Current Role of Artificial Intelligence in the Management of Esophageal Cancer
Abstract
1. Introduction
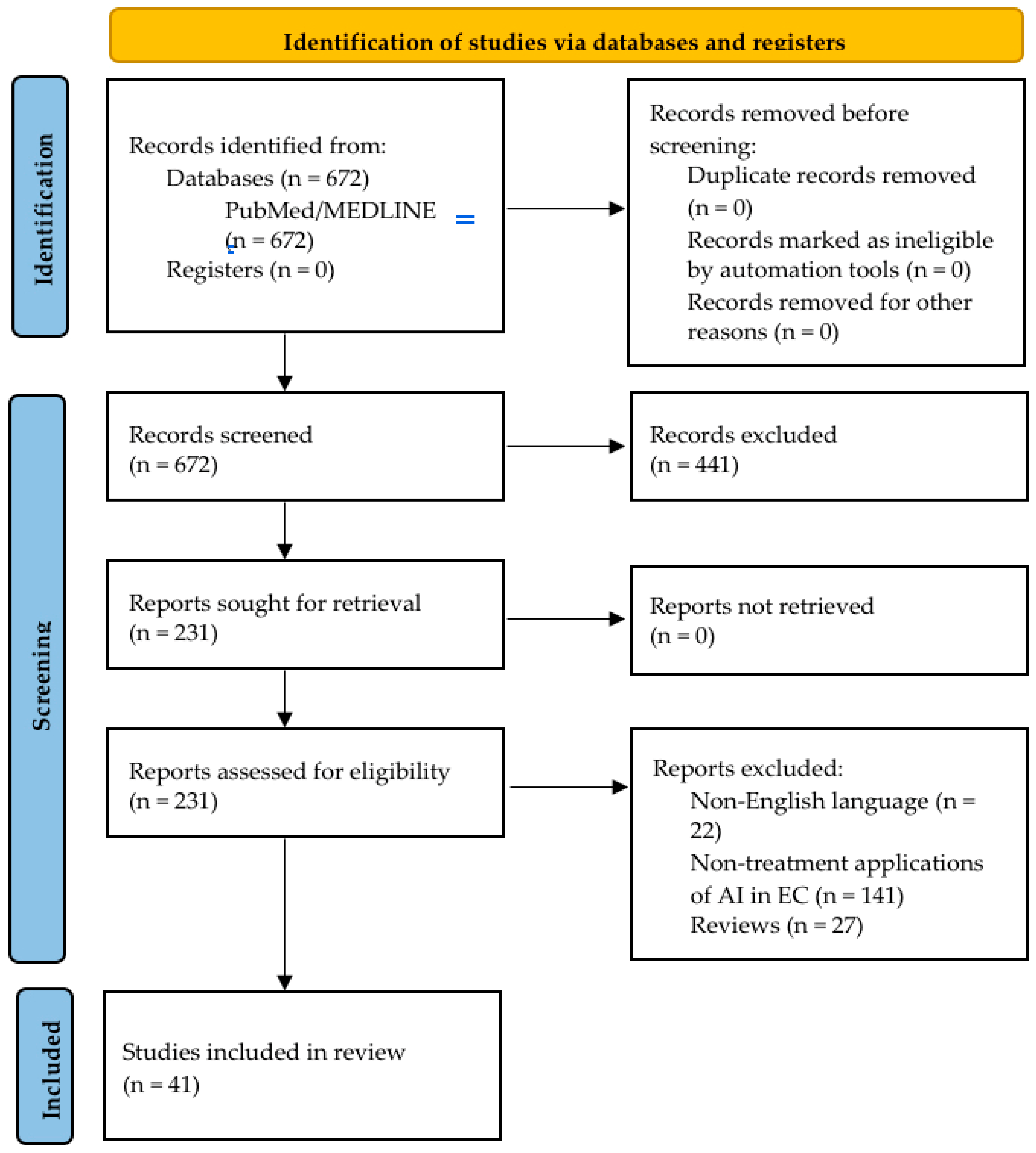
2. Materials and Methods
3. AI Applications in EC Treatment and Prognosis Prediction
3.1. AI in Treatment Decision and Planning
3.2. AI in EC Surgical Treatment
3.3. AI in Treatment Response Prediction
3.4. AI in EC Prognosis
4. Limitations
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| EC | Esophageal cancer |
| AI | Artificial Intelligence |
| ML | Machine learning |
| DL | Deep learning |
| AUC | Area under the curve |
| ANN | Artificial neural network |
| CTV | Clinical target volume |
| CNN | Convolutional neural network |
| pCR | Pathological complete response |
| ESCC | Esophageal squamous cell carcinoma |
| SUV | Standardized uptake value |
| PET | Positron emission tomography |
| PFS | Progression-free survival |
| OS | Overall survival |
| MRI | Magnetic resonance imaging |
| DFS | Disease-free survival |
References
- Collaborators CBDOC. The global, regional, and national burden of oesophageal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.Q.; Ma, Y.L.; Qin, Q.; Wang, P.H.; Luo, Y.; Xu, P.F.; Cui, Y. Epidemiology of esophageal cancer in 2020 and projections to 2030 and 2040. Thorac. Cancer 2023, 14, 3–11. [Google Scholar] [CrossRef]
- Arnold, M.; Laversanne, M.; Brown, L.M.; Devesa, S.S.; Bray, F. Predicting the Future Burden of Esophageal Cancer by Histological Subtype: International Trends in Incidence up to 2030. Am. J. Gastroenterol. 2017, 112, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Cancer Facts & Figures 2024; American Cancer Society: Atlanta, GA, USA, 2024. [Google Scholar]
- Global Burden of Disease Cancer Collaboration; Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548, Erratum in JAMA Oncol. 2017, 3, 418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Mu, W.; Dong, D.; Wei, J.; Fang, M.; Shao, L.; Zhou, Y.; He, B.; Zhang, S.; Liu, Z.; et al. The Applications of Artificial Intelligence in Digestive System Neoplasms: A Review. Health Data Sci. 2023, 3, 0005. [Google Scholar] [CrossRef]
- Moglia, A.; Georgiou, K.; Morelli, L.; Toutouzas, K.; Satava, R.M.; Cuschieri, A. Breaking down the silos of artificial intelligence in surgery: Glossary of terms. Surg. Endosc. 2022, 36, 7986–7997. [Google Scholar] [CrossRef]
- Garbarino, G.M.; Polici, M.; Caruso, D.; Laghi, A.; Mercantini, P.; Pilozzi, E.; van Berge Henegouwen, M.I.; Gisbertz, S.S.; van Grieken, N.C.T.; Berardi, E.; et al. Radiomics in Oesogastric Cancer: Staging and Prediction of Preoperative Treatment Response: A Narrative Review and the Results of Personal Experience. Cancers 2024, 16, 2664. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Guo, L.J.; Yuan, X.L.; Hu, B. Artificial intelligence-assisted esophageal cancer management: Now and future. World J. Gastroenterol. 2020, 26, 5256–5271. [Google Scholar] [CrossRef]
