Urinary and Plasma miRNAs in the Early Detection of Acute Kidney Injury and Their Possible Role as Therapeutic Targets
Abstract
1. Introduction
Aim of the Work
2. Material and Methods
3. MiRNAs
4. The Role of miRNAs in AKI
4.1. MiRNAs as Diagnostic Biomarkers and Therapeutic Targets
| miRNA | Effect | AKI Model | Origin of Sample | Reference |
|---|---|---|---|---|
| miR-21-5p | Decrease in serum inflammation, apoptosis and oxidative stress through RUNX1 | Sepsis-induced AKI | Serum and kidney tissue | Zhang et al., 2021 [24] |
| miR-374a-3p | Decrease in apoptosis and cytokines release through a down-regulation of SNHG5 through the axis TLR4/NF-ĸB | Sepsis-induced AKI | Serum | Wang et al., 2021 [25] |
| miR-29a and -10a-5p | Good predictors for 28-day survival | Sepsis-induced AKI | Serum | Huo et al., 2017 [26] |
| miR-30a, -30c, and -30e | >2-fold increase in contrast-induced AKI patients when compared with controls | Contrast-induced AKI | Plasma and kidney tissue | Gutiérrez-Escolano et al., 2015 [27] |
| miR-188, -30 and -30e | >1.5-fold increase in contrast-induced AKI patients when compared with controls | Contrast-induced AKI | Plasma and kidney tissue | Sun et al., 2016 [28] |
| miR-30c-5p and -192-5p | Significant increase in urine levels 2 h after cardiac surgery | Post-operative AKI | Urine | Zou et al., 2017 [29] |
| miR-21 | An increase in urine and plasma levels is associated with the development and progression of AKI and predicts the need for postoperative renal replacement therapy, 30-day in-hospital mortality and prolonged stay in hospital or in intensive care unit | Post-operative AKI | Urine and plasma | Du et al., 2013 [30] |
| miR-330-5p | Reduction of renal cell apoptosis in the setting of ischemic AKI, stimulated by long non-coding RNA 122049 | I/R injury | Serum and kidney tissue | Xiao et al., 2022 [32] |
| miR-21 | Antiapoptotic effect in delayed ischemic preconditioning in mice | I/R injury | Serum and kidney tissue | Xu et al., 2012 [31] |
| miR-17-5p | Antiapoptotic effect during hypoxia through the inhibition of the expression of DR6 | I/R injury | Kidney tissue | Hao et al., 2018 [33] |
| miR-874-3p | Attenuation of renal tubular epithelial cell injury and enhancement of repair mechanisms after kidney injury | Cisplatin induced AKI | Serum and kidney tissue | Yu et al., 2023 [39] |
| miR -24, -126, -494 and -687 | Regulation of inflammation, cell cycle and programmed cell death in the repair stages of AKI | AKI progression | Serum | Ren et al., 2018 [40] |
| miR-21 | Its upregulation promotes renal fibrosis through the suppression of peroxisome proliferator-activator receptor alpha and phosphatase and tensin homolog | AKI progression | Serum | Lv et al., 2018 [41] McClelland et al., 2015 [42] |
| miR-29a, -29b and -29c | Antifibrotic effect | Renal fibrosis in diabetic nephropathy | Kidney tissue | Wang et al., 2012 [43] |
