human chromosome 3p14-q13. J. Med. Genet. 2004, 41, 188–192. [Google Scholar] [CrossRef]
- Nejabat, M.; Naghash, P.; Dastsooz, H.; Mohammadi, S.; Alipour, M.; Fardaei, M. VSX1 and SOD1 Mutation Screening in Patients with Keratoconus in the South of Iran. J. Ophthalmic Vis. Res. 2017, 12, 135–140. [Google Scholar] [CrossRef] [PubMed]
- Udar, N.; Atilano, S.R.; Brown, D.J.; Holguin, B.; Small, K.; Nesburn, A.B.; Kenney, M.C. SOD1: A candidate gene for keratoconus. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3345–3351. [Google Scholar] [CrossRef]
- Tanwar, M.; Kumar, M.; Nayak, B.; Pathak, D.; Sharma, N.; Titiyal, J.S.; Dada, R. VSX1 gene analysis in keratoconus. Mol. Vis. 2010, 16, 2395–2401. [Google Scholar] [PubMed]
- Zhang, J.; Cai, B.; Ma, L.; Qin, Y.; Li, S.; Sun, C.; Liang, J.; Han, Y.; Zhuang, W. Clinical and genetic analysis VSX1 variants among families with keratoconus in northwest China. Front. Genet. 2023, 14, 1145426. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, B.; Li, X.; Li, M.; Wang, Y.; Dan, H.; Zhou, J.; Wei, Y.; Ge, K.; Li, P.; et al. The application and progression of CRISPR/Cas9 technology in ophthalmological diseases. Eye 2023, 37, 607–617. [Google Scholar] [CrossRef]
- Virolainen, S.J.; VonHandorf, A.; Viel, K.C.M.F.; Weirauch, M.T.; Kottyan, L.C. Gene-environment interactions and their impact on human health. Genes. Immun. 2023, 24, 1–11. [Google Scholar] [CrossRef]
- Balasubramanian, S.A.; Pye, D.C.; Willcox, M.D.P. Are proteinases the reason for keratoconus? Curr. Eye Res. 2010, 35, 185–191. [Google Scholar] [CrossRef]
- Alkozi, H.A.; Franco, R.; Pintor, J.J. Epigenetics in the eye: An overview of the most relevant ocular diseases. Front. Genet. 2017, 8, 144. [Google Scholar] [CrossRef]
- Upaphong, P.; Thonusin, C.; Wanichthanaolan, O.; Chattipakorn, N.; Chattipakorn, S.C. Consequences of exposure to particulate matter on the ocular surface: Mechanistic insights from cellular mechanisms to epidemiological findings. Environ. Pollut. 2024, 345, 123488. [Google Scholar] [CrossRef]
- Marsit, C.J. Influence of environmental exposure on human epigenetic regulation. J. Exp. Biol. 2015, 218, 71–79. [Google Scholar] [CrossRef]
- Cao, Q.; Xu, W.; Chen, W.; Peng, D.; Liu, Q.; Dong, J.; Reinach, P.S.; Yan, D. MicroRNA-184 negatively regulates corneal epithelial wound healing via targeting CDC25A, CARM1, and LASP1. Eye Vis. 2020, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Breton, C.V.; Landon, R.; Kahn, L.G.; Enlow, M.B.; Peterson, A.K.; Bastain, T.; Braun, J.; Comstock, S.S.; Duarte, C.S.; Hipwell, A.; et al. Exploring the evidence for epigenetic regulation of environmental influences on child health across generations. Commun. Biol. 2021, 4, 769. [Google Scholar] [CrossRef] [PubMed]
- Kather, J.N.; Friedrich, J.; Woik, N.; Sticht, C.; Gretz, N.; Hammes, H.-P.; Kroll, J. Angiopoietin-1 Is Regulated by miR-204 and Contributes to Corneal Neovascularization in KLEIP-Deficient Mice. Investig. Ophthalmol. Vis. Sci. 2014, 55, 4295–4303. [Google Scholar] [CrossRef]
- Liao, C.-H.; Tseng, C.-L.; Lin, S.-L.; Liang, C.-L.; Juo, S.-H.H. MicroRNA Therapy for Dry Eye Disease. J. Ocul. Pharmacol. Ther. 2022, 38, 125–132. [Google Scholar] [CrossRef]
- Kabza, M.; Karolak, J.A.; Rydzanicz, M.; Udziela, M.; Gasperowicz, P.; Ploski, R.; Szaflik, J.P.; Gajecka, M. Multiple Differentially Methylated Regions Specific to Keratoconus Explain Known Keratoconus Linkage Loci. Investig. Ophthalmol. Vis. Sci. 2019, 60, 1501–1509. [Google Scholar] [CrossRef]
- Kabza, M.; Karolak, J.A.; Rydzanicz, M.; Szcześniak, M.W.; Nowak, D.M.; Ginter-Matuszewska, B.; Polakowski, P.; Ploski, R.; Szaflik, J.P.; Gajecka, M. Collagen synthesis disruption and downregulation of core elements of TGF-β, Hippo, and Wnt pathways in keratoconus corneas. Eur. J. Hum. Genet. 2017, 25, 582–590. [Google Scholar] [CrossRef]
- Cuellar-Partida, G.; Springelkamp, H.; Lucas, S.E.M.; Yazar, S.; Hewitt, A.W.; Iglesias, A.I.; Montgomery, G.W.; Martin, N.G.; Pennell, C.E.; van Leeuwen, E.M.; et al. WNT10A exonic variant increases the risk of keratoconus by decreasing corneal thickness. Hum. Mol. Genet. 2015, 24, 5060–5068. [Google Scholar] [CrossRef]
- Vazirani, J.; Basu, S. Keratoconus: Current perspectives. Clin. Ophthalmol. 2013, 7, 2019–2030. [Google Scholar] [CrossRef]
- Sorkhabi, R.; Ghorbanihaghjo, A.; Taheri, N.; Ahoor, M.H. Tear film inflammatory mediators in patients with keratoconus. Int. Ophthalmol. 2015, 35, 467–472. [Google Scholar] [CrossRef]
- Nichani, P.A.H.; Solomon, B.; Trinh, T.; Mimouni, M.; Rootman, D.; Singal, N.; Chan, C.C. Investigating the role of inflammation in keratoconus: A retrospective analysis of 551 eyes. Eur. J. Ophthalmol. 2023, 33, 35–43. [Google Scholar] [CrossRef]