- Tsai, T.J.; Mukundan, A.; Chi, Y.S.; Tsao, Y.M.; Wang, Y.K.; Chen, T.H.; Wu, I.C.; Huang, C.W.; Wang, H.C. Intelligent Identification of Early Esophageal Cancer by Band-Selective Hyperspectral Imaging. Cancers 2022, 14, 4292. [Google Scholar] [CrossRef]
- Fang, Y.J.; Huang, C.W.; Karmakar, R.; Mukundan, A.; Tsao, Y.M.; Yang, K.Y.; Wang, H.C. Assessment of Narrow-Band Imaging Algorithm for Video Capsule Endoscopy Based on Decorrelated Color Space for Esophageal Cancer: Part II, Detection and Classification of Esophageal Cancer. Cancers 2024, 16, 572. [Google Scholar] [CrossRef]
- Fang, Y.J.; Mukundan, A.; Tsao, Y.M.; Huang, C.W.; Wang, H.C. Identification of Early Esophageal Cancer by Semantic Segmentation. J. Pers. Med. 2022, 12, 1204. [Google Scholar] [CrossRef] [PubMed]
- Thavanesan, N.; Bodala, I.; Walters, Z.; Ramchurn, S.; Underwood, T.J.; Vigneswaran, G. Machine learning to predict curative multidisciplinary team treatment decisions in oesophageal cancer. Eur. J. Surg. Oncol. 2023, 49, 106986. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, H.; Mehrshad, N.; Anvari, K. Intelligent modelling of oesophageal cancer treatment and its use to determine the dose of chemotherapy drug. J. Med. Eng. Technol. 2012, 36, 261–266. [Google Scholar] [CrossRef]
- Barragán-Montero, A.M.; Thomas, M.; Defraene, G.; Michiels, S.; Haustermans, K.; Lee, J.A.; Sterpin, E. Deep learning dose prediction for IMRT of esophageal cancer: The effect of data quality and quantity on model performance. Phys. Med. 2021, 83, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Pei, X.; Ge, N.; Zheng, C. Clinical Target Volume Auto-Segmentation of Esophageal Cancer for Radiotherapy After Radical Surgery Based on Deep Learning. Technol. Cancer Res. Treat. 2021, 20, 15330338211034284. [Google Scholar] [CrossRef]
- Jin, L.; Chen, Q.; Shi, A.; Wang, X.; Ren, R.; Zheng, A.; Song, P.; Zhang, Y.; Wang, N.; Wang, C.; et al. Deep Learning for Automated Contouring of Gross Tumor Volumes in Esophageal Cancer. Front. Oncol. 2022, 12, 892171. [Google Scholar] [CrossRef]
- Duan, Y.; Wang, J.; Wu, P.; Shao, Y.; Chen, H.; Wang, H.; Cao, H.; Gu, H.; Feng, A.; Huang, Y.; et al. AS-NeSt: A Novel 3D Deep Learning Model for Radiation Therapy Dose Distribution Prediction in Esophageal Cancer Treatment With Multiple Prescriptions. Int. J. Radiat. Oncol. Biol. Phys. 2024, 119, 978–989. [Google Scholar] [CrossRef]
- Zhu, C.; Mohan, R.; Lin, S.H.; Jun, G.; Yaseen, A.; Jiang, X.; Wang, Q.; Cao, W.; Hobbs, B.P. Identifying Individualized Risk Profiles for Radiotherapy-Induced Lymphopenia Among Patients With Esophageal Cancer Using Machine Learning. JCO Clin. Cancer Inform. 2021, 5, 1044–1053. [Google Scholar] [CrossRef]