| AKI Etiology | miRNA | Role in Clinical Practice |
|---|---|---|
| Sepsis | miR-21-5p, miR-374a-3p | Therapeutic targets |
| miR-29-a, miR-10a-5p | Mortality predictors | |
| Contrast-induced AKI | miR-30a, miR-30c miR-30e, miRNA-188 | Diagnostic biomarkers |
| miR-30c-5p, miR-192-5p | Diagnostic biomarkers | |
| Cardiac surgery | miR-21 | Predictor of AKI progression, the need of RRT, mortality and prolonged stay |
| Ischemia/Reperfusion injury | miR-330-5p miR-21, miR-17-5p | Therapeutic targets |
| Kidney transplant-associated AKI | miR-155, miR-125 | Diagnostic biomarkers |
| miR-210 | Diagnostic biomarker and therapeutic target |
4.2. MiRNAs Involved in Fibrosis Progression and CKD
5. Limitations and Clinical Challenges
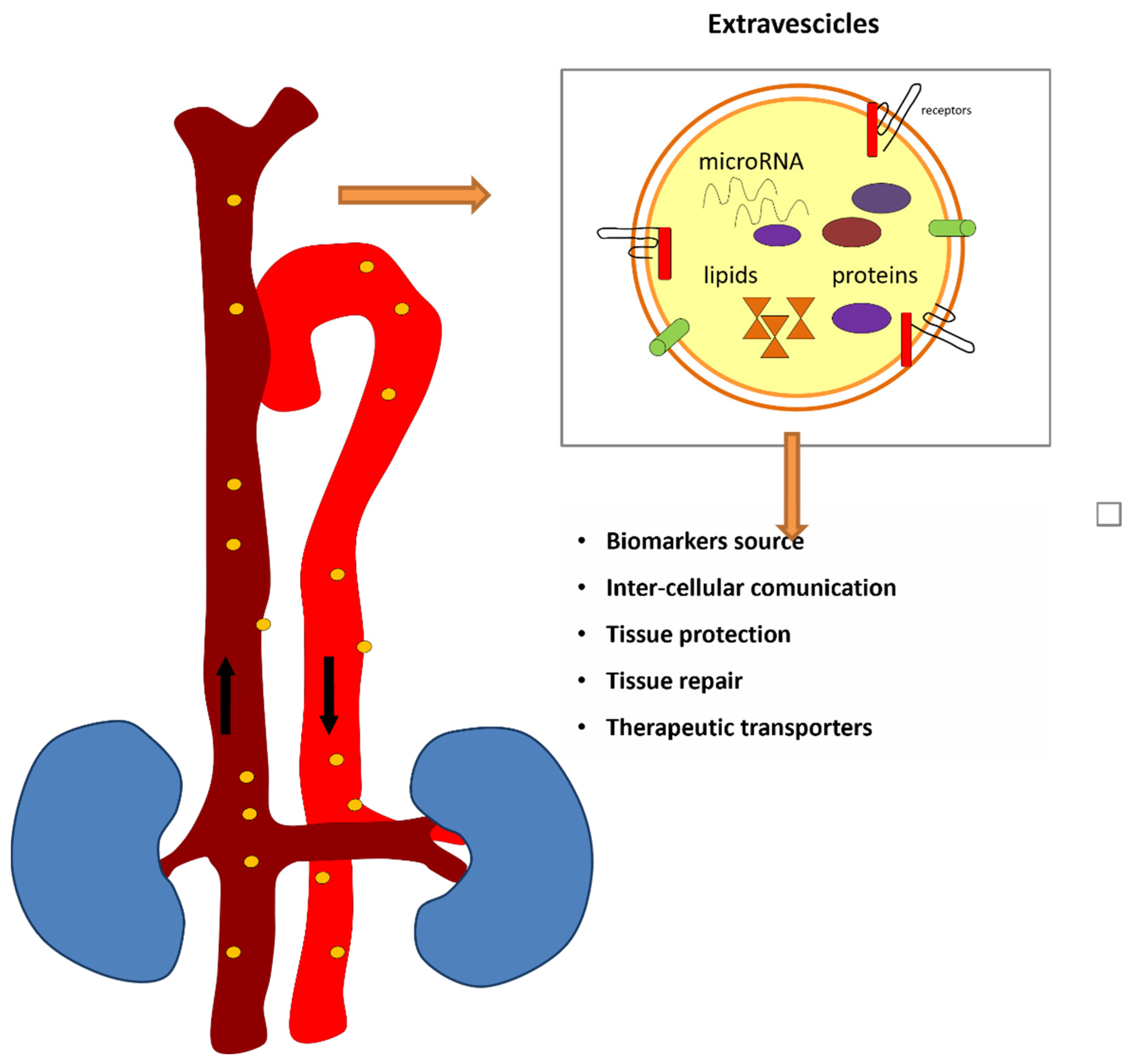
6. Extracellular Vesicles and Circulating miRNAs
7. Limitations and Clinical Challenges
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AKI | Acute kidney injury |
| CKD | Chronic kidney disease |
| IR | Ischemia–reperfusion |
| IRI | Ischemia–reperfusion injury |
| miRNA | MicroRNA |
| Evs | Extracellular vesicles |
| MVs | Microvesicles |
| piRNA | Piwi-interacting RNA |
| rasi-RNA | Repeat-associated small interfering RNA |
| si-RNA | Small interfering RNA |
| sco-RNA | Small cell osteosarcoma RNA |
| sn-RNA | Small nuclear RNA |
| sno-RNA | Small nucleolar RNA |
| ti-RNA | Transcription initiation RNA |
References
- Ülger, F.; Pehlivanlar Küçük, M.; Küçük, A.O.; İlkaya, N.K.; Murat, N.; Bilgiç, B.; Abanoz, H. Evaluation of Acute Kidney Injury (AKI) with RIFLE, AKIN, CK, and KDIGO in Critically Ill Trauma Patients. Eur. J. Trauma Emerg. Surg. 2018, 44, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Susantitaphong, P.; Cruz, D.N.; Cerda, J.; Abulfaraj, M.; Alqahtani, F.; Koulouridis, I.; Jaber, B.L.; Acute Kidney Injury Advisory Group of the American Society of Nephrology. World Incidence of AKI: A Meta-Analysis. Clin. J. Am. Soc. Nephrol. 2013, 8, 1482–1493. [Google Scholar] [CrossRef] [PubMed]
- Hoste, E.A.J.; Bagshaw, S.M.; Bellomo, R.; Cely, C.M.; Colman, R.; Cruz, D.N.; Edipidis, K.; Forni, L.G.; Gomersall, C.D.; Govil, D.; et al. Epidemiology of Acute Kidney Injury in Critically Ill Patients: The Multinational AKI-EPI Study. Intensive Care Med. 2015, 41, 1411–1423. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.F.; Forni, L.G. Acute Kidney Injury: Short-Term and Long-Term Effects. Crit. Care 2016, 20, 188. [Google Scholar] [CrossRef]
- Cerdá, J.; Lameire, N.; Eggers, P.; Pannu, N.; Uchino, S.; Wang, H.; Bagga, A.; Levin, A. Epidemiology of Acute Kidney Injury. Clin. J. Am. Soc. Nephrol. 2008, 3, 881–886. [Google Scholar] [CrossRef]
- Bhosale, S.J.; Kulkarni, A.P. Biomarkers in Acute Kidney Injury. Indian J. Crit. Care Med. 2020, 24, S90–S93. [Google Scholar] [CrossRef]