- Loh, I.-P.; Sherwin, T. Is Keratoconus an Inflammatory Disease? The Implication of Inflammatory Pathways. Ocul. Immunol. Inflamm. 2022, 30, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Khaled, M.L.; Helwa, I.; Drewry, M.; Seremwe, M.; Estes, A.; Liu, Y. Molecular and Histopathological Changes Associated with Keratoconus. Biomed. Res. Int. 2017, 2017, 7803029. [Google Scholar] [CrossRef]
- Teo, A.W.J.; Mansoor, H.; Sim, N.; Lin, M.T.-Y.; Liu, Y.-C. In Vivo Confocal Microscopy Evaluation in Patients with Keratoconus. J. Clin. Med. 2022, 11, 393. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, C.; Corbu, C.G.; Tanase, C.; Jonescu-Cuypers, C.; Nicula, C.; Dascalescu, D.; Cristea, M.; Voinea, L.-M. Inflammatory Biomarkers Profile as Microenvironmental Expression in Keratoconus. Dis. Markers 2016, 2016, 1243819. [Google Scholar] [CrossRef]
- Shetty, R.; Ghosh, A.; Lim, R.R.; Subramani, M.; Mihir, K.; R, R.A.; Ranganath, A.; Nagaraj, S.; Nuijts, R.M.M.A.; Beuerman, R.; et al. Elevated Expression of Matrix Metalloproteinase-9 and Inflammatory Cytokines in Keratoconus Patients Is Inhibited by Cyclosporine A. Investig. Ophthalmol. Vis. Sci. 2015, 56, 738–750. [Google Scholar] [CrossRef]
- Nabil, K.M.; Elhady, G.M.; Morsy, H. The Association Between Interleukin 1 Beta Promoter Polymorphisms And Keratoconus Incidence And Severity In An Egyptian Population. Clin. Ophthalmol. 2019, 13, 2217–2223. [Google Scholar] [CrossRef]
- Tseng, H.-C.; Lee, I.-T.; Lin, C.-C.; Chi, P.-L.; Cheng, S.-E.; Shih, R.-H.; Hsiao, L.-D.; Yang, C.-M. IL-1β promotes corneal epithelial cell migration by increasing MMP-9 expression through NF-κB- and AP-1-dependent pathways. PLoS ONE 2013, 8, e57955. [Google Scholar] [CrossRef]
- Balmus, I.-M.; Alexa, A.I.; Ciuntu, R.-E.; Danielescu, C.; Stoica, B.; Cojocaru, S.I.; Ciobica, A.; Cantemir, A. Oxidative stress markers dynamics in keratoconus patients’ tears before and after corneal collagen crosslinking procedure. Exp. Eye Res. 2020, 190, 107897. [Google Scholar] [CrossRef]
- Dammak, A.; Pastrana, C.; Martin-Gil, A.; Carpena-Torres, C.; Peral Cerda, A.; Simovart, M.; Alarma, P.; Huete-Toral, F.; Carracedo, G. Oxidative Stress in the Anterior Ocular Diseases: Diagnostic and Treatment. Biomedicines 2023, 11, 292. [Google Scholar] [CrossRef]
- Wojakowska, A.; Pietrowska, M.; Widlak, P.; Dobrowolski, D.; Wylęgała, E.; Tarnawska, D. Metabolomic signature discriminates normal human cornea from Keratoconus—A pilot GC/MS study. Molecules 2020, 25, 2933. [Google Scholar] [CrossRef]
- Lema, I.; Sobrino, T.; Durán, J.A.; Brea, D.; Díez-Feijoo, E. Subclinical keratoconus and inflammatory molecules from tears. Br. J. Ophthalmol. 2009, 93, 820–824. [Google Scholar] [CrossRef]
- Atilano, S.R.; Lee, D.H.; Fukuhara, P.S.; Chwa, M.; Nesburn, A.B.; Udar, N.; Kenney, M.C. Corneal Oxidative Damage in Keratoconus Cells due to Decreased Oxidant Elimination from Modified Expression Levels of SOD Enzymes, PRDX6, SCARA3, CPSF3, and FOXM1. J. Ophthalmic Vis. Res. 2019, 14, 62–70. [Google Scholar] [CrossRef]
- Biswas, S.K. Does the Interdependence between Oxidative Stress and Inflammation Explain the Antioxidant Paradox? Oxid. Med. Cell. Longev. 2016, 2016, 5698931. [Google Scholar] [CrossRef]
- Soiberman, U.; Foster, J.W.; Jun, A.S.; Chakravarti, S. Pathophysiology of Keratoconus: What Do We Know Today. Open Ophthalmol. J. 2017, 11, 252–261. [Google Scholar] [CrossRef]
- Mohaghegh, S.; Kangari, H.; Masoumi, S.J.; Bamdad, S.; Rahmani, S.; Abdi, S.; Fazil, N.; Shahbazi, S. Prevalence and risk factors of keratoconus (including oxidative stress biomarkers) in a cohort study of Shiraz university of medical science employees in Iran. BMC Ophthalmol. 2023, 23, 188. [Google Scholar] [CrossRef]
- Shetty, R.; Sharma, A.; Pahuja, N.; Chevour, P.; Padmajan, N.; Dhamodaran, K.; Jayadev, C.; Nuijts, R.M.M.A.; Ghosh, A.; Nallathambi, J. Oxidative stress induces dysregulated autophagy in corneal epithelium of keratoconus patients. PLoS ONE 2017, 12, e0184628. [Google Scholar] [CrossRef]
- Matthews, F.J.; Cook, S.D.; Majid, M.A.; Dick, A.D.; Smith, V.A. Changes in the balance of the tissue inhibitor of matrix metalloproteinases (TIMPs)-1 and -3 may promote keratocyte apoptosis in keratoconus. Exp. Eye Res. 2007, 84, 1125–1134. [Google Scholar] [CrossRef]
- Nowak-Wąs, M.; Wąs, P.; Czuba, Z.; Wojnicz, R.; Wyględowska-Promieńska, D. Expression of Tissue Inhibitors of Metalloproteinases (TIMP-1, TIMP-2, TIMP-3, TIMP-4) in Blood Serum of Patients with Keratoconus. J. Clin. Med. 2024, 13, 1168. [Google Scholar] [CrossRef]
- Kaldawy, R.M.; Wagner, J.; Ching, S.; Seigel, G.M. Evidence of apoptotic cell death in keratoconus. Cornea 2002, 21, 206–209. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J.; Pan, D.; Wang, X.; Xu, Y.; Yan, J.; Wang, L.; Yang, X.; Yang, M.; Liu, G.-P. Applications of multi-omics analysis in human diseases. MedComm 2023, 4, e315. [Google Scholar] [CrossRef]