- Sheng, L.; Zhuang, L.; Yang, J.; Zhang, D.; Chen, Y.; Zhang, J.; Wang, S.; Shan, G.; Du, X.; Bai, X. Radiation pneumonia predictive model for radiotherapy in esophageal carcinoma patients. BMC Cancer 2023, 23, 988. [Google Scholar] [CrossRef]
- Sato, K.; Fujita, T.; Matsuzaki, H.; Takeshita, N.; Fujiwara, H.; Mitsunaga, S.; Kojima, T.; Mori, K.; Daiko, H. Correction to: Real-time detection of the recurrent laryngeal nerve in thoracoscopic esophagectomy using artificial intelligence. Surg. Endosc. 2022, 36, 9483. [Google Scholar] [CrossRef]
- Maktabi, M.; Köhler, H.; Ivanova, M.; Jansen-Winkeln, B.; Takoh, J.; Niebisch, S.; Rabe, S.M.; Neumuth, T.; Gockel, I.; Chalopin, C. Tissue classification of oncologic esophageal resectates based on hyperspectral data. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Furube, T.; Takeuchi, M.; Kawakubo, H.; Maeda, Y.; Matsuda, S.; Fukuda, K.; Nakamura, R.; Kato, M.; Yahagi, N.; Kitagawa, Y. Automated artificial intelligence-based phase-recognition system for esophageal endoscopic submucosal dissection (with video). Gastrointest. Endosc. 2024, 99, 830–838. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Kawakubo, H.; Saito, K.; Maeda, Y.; Matsuda, S.; Fukuda, K.; Nakamura, R.; Kitagawa, Y. Automated Surgical-Phase Recognition for Robot-Assisted Minimally Invasive Esophagectomy Using Artificial Intelligence. Ann. Surg. Oncol. 2022, 29, 6847–6855. [Google Scholar] [CrossRef]
- Jung, J.O.; Pisula, J.I.; Bozek, K.; Popp, F.; Fuchs, H.F.; Schröder, W.; Bruns, C.J.; Schmidt, T. Prediction of postoperative complications after oesophagectomy using machine-learning methods. Br. J. Surg. 2023, 110, 1361–1366. [Google Scholar] [CrossRef]
- van Kooten, R.T.; Bahadoer, R.R.; Ter Buurkes de Vries, B.; Wouters, M.W.J.M.; Tollenaar, R.A.E.M.; Hartgrink, H.H.; Putter, H.; Dikken, J.L. Conventional regression analysis and machine learning in prediction of anastomotic leakage and pulmonary complications after esophagogastric cancer surgery. J. Surg. Oncol. 2022, 126, 490–501. [Google Scholar] [CrossRef]
- Klontzas, M.E.; Ri, M.; Koltsakis, E.; Stenqvist, E.; Kalarakis, G.; Boström, E.; Kechagias, A.; Schizas, D.; Rouvelas, I.; Tzortzakakis, A. Prediction of Anastomotic Leakage in Esophageal Cancer Surgery: A Multimodal Machine Learning Model Integrating Imaging and Clinical Data. Acad. Radiol. 2024, 31, 4878–4885. [Google Scholar] [CrossRef]