- Alge, J.L.; Arthur, J.M. Biomarkers of AKI: A Review of Mechanistic Relevance and Potential Therapeutic Implications. Clin. J. Am. Soc. Nephrol. 2015, 10, 147–155. [Google Scholar] [CrossRef]
- McMahon, B.A.; Koyner, J.L. Risk Stratification for Acute Kidney Injury: Are Biomarkers Enough? Adv. Chronic Kidney Dis. 2016, 23, 167–178. [Google Scholar] [CrossRef]
- Teo, S.H.; Endre, Z.H. Biomarkers in Acute Kidney Injury (AKI). Best Pract. Res. Clin. Anaesthesiol. 2017, 31, 331–344. [Google Scholar] [CrossRef]
- Han, W.K.; Wagener, G.; Zhu, Y.; Wang, S.; Lee, H.T. Urinary Biomarkers in the Early Detection of Acute Kidney Injury after Cardiac Surgery. Clin. J. Am. Soc. Nephrol. 2009, 4, 873–882. [Google Scholar] [CrossRef]
- Koyner, J.L.; Bennett, M.R.; Worcester, E.M.; Ma, Q.; Raman, J.; Jeevanandam, V.; Kasza, K.E.; O’Connor, M.F.; Konczal, D.J.; Trevino, S.; et al. Urinary Cystatin C as an Early Biomarker of Acute Kidney Injury Following Adult Cardiothoracic Surgery. Kidney Int. 2008, 74, 1059–1069. [Google Scholar] [CrossRef] [PubMed]
- Kashani, K.; Al-Khafaji, A.; Ardiles, T.; Artigas, A.; Bagshaw, S.M.; Bell, M.; Bihorac, A.; Birkhahn, R.; Cely, C.M.; Chawla, L.S.; et al. Discovery and Validation of Cell Cycle Arrest Biomarkers in Human Acute Kidney Injury. Crit. Care 2013, 17, R25. [Google Scholar] [CrossRef] [PubMed]
- Brandenburger, T.; Salgado Somoza, A.; Devaux, Y.; Lorenzen, J.M. Noncoding RNAs in Acute Kidney Injury. Kidney Int. 2018, 94, 870–881. [Google Scholar] [CrossRef]
- Hüttenhofer, A.; Mayer, G. Circulating MiRNAs as Biomarkers of Kidney Disease. Clin. Kidney J. 2017, 10, 27–29. [Google Scholar] [CrossRef]
- Trionfini, P.; Benigni, A.; Remuzzi, G. MicroRNAs in Kidney Physiology and Disease. Nat. Rev. Nephrol. 2015, 11, 23–33. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Li, Y.-F.; Jing, Y.; Hao, J.; Frankfort, N.C.; Zhou, X.; Shen, B.; Liu, X.; Wang, L.; Li, R. MicroRNA-21 in the Pathogenesis of Acute Kidney Injury. Protein Cell 2013, 4, 813–819. [Google Scholar] [CrossRef]
- Saliminejad, K.; KhorramKhorshid, H.R.; SoleymaniFard, S.; Ghaffari, S.H. An Overview of MicroRNAs: Biology, Functions, Therapeutics, and Analysis Methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Wang, F.; Chen, C.; Wang, D. Circulating MicroRNAs in Cardiovascular Diseases: From Biomarkers to Therapeutic Targets. Front. Med. 2014, 8, 404–418. [Google Scholar] [CrossRef]
- Pavkovic, M.; Vaidya, V.S. MicroRNAs and Drug-Induced Kidney Injury. Pharmacol. Ther. 2016, 163, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.F.; Bekele, S.; O’Dwyer, M.J.; Prowle, J.R. MicroRNAs in Acute Kidney Injury. Nephron 2018, 140, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Connor, K.L.; Denby, L. MicroRNAs as Non-Invasive Biomarkers of Renal Disease. Nephrol. Dial. Transplant. 2021, 36, 428–429. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, H.; Liu, W.; Liu, S.; Wang, X.Y.; Diao, Z.L.; Zhang, A.H.; Guo, W.; Han, X.; Dong, X.; et al. Endothelial Progenitor Cells-Derived Exosomal MicroRNA-21-5p Alleviates Sepsis-Induced Acute Kidney Injury by Inhibiting RUNX1 Expression. Cell Death Dis. 2021, 12, 335. [Google Scholar] [CrossRef]