- Kim, D.-H.; Kim, Y.-S.; Son, N.-I.; Kang, C.-K.; Kim, A.-R. Recent omics technologies and their emerging applications for personalised medicine. IET Syst. Biol. 2017, 11, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Delpierre, C.; Lefèvre, T. Precision and personalized medicine: What their current definition says and silences about the model of health they promote. Implication for the development of personalized health. Front. Sociol. 2023, 8, 1112159. [Google Scholar] [CrossRef]
- Tebani, A.; Afonso, C.; Marret, S.; Bekri, S. Omics-Based Strategies in Precision Medicine: Toward a Paradigm Shift in Inborn Errors of Metabolism Investigations. Int. J. Mol. Sci. 2016, 17, 1555. [Google Scholar] [CrossRef]
- Gasperskaja, E.; Kučinskas, V. The most common technologies and tools for functional genome analysis. Acta Medica Litu. 2017, 24, 1–11. [Google Scholar] [CrossRef]
- Donath, X.; Saint-Martin, C.; Dubois-Laforgue, D.; Rajasingham, R.; Mifsud, F.; Ciangura, C.; Timsit, J.; Bellanné-Chantelot, C. Next-generation sequencing identifies monogenic diabetes in 16% of patients with late adolescence/adult-onset diabetes selected on a clinical basis: A cross-sectional analysis. BMC Med. 2019, 17, 132. [Google Scholar] [CrossRef]
- Orsini, A.; Diquigiovanni, C.; Bonora, E. Omics Technologies Improving Breast Cancer Research and Diagnostics. Int. J. Mol. Sci. 2023, 24, 2690. [Google Scholar] [CrossRef]
- Mu, A.; Klare, W.P.; Baines, S.L.; Ignatius Pang, C.N.; Guérillot, R.; Harbison-Price, N.; Keller, N.; Wilksch, J.; Nhu, N.T.K.; Phan, M.-D.; et al. Integrative omics identifies conserved and pathogen-specific responses of sepsis-causing bacteria. Nat. Commun. 2023, 14, 1530. [Google Scholar] [CrossRef]
- Hamel, A.R.; Yan, W.; Rouhana, J.M.; Monovarfeshani, A.; Jiang, X.; Mehta, P.A.; Advani, J.; Luo, Y.; Liang, Q.; Rajasundaram, S.; et al. Integrating genetic regulation and single-cell expression with GWAS prioritizes causal genes and cell types for glaucoma. Nat. Commun. 2024, 15, 396. [Google Scholar] [CrossRef]
- Emilsson, V.; Gudmundsson, E.F.; Jonmundsson, T.; Jonsson, B.G.; Twarog, M.; Gudmundsdottir, V.; Li, Z.; Finkel, N.; Poor, S.; Liu, X.; et al. A proteogenomic signature of age-related macular degeneration in blood. Nat. Commun. 2022, 13, 3401. [Google Scholar] [CrossRef]
- Fineide, F.A.; Tashbayev, B.; Elgstøen, K.B.P.; Sandås, E.M.; Rootwelt, H.; Hynne, H.; Chen, X.; Ræder, S.; Vehof, J.; Dartt, D.; et al. Tear and Saliva Metabolomics in Evaporative Dry Eye Disease in Females. Metabolites 2023, 13, 1125. [Google Scholar] [CrossRef]
- Yehia, L.; Eng, C. Genetics and genomics in healthcare: The future is now. Singap. Med. J. 2023, 64, 4–6. [Google Scholar] [CrossRef]
- Guigo, R.; de Hoon, M. Recent advances in functional genome analysis. F1000Research 2018, 7, 1968. [Google Scholar] [CrossRef]
- Molster, C.M.; Bowman, F.L.; Bilkey, G.A.; Cho, A.S.; Burns, B.L.; Nowak, K.J.; Dawkins, H.J.S. The Evolution of Public Health Genomics: Exploring Its Past, Present, and Future. Front. Public Heal. 2018, 6, 247. [Google Scholar] [CrossRef]
- Hardcastle, A.J.; Liskova, P.; Bykhovskaya, Y.; McComish, B.J.; Davidson, A.E.; Inglehearn, C.F.; Li, X.; Choquet, H.; Habeeb, M.; Lucas, S.E.M.; et al. A multi-ethnic genome-wide association study implicates collagen matrix integrity and cell differentiation pathways in keratoconus. Commun. Biol. 2021, 4, 266. [Google Scholar] [CrossRef]
- Niazi, S.; Moshirfar, M.; Alizadeh, F.; Doroodgar, F.; Baradaran-Rafii, A.; Filutowski, O.; Niazi, F.; Ambrósio, R.J. Association of 2 Lysyl Oxidase Gene Single Nucleotide Polymorphisms with Keratoconus: A Nationwide Registration Study. Ophthalmol. Sci. 2023, 3, 100247. [Google Scholar] [CrossRef]
- Hosoda, Y.; Miyake, M.; Meguro, A.; Tabara, Y.; Iwai, S.; Ueda-Arakawa, N.; Nakano, E.; Mori, Y.; Yoshikawa, M.; Nakanishi, H.; et al. Keratoconus-susceptibility gene identification by corneal thickness genome-wide association study and artificial intelligence IBM Watson. Commun. Biol. 2020, 3, 410. [Google Scholar] [CrossRef]
- He, W.; Han, X.; Ong, J.-S.; Hewitt, A.W.; Mackey, D.A.; Gharahkhani, P.; MacGregor, S. Association of Novel Loci With Keratoconus Susceptibility in a Multitrait Genome-Wide Association Study of the UK Biobank Database and Canadian Longitudinal Study on Aging. JAMA Ophthalmol. 2022, 140, 568–576. [Google Scholar] [CrossRef]
- Postel, M.D.; Culver, J.O.; Ricker, C.; Craig, D.W. Transcriptome analysis provides critical answers to the “variants of uncertain significance” conundrum. Hum. Mutat. 2022, 43, 1590–1608. [Google Scholar] [CrossRef]
- Kolobkov, D.S.; Sviridova, D.A.; Abilev, S.K.; Kuzovlev, A.N.; Salnikova, L.E. Genes and Diseases: Insights from Transcriptomics Studies. Genes 2022, 13, 1168. [Google Scholar] [CrossRef]
- Kukurba, K.R.; Montgomery, S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Lupasco, T.; He, Z.; Cassagne, M.; Sagnial, T.; Brion, L.; Fournié, P.; Gain, P.; Thuret, G.; Allouche, M.; Malecaze, F.; et al. Corneal epithelium in keratoconus underexpresses active NRF2 and a subset of oxidative stress-related genes. PLoS ONE 2022, 17, e0273807. [Google Scholar] [CrossRef] [PubMed]