- Bolourani, S.; Tayebi, M.A.; Diao, L.; Wang, P.; Patel, V.; Manetta, F.; Lee, P.C. Using machine learning to predict early readmission following esophagectomy. J. ThoracCardiovasc. Surg. 2021, 161, 1926–1939.e8, Erratum in J. Thorac. Cardiovasc. Surg. 2020. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, C.; Yang, H.; Ho, J.W.K.; Wen, J.; Han, L.; Chiu, K.W.H.; Fu, J.; Vardhanabhuti, V. Assessment of Intratumoral and Peritumoral Computed Tomography Radiomics for Predicting Pathological Complete Response to Neoadjuvant Chemoradiation in Patients With Esophageal Squamous Cell Carcinoma. JAMA Netw. Open. 2020, 3, e2015927. [Google Scholar] [CrossRef]
- Hu, Y.; Xie, C.; Yang, H.; Ho, J.W.K.; Wen, J.; Han, L.; Lam, K.O.; Wong, I.Y.H.; Law, S.Y.K.; Chiu, K.W.H.; et al. Computed tomography-based deep-learning prediction of neoadjuvant chemoradiotherapy treatment response in esophageal squamous cell carcinoma. Radiother. Oncol. 2021, 154, 6–13. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, X.; Zeng, J.; Liu, C.; Shen, W.; Sun, X.; Lin, Q.; Fang, J.; Chen, Q.; Ji, Y. Using clinical and radiomic feature-based machine learning models to predict pathological complete response in patients with esophageal squamous cell carcinoma receiving neoadjuvant chemoradiation. Eur. Radiol. 2023, 33, 8554–8563. [Google Scholar] [CrossRef]
- Li, X.; Gao, H.; Zhu, J.; Huang, Y.; Zhu, Y.; Huang, W.; Li, Z.; Sun, K.; Liu, Z.; Tian, J.; et al. 3D Deep Learning Model for the Pretreatment Evaluation of Treatment Response in Esophageal Carcinoma: A Prospective Study (ChiCTR2000039279). Int. J. Radiat. Oncol. Biol. Phys. 2021, 111, 926–935. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yao, W.; Xu, B.C.; Lei, Y.Y.; Guo, Q.K.; Liu, L.Z.; Li, H.J.; Xu, M.; Yan, J.; Chang, D.D.; et al. Predicting response to immunotherapy plus chemotherapy in patients with esophageal squamous cell carcinoma using non-invasive Radiomic biomarkers. BMC Cancer 2021, 21, 1167. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.-Y.; Pang, C.-L.; Chan, B.; Wong, E.Y.-Y.; Dou, Q.; Vardhanabhuti, V. Machine Learning and Radiomics Applications in Esophageal Cancers Using Non-Invasive Imaging Methods—A Critical Review of Literature. Cancers 2021, 13, 2469. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, Q.; Ji, C.; Sun, Y.; Zhang, S.; Hua, M.; Liu, X.; Pan, S.; Hu, W.; Ma, Y.; et al. An artificial neural network-based radiomics model for predicting the radiotherapy response of advanced esophageal squamous cell carcinoma patients: A multicenter study. Sci. Rep. 2023, 13, 8673. [Google Scholar] [CrossRef]
- Jin, X.; Zheng, X.; Chen, D.; Jin, J.; Zhu, G.; Deng, X.; Han, C.; Gong, C.; Zhou, Y.; Liu, C.; et al. Prediction of response after chemoradiation for esophageal cancer using a combination of dosimetry and CT radiomics. Eur. Radiol. 2019, 29, 6080–6088. [Google Scholar] [CrossRef]