- Wang, M.; Wei, J.; Shang, F.; Zang, K.; Zhang, P. Down-Regulation of LncRNA SNHG5 Relieves Sepsis-Induced Acute Kidney Injury by Regulating the MiR-374a-3p/TLR4/NF-ΚB Pathway. J. Biochem. 2021, 169, 575–583. [Google Scholar] [CrossRef]
- Huo, R.; Dai, M.; Fan, Y.; Zhou, J.-Z.; Li, L.; Zu, J. Predictive Value of MiRNA-29a and MiRNA-10a-5p for 28-Day Mortality in Patients with Sepsis-Induced Acute Kidney Injury. Nan Fang Yi Ke Da Xue Xue Bao 2017, 37, 646–651. [Google Scholar] [CrossRef]
- Gutiérrez-Escolano, A.; Santacruz-Vázquez, E.; Gómez-Pérez, F. Dysregulated MicroRNAs Involved in Contrast-Induced Acute Kidney Injury in Rat and Human. Ren. Fail. 2015, 37, 1498–1506. [Google Scholar] [CrossRef]
- Sun, S.-Q.; Zhang, T.; Ding, D.; Zhang, W.-F.; Wang, X.-L.; Sun, Z.; Hu, L.-H.; Qin, S.-Y.; Shen, L.-H.; He, B. Circulating MicroRNA-188, -30a, and -30e as Early Biomarkers for Contrast-Induced Acute Kidney Injury. J. Am. Heart Assoc. 2016, 5, e004138. [Google Scholar] [CrossRef]
- Zou, Y.-F.; Wen, D.; Zhao, Q.; Shen, P.-Y.; Shi, H.; Zhao, Q.; Chen, Y.-X.; Zhang, W. Urinary MicroRNA-30c-5p and MicroRNA-192-5p as Potential Biomarkers of Ischemia-Reperfusion-Induced Kidney Injury. Exp. Biol. Med. 2017, 242, 657–667. [Google Scholar] [CrossRef]
- Du, J.; Cao, X.; Zou, L.; Chen, Y.; Guo, J.; Chen, Z.; Hu, S.; Zheng, Z. MicroRNA-21 and Risk of Severe Acute Kidney Injury and Poor Outcomes after Adult Cardiac Surgery. PLoS ONE 2013, 8, e63390. [Google Scholar] [CrossRef]
- Xu, X.; Kriegel, A.J.; Liu, Y.; Usa, K.; Mladinov, D.; Liu, H.; Fang, Y.; Ding, X.; Liang, M. Delayed Ischemic Preconditioning Contributes to Renal Protection by Upregulation of MiR-21. Kidney Int. 2012, 82, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, X.; Qiu, S.; Wang, Y.; Zhang, D. LncRNA 122049 Suppresses Apoptosis of Renal Tubular Epithelial Cells in Ischemic AKI by Targeting the MiR-330-5p/ELK1 Axis. FASEB J. 2022, 36, e22395. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Wei, Q.; Mei, S.; Li, L.; Su, Y.; Mei, C.; Dong, Z. Induction of MicroRNA-17-5p by P53 Protects against Renal Ischemia-Reperfusion Injury by Targeting Death Receptor 6. Kidney Int. 2017, 91, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Wilflingseder, J.; Regele, H.; Perco, P.; Kainz, A.; Soleiman, A.; Mühlbacher, F.; Mayer, B.; Oberbauer, R. miRNA Profiling Discriminates Types of Rejection and Injury in Human Renal Allografts. Transplantation 2013, 95, 835–841. [Google Scholar] [CrossRef]
- Wilflingseder, J.; Sunzenauer, J.; Toronyi, E.; Heinzel, A.; Kainz, A.; Mayer, B.; Perco, P.; Telkes, G.; Langer, R.M.; Oberbauer, R. Molecular Pathogenesis of Post-Transplant Acute Kidney Injury: Assessment of Whole-Genome mRNA and miRNA Profiles. PLoS ONE 2014, 9, e104164. [Google Scholar] [CrossRef]