- Shinde, V.; Hu, N.; Mahale, A.; Maiti, G.; Daoud, Y.; Eberhart, C.G.; Maktabi, A.; Jun, A.S.; Al-Swailem, S.A.; Chakravarti, S. RNA sequencing of corneas from two keratoconus patient groups identifies potential biomarkers and decreased NRF2-antioxidant responses. Sci. Rep. 2020, 10, 9907. [Google Scholar] [CrossRef]
- You, J.; Corley, S.M.; Wen, L.; Hodge, C.; Höllhumer, R.; Madigan, M.C.; Wilkins, M.R.; Sutton, G. RNA-Seq analysis and comparison of corneal epithelium in keratoconus and myopia patients. Sci. Rep. 2018, 8, 389. [Google Scholar] [CrossRef]
- Stachon, T.; Nastaranpour, M.; Seitz, B.; Meese, E.; Latta, L.; Taneri, S.; Ardjomand, N.; Szentmáry, N.; Ludwig, N. Altered Regulation of mRNA and miRNA Expression in Epithelial and Stromal Tissue of Keratoconus Corneas. Investig. Ophthalmol. Vis. Sci. 2022, 63, 7. [Google Scholar] [CrossRef] [PubMed]
- McMonnies, C.W. Inflammation and keratoconus. Optom. Vis. Sci. 2015, 92, e35–e41. [Google Scholar] [CrossRef]
- Dou, S.; Wang, Q.; Zhang, B.; Wei, C.; Wang, H.; Liu, T.; Duan, H.; Jiang, H.; Liu, M.; Qi, X.; et al. Single-cell atlas of keratoconus corneas revealed aberrant transcriptional signatures and implicated mechanical stretch as a trigger for keratoconus pathogenesis. Cell Discov. 2022, 8, 66. [Google Scholar] [CrossRef]
- Krok, M.; Wróblewska-Czajka, E.; Łach-Wojnarowicz, O.; Bronikowska, J.; Czuba, Z.P.; Wylęgała, E.; Dobrowolski, D. Analysis of Cytokine and Chemokine Level in Tear Film in Keratoconus Patients before and after Corneal Cross-Linking (CXL) Treatment. Int. J. Mol. Sci. 2024, 25, 1052. [Google Scholar] [CrossRef]
- Sharif, R.; Khaled, M.L.; McKay, T.B.; Liu, Y.; Karamichos, D. Transcriptional profiling of corneal stromal cells derived from patients with keratoconus. Sci. Rep. 2019, 9, 12567. [Google Scholar] [CrossRef]
- Niu, X.; Xu, M.; Zhu, J.; Zhang, S.; Yang, Y. Identification of the immune-associated characteristics and predictive biomarkers of keratoconus based on single-cell RNA-sequencing and bulk RNA-sequencing. Front. Immunol. 2023, 14, 1220646. [Google Scholar] [CrossRef]
- D’Souza, S.; Nair, A.P.; Sahu, G.R.; Vaidya, T.; Shetty, R.; Khamar, P.; Mullick, R.; Gupta, S.; Dickman, M.M.; Nuijts, R.M.M.A.; et al. Keratoconus patients exhibit a distinct ocular surface immune cell and inflammatory profile. Sci. Rep. 2021, 11, 20891. [Google Scholar] [CrossRef]
- Wolf, J.; Boneva, S.; Schlecht, A.; Lapp, T.; Auw-Haedrich, C.; Lagrèze, W.; Agostini, H.; Reinhard, T.; Schlunck, G.; Lange, C. The Human Eye Transcriptome Atlas: A searchable comparative transcriptome database for healthy and diseased human eye tissue. Genomics 2022, 114, 110286. [Google Scholar] [CrossRef]
- Gobena, S.; Admassu, B.; Kinde, M.Z.; Gessese, A.T. Proteomics and Its Current Application in Biomedical Area: Concise Review. Sci. World J. 2024, 2024, 4454744. [Google Scholar] [CrossRef]
- Gou, W.; Yue, L.; Tang, X.-Y.; Wu, Y.-Y.; Cai, X.; Shuai, M.; Miao, Z.; Fu, Y.; Chen, H.; Jiang, Z.; et al. Circulating Proteome and Progression of Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2022, 107, 1616–1625. [Google Scholar] [CrossRef]
- Kline, R.A.; Lößlein, L.; Kurian, D.; Aguilar Martí, J.; Eaton, S.L.; Court, F.A.; Gillingwater, T.H.; Wishart, T.M. An Optimized Comparative Proteomic Approach as a Tool in Neurodegenerative Disease Research. Cells 2022, 11, 2653. [Google Scholar] [CrossRef]
- Starodubtseva, N.L.; Tokareva, A.O.; Rodionov, V.V.; Brzhozovskiy, A.G.; Bugrova, A.E.; Chagovets, V.V.; Kometova, V.V.; Kukaev, E.N.; Soares, N.C.; Kovalev, G.I.; et al. Integrating Proteomics and Lipidomics for Evaluating the Risk of Breast Cancer Progression: A Pilot Study. Biomedicines 2023, 11, 1786. [Google Scholar] [CrossRef]
- de Freitas Campos, C.; Cole, N.; Van Dyk, D.; Walsh, B.J.; Diakos, P.; Almeida, D.; Torrecilhas, A.; Laus, J.L.; Willcox, M.D.P. Proteomic analysis of dog tears for potential cancer markers. Res. Vet. Sci. 2008, 85, 349–352. [Google Scholar] [CrossRef]
- You, J.; Willcox, M.D.; Madigan, M.C.; Wasinger, V.; Schiller, B.; Walsh, B.J.; Graham, P.H.; Kearsley, J.H.; Li, Y. Tear Fluid Protein Biomarkers. Adv. Clin. Chem. 2013, 62, 151–196. [Google Scholar] [CrossRef]
- Aass, C.; Norheim, I.; Eriksen, E.F.; Thorsby, P.M.; Pepaj, M. Single unit filter-aided method for fast proteomic analysis of tear fluid. Anal. Biochem. 2015, 480, 1–5. [Google Scholar] [CrossRef]
- Azkargorta, M.; Soria, J.; Acera, A.; Iloro, I.; Elortza, F. Human tear proteomics and peptidomics in ophthalmology: Toward the translation of proteomic biomarkers into clinical practice. J. Proteom. 2017, 150, 359–367. [Google Scholar] [CrossRef]
- Ghosh, A.; Zhou, L.; Ghosh, A.; Shetty, R.; Beuerman, R. Proteomic and gene expression patterns of keratoconus. Indian J. Ophthalmol. 2013, 61, 389–391. [Google Scholar] [CrossRef]