- Yap, W.K.; Hsiao, I.T.; Yap, W.L.; Tsai, T.Y.; Lu, Y.A.; Yang, C.K.; Peng, M.T.; Su, E.L.; Cheng, S.C. A Radiotherapy Dose Map-Guided Deep Learning Method for Predicting Pathological Complete Response in Esophageal Cancer Patients after Neoadjuvant Chemoradiotherapy Followed by Surgery. Biomedicines 2023, 11, 3072. [Google Scholar] [CrossRef]
- Schollaert, P.; Crott, R.; Bertrand, C.; D’Hondt, L.; Borght, T.V.; Krug, B. A systematic review of the predictive value of (18)FDG-PET in esophageal and esophagogastric junction cancer after neoadjuvant chemoradiation on the survival outcome stratification. J. Gastrointest. Surg. 2014, 18, 894–905. [Google Scholar] [CrossRef]
- Zhu, W.; Xing, L.; Yue, J.; Sun, X.; Sun, X.; Zhao, H.; Yu, J. Prognostic significance of SUV on PET/CT in patients with localised oesophagogastric junction cancer receiving neoadjuvant chemotherapy/chemoradiation: A systematic review and meta-analysis. Br. J. Radiol. 2012, 85, e694–e701. [Google Scholar] [CrossRef]
- Ypsilantis, P.P.; Siddique, M.; Sohn, H.M.; Davies, A.; Cook, G.; Goh, V.; Montana, G. Predicting Response to Neoadjuvant Chemotherapy with PET Imaging Using Convolutional Neural Networks. PLoS ONE 2015, 10, e0137036. [Google Scholar] [CrossRef]
- Rishi, A.; Zhang, G.G.; Yuan, Z.; Sim, A.J.; Song, E.Y.; Moros, E.G.; Tomaszewski, M.R.; Latifi, K.; Pimiento, J.M.; Fontaine, J.P.; et al. Pretreatment CT and 18 F-FDG PET-based radiomic model predicting pathological complete response and loco-regional control following neoadjuvant chemoradiation in oesophageal cancer. J. Med. Imaging Radiat. Oncol. 2021, 65, 102–111. [Google Scholar] [CrossRef]
- Qi, W.X.; Li, S.; Xiao, J.; Li, H.; Chen, J.; Zhao, S. A machine learning approach using 18F-FDG PET and enhanced CT scan-based radiomics combined with clinical model to predict pathological complete response in ESCC patients after neoadjuvant chemoradiotherapy and anti-PD-1 inhibitors. Front. Immunol. 2024, 15, 1351750. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, D.; Murakami, Y.; Tani, S.; Nagata, Y. A prediction model for pathological findings after neoadjuvant chemoradiotherapy for resectable locally advanced esophageal squamous cell carcinoma based on endoscopic images using deep learning. Br. J. Radiol. 2022, 95, 20210934. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, S.; Irino, T.; Okamura, A.; Mayanagi, S.; Booka, E.; Takeuchi, M.; Kawakubo, H.; Takeuchi, H.; Watanabe, M.; Kitagawa, Y. Endoscopic Evaluation of Pathological Complete Response Using Deep Neural Network in Esophageal Cancer Patients Who Received Neoadjuvant Chemotherapy-Multicenter Retrospective Study from Four Japanese Esophageal Centers. Ann. Surg. Oncol. 2023, 30, 7472–7480. [Google Scholar] [CrossRef]