- Khalid, U.; Newbury, L.J.; Simpson, K.; Jenkins, R.H.; Bowen, T.; Bates, L.; Sheerin, N.S.; Chavez, R.; Fraser, D.J. A Urinary microRNA Panel that is an Early Predictive Biomarker of Delayed Graft Function Following Kidney Transplantation. Sci. Rep. 2019, 9, 3584. [Google Scholar] [CrossRef]
- Cooper, J.E.; Wiseman, A.C. Acute Kidney Injury in Kidney Transplantation. Curr. Opin. Nephrol. Hypertens. 2013, 22, 698–703. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Volkmann, I.; Fiedler, J.; Schmidt, M.; Scheffner, I.; Haller, H.; Gwinner, W.; Thum, T. Urinary miR-210 as a Mediator of Acute T-Cell Mediated Rejection in Renal Allograft Recipients. Am. J. Transplant. 2011, 11, 2221–2227. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, M.; Guo, Q.; Shen, L.; Liu, X.; Pan, J.; Zhang, Y.; Xu, T.; Zhang, D.; Wei, G. Human Umbilical Cord Mesenchymal Stem Cell Exosome-Derived MiR-874-3p Targeting RIPK1/PGAM5 Attenuates Kidney Tubular Epithelial Cell Damage. Cell Mol. Biol. Lett. 2023, 28, 12. [Google Scholar] [CrossRef]
- Ren, G.-L.; Zhu, J.; Li, J.; Meng, X.-M. Noncoding RNAs in Acute Kidney Injury. J. Cell. Physiol. 2019, 234, 2266–2276. [Google Scholar] [CrossRef]
- Lv, W.; Fan, F.; Wang, Y.; Gonzalez-Fernandez, E.; Wang, C.; Yang, L.; Booz, G.W.; Roman, R.J. Therapeutic Potential of MicroRNAs for the Treatment of Renal Fibrosis and CKD. Physiol. Genom. 2018, 50, 20–34. [Google Scholar] [CrossRef] [PubMed]
- McClelland, A.D.; Herman-Edelstein, M.; Komers, R.; Jha, J.C.; Winbanks, C.E.; Hagiwara, S.; Gregorevic, P.; Kantharidis, P.; Cooper, M.E. MiR-21 Promotes Renal Fibrosis in Diabetic Nephropathy by Targeting PTEN and SMAD7. Clin. Sci. 2015, 129, 1237–1249. [Google Scholar] [CrossRef]
- Wang, B.; Komers, R.; Carew, R.; Winbanks, C.E.; Xu, B.; Herman-Edelstein, M.; Koh, P.; Thomas, M.; Jandeleit-Dahm, K.; Gregorevic, P.; et al. Suppression of MicroRNA-29 Expression by TGF-Β1 Promotes Collagen Expression and Renal Fibrosis. J. Am. Soc. Nephrol. 2012, 23, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Bang, C.; Thum, T. Circulating MicroRNAs as Biomarkers and Potential Paracrine Mediators of Cardiovascular Disease. Circ. Cardiovasc. Genet. 2010, 3, 484–488. [Google Scholar] [CrossRef]
- Lorenzen, J.M.; Batkai, S.; Thum, T. Regulation of Cardiac and Renal Ischemia–Reperfusion Injury by MicroRNAs. Free Radic. Biol. Med. 2013, 64, 78–84. [Google Scholar] [CrossRef]
- Kumar, M.A.; Baba, S.K.; Sadida, H.Q.; Marzooqi, S.A.; Jerobin, J.; Altemani, F.H.; Algehainy, N.; Alanazi, M.A.; Abou-Samra, A.B.; Kumar, R.; et al. Extracellular Vesicles as Tools and Targets in Therapy for Diseases. Signal Transduct. Target. Ther. 2024, 9, 27. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/Microvesicles as a Mechanism of Cell-to-Cell Communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef]