- Ghosh, R.; Gilda, J.E.; Gomes, A.V. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev. Proteom. 2014, 11, 549–560. [Google Scholar] [CrossRef] [PubMed]
- López-López, M.; Regueiro, U.; Bravo, S.B.; Chantada-Vázquez, M.D.P.; Varela-Fernández, R.; Ávila-Gómez, P.; Hervella, P.; Lema, I. Tear Proteomics in Keratoconus: A Quantitative SWATH-MS Analysis. Investig. Ophthalmol. Vis. Sci. 2021, 62, 30. [Google Scholar] [CrossRef] [PubMed]
- Lema, I.; Brea, D.; Rodríguez-González, R.; Díez-Feijoo, E.; Sobrino, T. Proteomic analysis of the tear film in patients with keratoconus. Mol. Vis. 2010, 16, 2055–2061. [Google Scholar]
- Gijs, M.; Adelaar, T.I.; Vergouwen, D.P.C.; Visser, N.; Dickman, M.M.; Ollivier, R.C.I.; Berendschot, T.T.J.M.; Nuijts, R.M.M.A. Tear Fluid Inflammatory Proteome Analysis Highlights Similarities Between Keratoconus and Allergic Conjunctivitis. Investig. Ophthalmol. Vis. Sci. 2023, 64, 9. [Google Scholar] [CrossRef]
- Shinde, V.; Hu, N.; Renuse, S.; Mahale, A.; Pandey, A.; Eberhart, C.; Stone, D.; Al-Swailem, S.A.; Maktabi, A.; Chakravarti, S. Mapping Keratoconus Molecular Substrates by Multiplexed High-Resolution Proteomics of Unpooled Corneas. OMICS 2019, 23, 583–597. [Google Scholar] [CrossRef]
- López-López, M.; Regueiro, U.; Bravo, S.B.; Chantada-Vázquez, M.D.P.; Pena, C.; Díez-Feijoo, E.; Hervella, P.; Lema, I. Shotgun Proteomics for the Identification and Profiling of the Tear Proteome of Keratoconus Patients. Investig. Ophthalmol. Vis. Sci. 2022, 63, 12. [Google Scholar] [CrossRef]
- Li, X.; Chen, K.; Wang, Z.; Li, J.; Wang, X.; Xie, C.; Tong, J.; Shen, Y. The mTOR signalling in corneal diseases: A recent update. Biochem. Pharmacol. 2023, 213, 115620. [Google Scholar] [CrossRef]
- McKay, T.B.; Priyadarsini, S.; Rowsey, T.; Karamichos, D. Arginine Supplementation Promotes Extracellular Matrix and Metabolic Changes in Keratoconus. Cells 2021, 10, 2076. [Google Scholar] [CrossRef]
- Rozek, L.S.; Dolinoy, D.C.; Sartor, M.A.; Omenn, G.S. Epigenetics: Relevance and implications for public health. Annu. Rev. Public Health 2014, 35, 105–122. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, Q.; Chang, C. Epigenetics in Health and Disease. Adv. Exp. Med. Biol. 2020, 1253, 3–55. [Google Scholar] [CrossRef]
- Alegría-Torres, J.A.; Baccarelli, A.; Bollati, V. Epigenetics and lifestyle. Epigenomics 2011, 3, 267–277. [Google Scholar] [CrossRef] [PubMed]
- Hardy, T.M.; Tollefsbol, T.O. Epigenetic diet: Impact on the epigenome and cancer. Epigenomics 2011, 3, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, R.B.; Miller, C.M.; Martinez-Caballero, J.D.; Ramos, I. Epigenetic Control of Innate Immunity: Consequences of Acute Respiratory Virus Infection. Viruses 2024, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Yeon, Y.; Lee, W.J.; Shin, Y.U.; Cho, H.; Sung, Y.K.; Kim, D.R.; Lim, H.W.; Kang, M.H. Comparison of microRNA expression in tears of normal subjects and Sjögren syndrome patients. Investig. Ophthalmol. Vis. Sci. 2019, 60, 4889–4895. [Google Scholar] [CrossRef]
- Kowluru, R.A. Cross Talks between Oxidative Stress, Inflammation and Epigenetics in Diabetic Retinopathy. Cells 2023, 12, 300. [Google Scholar] [CrossRef]
- Puigoriol-Illamola, D.; Martínez-Damas, M.; Griñán-Ferré, C.; Pallàs, M. Chronic Mild Stress Modified Epigenetic Mechanisms Leading to Accelerated Senescence and Impaired Cognitive Performance in Mice. Int. J. Mol. Sci. 2020, 21, 1154. [Google Scholar] [CrossRef]
- McMonnies, C.W. Epigenetic Mechanisms Might Help Explain Environmental Contributions to the Pathogenesis of Keratoconus. Eye Contact Lens 2014, 40, 371–375. [Google Scholar]
- Jiang, X.; Dellepiane, N.; Pairo-Castineira, E.; Boutin, T.; Kumar, Y.; Bickmore, W.A.; Vitart, V. Fine-mapping and cell-specific enrichment at corneal resistance factor loci prioritize candidate causal regulatory variants. Commun. Biol. 2020, 3, 762. [Google Scholar] [CrossRef]
- Drewry, M.; Helwa, I.; Allingham, R.R.; Hauser, M.A.; Liu, Y. miRNA Profile in Three Different Normal Human Ocular Tissues by miRNA-Seq. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3731–3739. [Google Scholar] [CrossRef]
- An, J.; Chen, X.; Chen, W.; Liang, R.; Reinach, P.S.; Yan, D.; Tu, L. MicroRNA Expression Profile and the Role of miR-204 in Corneal Wound Healing. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3673–3683. [Google Scholar] [CrossRef]
- Stunf Pukl, S. Are miRNAs Dynamic Biomarkers in Keratoconus? A Review of the Literature. Genes 2022, 13, 588. [Google Scholar] [CrossRef] [PubMed]
- Kalaimani, L.; Devarajan, B.; Subramanian, U.; Ayyasamy, V.; Namperumalsamy, V.P.; Veerappan, M.; Chidambaranathan, G.P. MicroRNA Profiling of Highly Enriched Human Corneal Epithelial Stem Cells by Small RNA Sequencing. Sci. Rep. 2020, 10, 7418. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, Y.; Zhang, X.; Ullah, R.; Tong, J.; Shen, Y. The role of the PI3K/AKT signalling pathway in the corneal epithelium: Recent updates. Cell Death Dis. 2022, 13, 513. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, J.; Tan, Y.; Wang, Y. Unraveling the mechanobiology of cornea: From bench side to the clinic. Front. Bioeng. Biotechnol. 2022, 10, 953590. [Google Scholar] [CrossRef]