- Matsuda, S.; Irino, T.; Kawakubo, H.; Takeuchi, M.; Nishimura, E.; Hisaoka, K.; Sano, J.; Kobayashi, R.; Fukuda, K.; Nakamura, R.; et al. Evaluation of Endoscopic Response Using Deep Neural Network in Esophageal Cancer Patients Who Received Neoadjuvant Chemotherapy. Ann. Surg. Oncol. 2023, 30, 3733–3742. [Google Scholar] [CrossRef]
- Brierley, J.D.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- Sato, F.; Shimada, Y.; Selaru, F.M.; Shibata, D.; Maeda, M.; Watanabe, G.; Mori, Y.; Stass, S.A.; Imamura, M.; Meltzer, S.J. Prediction of survival in patients with esophageal carcinoma using artificial neural networks. Cancer 2005, 103, 1596–1605. [Google Scholar] [CrossRef]
- Mofidi, R.; Deans, C.; Duff, M.D.; de Beaux, A.C.; Paterson Brown, S. Prediction of survival from carcinoma of oesophagus and oesophago-gastric junction following surgical resection using an artificial neural network. Eur. J. Surg. Oncol. 2006, 32, 533–539. [Google Scholar] [CrossRef]
- Larue, R.T.H.M.; Klaassen, R.; Jochems, A.; Leijenaar, R.T.H.; Hulshof, M.C.C.M.; van Berge Henegouwen, M.I.; Schreurs, W.M.J.; Sosef, M.N.; van Elmpt, W.; van Laarhoven, H.W.M.; et al. Pre-treatment CT radiomics to predict 3-year overall survival following chemoradiotherapy of esophageal cancer. Acta Oncol. 2018, 57, 1475–1481. [Google Scholar] [CrossRef]
- Luo, H.S.; Chen, Y.Y.; Huang, W.Z.; Wu, S.X.; Huang, S.F.; Xu, H.Y.; Xue, R.L.; Du, Z.S.; Li, X.Y.; Lin, L.X.; et al. Development and validation of a radiomics-based model to predict local progression-free survival after chemo-radiotherapy in patients with esophageal squamous cell cancer. Radiat. Oncol. 2021, 16, 201. [Google Scholar] [CrossRef]
- Cui, Y.; Li, Z.; Xiang, M.; Han, D.; Yin, Y.; Ma, C. Machine learning models predict overall survival and progression free survival of non-surgical esophageal cancer patients with chemoradiotherapy based on CT image radiomics signatures. Radiat. Oncol. 2022, 17, 212. [Google Scholar] [CrossRef]
- Wang, J.; Zeng, J.; Li, H.; Yu, X. A Deep Learning Radiomics Analysis for Survival Prediction in Esophageal Cancer. J. Healthc. Eng. 2022, 2022, 4034404. [Google Scholar] [CrossRef]
- Zhang, H.; Jiang, X.; Yu, Q.; Yu, H.; Xu, C. A novel staging system based on deep learning for overall survival in patients with esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2023, 149, 8935–8944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Ye, B.; Wu, L.; Ni, S.; Li, Y.; Wang, Q.; Zhang, P.; Wang, D. Machine learningbased prediction of survival prognosis in esophageal squamous cell carcinoma. Sci. Rep. 2023, 13, 13532. [Google Scholar] [CrossRef]