- Schiffelers, R.; Kooijmans, S.; Vader, P.; van Dommelen, S.; van Solinge, W. Exosome Mimetics: A Novel Class of Drug Delivery Systems. Int. J. Nanomed. 2012, 7, 1525. [Google Scholar] [CrossRef]
- Jin, C.; Wu, P.; Wu, W.; Chen, W.; Liu, W.; Zhu, Y.; Wu, Q.; Chen, B.; Ji, C.; Qian, H. Therapeutic Role of hucMSC-sEV-enriched miR-13896 in Cisplatin-Induced Acute Kidney Injury through M2 Macrophage Polarization. Cell Biol. Toxicol. 2025, 41, 50. [Google Scholar] [CrossRef]
- Zhu, Z.; Wang, D.; Lu, X.; Jiang, T.; Zhang, L.; Chen, M.; Chen, S. Platelet-derived Extracellular Vesicles are Associated with Kidney Injury in Patients with Urosepsis. Mol. Cell. Probes 2024, 73, 101949. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Ma, W.; Wang, L.; Long, C.; Chen, S.; Liu, J.; Qian, Y.; Zhao, J.; Zhou, C.; Jia, R. Physically Engineered Extracellular Vesicles Targeted Delivering miR-21-5p to Promote Renoprotection after Renal Ischemia-Reperfusion Injury. Mater. Today Bio 2025, 31, 101528. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Kim, H.; Kim, J.; Kim, S.; Park, T.S.; Kim, T.M. Extracellular Vesicles from Induced Pluripotent Stem Cell-derived Mesenchymal Stem Cells Enhance the Recovery of Acute Kidney Injury. Cytotherapy 2024, 26, 51–62. [Google Scholar] [CrossRef]
- Yavuz, H.; Weder, M.M.; Erdbrügger, U. Extracellular Vesicles in Acute Kidney Injury. Nephron 2023, 147, 48–51. [Google Scholar] [CrossRef]
- Chen, G.; Li, X.; Zhou, X.; Li, Y.; Yu, H.; Peng, X.; Bai, X.; Zhang, C.; Feng, Z.; Mei, Y.; et al. Extracellular Vesicles Secreted from Mesenchymal Stem Cells Ameliorate Renal Ischemia Reperfusion Injury by Delivering miR-100-5p Targeting FKBP5/AKT Axis. Sci. Rep. 2024, 14, 6720. [Google Scholar] [CrossRef]
- Wang, X.; Kim, C.S.; Adams, B.C.; Wilkinson, R.; Hill, M.M.; Shah, A.K.; Mohamed, A.; Dutt, M.; Ng, M.S.Y.; Ungerer, J.P.J.; et al. Human Proximal Tubular Epithelial Cell-derived Small Extracellular Vesicles Mediate Synchronized Tubular Ferroptosis in Hypoxic Kidney Injury. Redox Biol. 2024, 70, 103042. [Google Scholar] [CrossRef]
- Jeon, J.S.; Kim, E.; Bae, Y.U.; Yang, W.M.; Lee, H.; Kim, H.; Noh, H.; Han, D.C.; Ryu, S.; Kwon, S.H. microRNA in Extracellular Vesicles Released by Damaged Podocytes Promote Apoptosis of Renal Tubular Epithelial Cells. Cells 2020, 9, 1409. [Google Scholar] [CrossRef]
- Xie, L.; Zhang, K.; Pan, K.; Su, X.; Zhao, X.; Li, R.; Wang, Y.; Pang, H.; Fu, E.; Li, Z. Engineered Extracellular Vesicles Promote the Repair of Acute Kidney Injury by Modulating Regulatory T Cells and the Immune Microenvironment. J. Transl. Med. 2025, 23, 304. [Google Scholar] [CrossRef]
- Lundy, D.J.; Szomolay, B.; Liao, C.T. Systems Approaches to Cell Culture-Derived Extracellular Vesicles for Acute Kidney Injury Therapy: Prospects and Challenges. Function 2024, 5, zqae012. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, H.; Zhang, L.; Yao, P.; Wang, S.; Yang, Q. Nanosystems for Oxidative Stress Regulation in the Anti-inflammatory Therapy of Acute Kidney Injury. Front. Bioeng. Biotechnol. 2023, 11, 1120148. [Google Scholar] [CrossRef]