- Urbich, C.; Kuehbacher, A.; Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 2008, 79, 581–588. [Google Scholar] [CrossRef]
- Agarwal, A.; Kansal, V.; Farooqi, H.; Singh, V.K.; Prasad, R. Differentially deregulated microRNAs contribute to ultraviolet radiation-induced photocarcinogenesis through immunomodulation: An-analysis of microRNAs expression profiling. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sieland, J.; Niederer, D.; Engeroff, T.; Vogt, L.; Troidl, C.; Schmitz-Rixen, T.; Banzer, W.; Troidl, K. Changes in miRNA expression in patients with peripheral arterial vascular disease during moderate- and vigorous-intensity physical activity. Eur. J. Appl. Physiol. 2023, 123, 645–654. [Google Scholar] [CrossRef]
- Burgos, K.; Malenica, I.; Metpally, R.; Courtright, A.; Rakela, B.; Beach, T.; Shill, H.; Adler, C.; Sabbagh, M.; Villa, S.; et al. Profiles of extracellular miRNA in cerebrospinal fluid and serum from patients with Alzheimer’s and Parkinson’s diseases correlate with disease status and features of pathology. PLoS ONE 2014, 9, e94839. [Google Scholar] [CrossRef]
- Ramanjaneya, M.; Priyanka, R.; Bensila, M.; Jerobin, J.; Pawar, K.; Sathyapalan, T.; Abou-Samra, A.B.; Halabi, N.M.; Moin, A.S.M.; Atkin, S.L.; et al. MiRNA and associated inflammatory changes from baseline to hypoglycemia in type 2 diabetes. Front. Endocrinol. 2022, 13, 917041. [Google Scholar] [CrossRef]
- Pilson, Q.; Smith, S.; Jefferies, C.A.; Ní Gabhann-Dromgoole, J.; Murphy, C.C. miR-744-5p contributes to ocular inflammation in patients with primary Sjogrens Syndrome. Sci. Rep. 2020, 10, 7484. [Google Scholar] [CrossRef]
- Zhang, Y.; Che, D.; Cao, Y.; Yue, Y.; He, T.; Zhu, Y.; Zhou, J. MicroRNA Profiling in the Aqueous Humor of Keratoconus Eyes. Transl. Vis. Sci. Technol. 2022, 11, 5. [Google Scholar] [CrossRef] [PubMed]
- Benavides-Aguilar, J.A.; Morales-Rodríguez, J.I.; Ambriz-González, H.; Ruiz-Manriquez, L.M.; Banerjee, A.; Pathak, S.; Duttaroy, A.K.; Paul, S. The regulatory role of microRNAs in common eye diseases: A brief review. Front. Genet. 2023, 14, 1152110. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Covarrubias, V.; Martínez-Martínez, E.; Del Bosque-Plata, L. The Potential of Metabolomics in Biomedical Applications. Metabolites 2022, 12, 194. [Google Scholar] [CrossRef]
- Letertre, M.P.M.; Giraudeau, P.; de Tullio, P. Nuclear Magnetic Resonance Spectroscopy in Clinical Metabolomics and Personalized Medicine: Current Challenges and Perspectives. Front. Mol. Biosci. 2021, 8, 698337. [Google Scholar] [CrossRef]
- Walker, D.I.; Valvi, D.; Rothman, N.; Lan, Q.; Miller, G.W.; Jones, D.P. The metabolome: A key measure for exposome research in epidemiology. Curr. Epidemiol. Rep. 2019, 6, 93–103. [Google Scholar]
- Hong, M.; Tong, L.; Mehta, J.S.; Ong, H.S. Impact of Exposomes on Ocular Surface Diseases. Int. J. Mol. Sci. 2023, 24, 1273. [Google Scholar] [CrossRef]
- Karamichos, D.; Zieske, J.D.; Sejersen, H.; Sarker-Nag, A.; Asara, J.M.; Hjortdal, J. Tear metabolite changes in keratoconus. Exp. Eye Res. 2015, 132, 1–8. [Google Scholar] [CrossRef]
- Daphne Teh, A.L.; Jayapalan, J.J.; Loke, M.F.; Wan Abdul Kadir, A.J.; Subrayan, V. Identification of potential serum metabolic biomarkers for patient with keratoconus using untargeted metabolomics approach. Exp. Eye Res. 2021, 211, 108734. [Google Scholar] [CrossRef]
- Karolak, J.A.; Ginter-Matuszewska, B.; Tomela, K.; Kabza, M.; Nowak-Malczewska, D.M.; Rydzanicz, M.; Polakowski, P.; Szaflik, J.P.; Gajecka, M. Further evaluation of differential expression of keratoconus candidate genes in human corneas. PeerJ 2020, 8, e9793. [Google Scholar] [CrossRef]
- Andrade, F.E.C.; Covre, J.L.; Ramos, L.; Hazarbassanov, R.M.; Dos Santos, M.S.; Campos, M.; Gomes, J.Á.P.; Gil, C.D. Evaluation of galectin-1 and galectin-3 as prospective biomarkers in keratoconus. Br. J. Ophthalmol. 2018, 102, 700–707. [Google Scholar] [CrossRef]
- Sharif, R.; Hjortdal, J.; Sejersen, H.; Frank, G.; Karamichos, D. Human in vitro Model Reveals the Effects of Collagen Cross-linking on Keratoconus Pathogenesis. Sci. Rep. 2017, 7, 12517. [Google Scholar] [CrossRef]
- Balogun, M.M.; Fashola, M.B. Association between keratoconus and allergic conjunctivitis in children attending a Tertiary Hospital in Nigeria. Rom. J. Ophthalmol. 2023, 67, 134–139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Deng, Y.; Li, S.; Du, X.; Zhao, X.; Zhang, T.; Yuan, J. Corneal biomechanical changes in allergic conjunctivitis. Eye Vis. 2021, 8, 17. [Google Scholar] [CrossRef]
- Yu, H.; Dou, S.; Wang, H.; Sun, Y.; Qu, J.; Liu, T.; Liu, X.; Wei, C.; Gao, H. Role of m(6)A methyltransferase METTL3 in keratoconus pathogenesis. Exp. Eye Res. 2025, 251, 110207. [Google Scholar] [CrossRef]
- Neveu, V.; Nicolas, G.; Amara, A.; Salek, R.M.; Scalbert, A. The human microbial exposome: Expanding the Exposome-Explorer database with gut microbial metabolites. Sci. Rep. 2023, 13, 1946. [Google Scholar] [CrossRef]
- Akkaya, S.; Ulusoy, D.M. Serum Vitamin D Levels in Patients with Keratoconus. Ocul. Immunol. Inflamm. 2020, 28, 348–353. [Google Scholar] [CrossRef]
- Markoulli, M.; Ahmad, S.; Arcot, J.; Arita, R.; Benitez-Del-Castillo, J.; Caffery, B.; Downie, L.E.; Edwards, K.; Flanagan, J.; Labetoulle, M.; et al. TFOS Lifestyle: Impact of nutrition on the ocular surface. Ocul. Surf. 2023, 29, 226–271. [Google Scholar] [CrossRef]
- Koithan, M.; Devika, J. New Approaches to Nutritional Therapy. J. Nurse Pract. 2010, 6, 805–806. [Google Scholar] [CrossRef][Green Version]
- Fucito, M.; Spedicato, M.; Felletti, S.; Yu, A.C.; Busin, M.; Pasti, L.; Franchina, F.A.; Cavazzini, A.; De Luca, C.; Catani, M. A Look into Ocular Diseases: The Pivotal Role of Omics Sciences in Ophthalmology Research. ACS Meas. Sci. Au 2024, 4, 247–259. [Google Scholar] [CrossRef]
- Shetty, R.; D’Souza, S.; Khamar, P.; Ghosh, A.; Nuijts, R.M.M.A.; Sethu, S. Biochemical Markers and Alterations in Keratoconus. Asia-Pac. J. Ophthalmol. 2020, 9, 533–540. [Google Scholar] [CrossRef]
- Kanclerz, P.; Khoramnia, R.; Wang, X. Current Developments in Corneal Topography and Tomography. Diagnostics 2021, 11, 1466. [Google Scholar] [CrossRef] [PubMed]
- Niazi, S.; Gatzioufas, Z.; Doroodgar, F.; Findl, O.; Baradaran-Rafii, A.; Liechty, J.; Moshirfar, M. Keratoconus: Exploring fundamentals and future perspectives—A comprehensive systematic review. Ther. Adv. Ophthalmol. 2024, 16, 25158414241232256. [Google Scholar] [CrossRef] [PubMed]
- Castro-Luna, G.; Pérez-Rueda, A. A predictive model for early diagnosis of keratoconus. BMC Ophthalmol. 2020, 20, 263. [Google Scholar] [CrossRef] [PubMed]

| Type of Analysis | Methodology Employed | Population Age (Mean) | Sample | Findings | Reference |
|---|---|---|---|---|---|
| RNA-seq | Corneal tissue/ Blood samples | 20.14 ± 1.81/16.00 ± 2.27 | Control/KC | Leukocyte cell–cell adhesion, regulation of lymphocyte activation, adaptative immune response, and humoral immune response. | [16] |
| qPCR | Corneal epitheliums | 28.5 ± 5.7/22 ± 4.2/45 ± 7 | Control/KC early/KC advanced | In early and advanced KC, they found downregulation in genes related to oxidative stress. In addition, there was an imbalance in apoptosis, proliferation, and differentiation pathways. | [91] |
| Microarrays/qPCR | Corneal epitheliums | 32.6 ± 9.4/26.6 ± 1.6 | Control/KC | The biological processes related with KC were leukocyte migration, cell chemotaxis, response to chemokine, and calcium ion homeostasis. | [19] |
| RNA-seq | Normal primary human corneal fibroblast (N-HCF) and primary corneal fibroblast cells of II-1 (II-1-HCF) | 28 ± 0/55.5 ± 0.5 | Control/KC | The analysis revealed collagen catabolic processes, collagen trimer, biosynthesis, assembly of collagen fibrils, and the proteinaceous ECM. | [18] |
| RNA-seq | Corneal epitheliums | 57.4 ± 14.7/30 ± 14.2 | Control/KC | IL-6 signaling and immune and inflammatory responses were significant in both patient groups. In addition, NRF2 (regulated oxidative stress response) was significantly decreased in samples. | [92] |
| RNA-seq | Corneal epitheliums | 61.87 ± 12.78/31.87 ± 8.55 | Control/KC early/KC advanced | Downregulated genes were in different pathways: including pathways related to collagen synthesis and modification, as well as extracellular matrix organization and focal adhesion. In addition, they found downregulation in TGF-β and WNT signaling pathways. | [46] |
| Microarrays/qPCR | Human corneal epithelial (HCE) and human corneal stromal fibroblast cells (HCF) | 22.4 ± 2.44/22.23 ± 1.99 | Control/ KC | A differential expression pattern of autophagy pathway (LC3A, RAB7, LAMP1), oxidative stress, and mTOR were found to be dysregulated. | [66] |
| RNA-seq | Corneal epithelium | 32 ± 6.16/27 ± 8.08 | Control (myopia)/ KC | Compared with control (myopia), in KC samples they found downregulated genes: cell–cell communication, cell signaling, WNT, and Notch1 signaling pathways. | [93] |
| Microarrays | Corneal epithelium and the stroma tissue | 75.0 ± 11.2/53.95 ± 12.1 | Control/KC | They found several genes known to play a role in KC: including AQP5, S100A8, EGLN3, EGFR1, and SFRP1. These genes are associated in metabolic inflammation and the MAPK pathway. | [94] |
| Type of Analysis | Methodology Employed | Population Age (Mean) | Sample | Findings | Reference |
|---|---|---|---|---|---|