- Gao, L.; Zhong, X.; Jin, J.; Li, J.; Meng, X.M. Potential Targeted Therapy and Diagnosis Based on Novel Insight into Growth Factors, Receptors, and Downstream Effectors in Acute Kidney Injury and Acute Kidney Injury-Chronic Kidney Disease Progression. Signal Transduct. Target. Ther. 2020, 5, 9. [Google Scholar] [CrossRef] [PubMed]
- Kosanović, M.; Milutinovic, B.; Glamočlija, S.; Morlans, I.M.; Ortiz, A.; Bozic, M. Extracellular Vesicles and Acute Kidney Injury: Potential Therapeutic Avenue for Renal Repair and Regeneration. Int. J. Mol. Sci. 2022, 23, 3792. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Farber, E.L.; Rapoport, A.L.; Tejada, D.; Deniskin, R.; Akhmedov, N.B.; Farber, D.B. Transfer of MicroRNAs by Embryonic Stem Cell Microvesicles. PLoS ONE 2009, 4, e4722. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T.S. Challenges and Opportunities in Exosome Research-Perspectives from Biology, Engineering, and Cancer Therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef]
- Ciferri, M.C.; Quarto, R.; Tasso, R. Extracellular Vesicles as Biomarkers and Therapeutic Tools: From Pre-Clinical to Clinical Applications. Biology 2021, 10, 359. [Google Scholar] [CrossRef]
- Sonbhadra, S.; Mehak; Pandey, L.M. Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics. Biosensors 2023, 13, 802. [Google Scholar] [CrossRef]
- Lorite, P.; Domínguez, J.N.; Palomeque, T.; Torres, M.I. Extracellular Vesicles: Advanced Tools for Disease Diagnosis, Monitoring, and Therapies. Int. J. Mol. Sci. 2024, 26, 189. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clementi, A.; Virzì, G.M.; Ronco, C.; Monciino, P.; Zanella, M. Urinary and Plasma miRNAs in the Early Detection of Acute Kidney Injury and Their Possible Role as Therapeutic Targets. J. Clin. Med. 2025, 14, 2306. https://doi.org/10.3390/jcm14072306
Clementi A, Virzì GM, Ronco C, Monciino P, Zanella M. Urinary and Plasma miRNAs in the Early Detection of Acute Kidney Injury and Their Possible Role as Therapeutic Targets. Journal of Clinical Medicine. 2025; 14(7):2306. https://doi.org/10.3390/jcm14072306
Chicago/Turabian StyleClementi, Anna, Grazia Maria Virzì, Claudio Ronco, Paola Monciino, and Monica Zanella. 2025. "Urinary and Plasma miRNAs in the Early Detection of Acute Kidney Injury and Their Possible Role as Therapeutic Targets" Journal of Clinical Medicine 14, no. 7: 2306. https://doi.org/10.3390/jcm14072306
APA StyleClementi, A., Virzì, G. M., Ronco, C., Monciino, P., & Zanella, M. (2025). Urinary and Plasma miRNAs in the Early Detection of Acute Kidney Injury and Their Possible Role as Therapeutic Targets. Journal of Clinical Medicine, 14(7), 2306. https://doi.org/10.3390/jcm14072306