| LC-MS/MS | Tears were collected with Schirmer strip | 43.96 ± 6.94/44.88 ± 5.01 | Control/KC | They showed upregulation in plastin 3, DNA dC→dU-editing enzyme APOBEC-3A1, tubulin α1C chain, 6-phosphogluconate dehydrogenase, decarboxylating, cofilin 1, annexin A2, and annexin A1. Downregulation was shown in serotransferrin, serum albumin, vitamin D binding protein, α1-acid glycoprotein 1, transthyretin, α2-HS-glycoprotein, hemopexin, angiotensinogen, latexin, heat shock cognate 71-kDa protein, and ρ GDP-dissociation inhibitor 1. | [112] |
| MALDI-TOF MS | Tears were collected with Schirmer strip | 30.8 ± 7.6/31.3 ± 7.8 | Control/KC | The MS analysis revealed a difference in the IGKC (Immunoglobulin Kappa Chain) protein, lactoferrin, and zinc-α2-glycoprotein (ZAG). | [113] |
| PEA | Tears were collected with Schirmer strip | 35.5 ± 14.9/36.9 ± 13.5 | AC/KC | There were notable differences between AC and KC in MLN and PTH1R, five-fold for IFNG and TPSAB1, and four-fold for IL17F, ITGA6, and ISM1. Additionally, they identified biological processes associated with the regulation of gene expression, cell population proliferation, transcription by RNA polymerase II, ERK1/2 cascade, and immune responses in AC and KC. | [114] |
| LC-MS/MS | Corneal tissue | 62.4 ± 12.4/27.5 ± 8.04 | Control/KC | Of 3132 proteins, 205 were modified with proline oxidation and all of the detected collagens fell. The associated biological processes were EIF2 signaling, complement system, regulation of eIF4 and p70S6K signaling, acute-phase response signaling, and mTOR signaling. | [115] |
| Type of Analysis | Methodology Employed | Population Age (Mean) | Sample | Findings | Reference |
|---|---|---|---|---|---|
| Epigenomic | Corneal epithelium | 52 ± 24.92/41.6 ± 7.81 | Control/KC | Methylated regions were found in linkage loci (3p14.3, 5q35.2, 13q32.3, 15q24.1, and 20p13) and DNA methylation changes in WNT3 and WNT5A. | [45] |
| Microarrays | Corneal epithelium and the stroma tissue | 75.0 ± 11.2/53.95 ± 12.1 | Control/KC | Downregulated miRNAs in KC epithelium or KC stroma were miR-634 and miR-936. In addition, the most upregulated miRNAs were miR-211-3p, miR184, and miR-135b-5p. | [94] |
| ATAC-seq | Immortalized corneal epithelial and keratocyte cell lines | KC | They reported fine-mapping identifies a subset of 55 highly likely causal variants, 91% of which are non-coding. They identified regions of open chromatin, for example, locus 101 associated with DPF3 gene a transcription regulator, histone acetyl-lysine reader. | [128] |
| Type of Analysis | Methodology Employed | Population Age (Mean) | Sample | Findings | References |
|---|---|---|---|---|---|
| GC/MS | 54.5/50.5 | Control/KC | They identified 13 metabolites whose levels differentiated between groups of samples. Downregulation of several carboxylic acids, fatty acids, and steroids was observed in KC. These metabolites were associated with lipid metabolism and energy production. | [60] | |
| LC-MS/MS | Tears were collected in capillary glass | 38 ± 7.02/29.7 ± 9.27 | Control/KC | A total of 296 different metabolites were identified, glycolysis and gluconeogenesis had significant changes, such as 3-phosphoglycerate and 1,3 diphopshateglycerate. As a result, the citric acid cycle (TCA) changes in isocitrate, aconitate, malate, and acetylphosphate were shown. | [147] |
| LC-Q-ToF/MS | Serum | 31.5 | Control/KC | Upregulated dehydroepiandrosterone sulfate from the steroidal hormone synthesis pathway and metabolic pathways associated with prostaglandin F2α, prostaglandin A2. | [148] |
| LC-MS | Epithelialized rabbit corneas | Corneal epithelium of rabbits/WST-D/NIR crosslinking/RF-D/UVA crosslinking/received no further treatment. | The pathways were significantly downregulated in RHOB GTPase cycle, repression of retinoic acid–induced cell differentiation, creatine metabolism, activation of the phototransduction cascade, GP1b-IX-V activation signaling. Another significantly downregulated pathway was the activation of matrix metalloproteinases (MMPs) related to ECM organization, which included TIMP-2 and MMP-2. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durán-Cristiano, S.C.; Bustamante-Arias, A.; Fernandez, G.J.; Martin-Gil, A.; Carracedo, G. Omics in Keratoconus: From Molecular to Clinical Practice. J. Clin. Med. 2025, 14, 2459. https://doi.org/10.3390/jcm14072459
Durán-Cristiano SC, Bustamante-Arias A, Fernandez GJ, Martin-Gil A, Carracedo G. Omics in Keratoconus: From Molecular to Clinical Practice. Journal of Clinical Medicine. 2025; 14(7):2459. https://doi.org/10.3390/jcm14072459
Chicago/Turabian StyleDurán-Cristiano, Sandra Carolina, Andres Bustamante-Arias, Geysson Javier Fernandez, Alba Martin-Gil, and Gonzalo Carracedo. 2025. "Omics in Keratoconus: From Molecular to Clinical Practice" Journal of Clinical Medicine 14, no. 7: 2459. https://doi.org/10.3390/jcm14072459
APA StyleDurán-Cristiano, S. C., Bustamante-Arias, A., Fernandez, G. J., Martin-Gil, A., & Carracedo, G. (2025). Omics in Keratoconus: From Molecular to Clinical Practice. Journal of Clinical Medicine, 14(7), 2459. https://doi.org/10.3390/jcm14072459