- Chu, F.; Liu, Y.; Liu, Q.; Li, W.; Jia, Z.; Wang, C.; Wang, Z.; Lu, S.; Li, P.; Zhang, Y.; et al. Development and validation of MRI-based radiomics signatures models for prediction of disease-free survival and overall survival in patients with esophageal squamous cell carcinoma. Eur. Radiol. 2022, 32, 5930–5942. [Google Scholar] [CrossRef] [PubMed]
- Paver, E.C.; Cooper, W.A.; Colebatch, A.J.; Ferguson, P.M.; Hill, S.K.; Lum, T.; Shin, J.S.; O’Toole, S.; Anderson, L.; Scolyer, R.A.; et al. Programmed death ligand-1 (PD-L1) as a predictive marker for immunotherapy in solid tumours: A guide to immunohistochemistry implementation and interpretation. Pathology 2021, 53, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, K.; Wang, T.; Li, M.; Li, B.; Li, S.; Yuan, L. The Combination Options and Predictive Biomarkers of PD-1/PD-L1 Inhibitors in Esophageal Cancer. Front. Oncol. 2020, 10, 300. [Google Scholar] [CrossRef]
- Chen, H.; Luo, J.; Guo, J. Construction and Validation of a 7-Immune Gene Model for Prognostic Assessment of Esophageal Carcinoma. Med. Sci. Monit. 2020, 26, e927392. [Google Scholar] [CrossRef]
- Li, B.; Qin, W.; Yang, L.; Li, H.; Jiang, C.; Yao, Y.; Cheng, S.; Zou, B.; Fan, B.; Dong, T.; et al. From pixels to patient care: Deep learning-enabled pathomics signature offers precise outcome predictions for immunotherapy in esophageal squamous cell cancer. J. Transl. Med. 2024, 22, 195. [Google Scholar] [CrossRef]
- Abas Mohamed, Y.; Ee Khoo, B.; Shahrimie Mohd Asaari, M.; Ezane Aziz, M.; Rahiman Ghazali, F. Decoding the black box: Explainable AI (XAI) for cancer diagnosis, prognosis, and treatment planning-A state-of-the art systematic review. Int. J. Med. Inform. 2025, 193, 105689. [Google Scholar] [CrossRef]
- Pearl, J. Causal inference in statistics: An overview. Statist. Surv. 2009, 3, 96–146. [Google Scholar] [CrossRef]
- Tu, J.X.; Lin, X.T.; Ye, H.Q.; Yang, S.L.; Deng, L.F.; Zhu, R.L.; Wu, L.; Zhang, X.Q. Global research trends of artificial intelligence applied in esophageal carcinoma: A bibliometric analysis (2000–2022) via CiteSpace and VOSviewer. Front. Oncol. 2022, 12, 972357. [Google Scholar] [CrossRef]
- Gerke, S.; Minssen, T.; Cohen, G.; Bohr, A.; Memarzadeh, K. Chapter 12-Ethical and legal challenges of artificial intelligence-driven healthcare. In Artificial Intelligence in Healthcare; Academic Press: Cambridge, MA, USA, 2020; pp. 295–336. [Google Scholar]
| Author | Year | Sample Size | Study Design | Objective | Results |
|---|---|---|---|---|---|
| Sato K et al. [21] | 2022 | 3000 images from 20 thoracoscopic videos and 40 images from 8 thoracoscopic videos | Retrospective | Development of a DL-based AI image model for identifying the location of the recurrent laryngeal nerve and assess if it reduces the incidence of recurrent laryngeal nerve paralysis | The model had an average dice coefficient of 0.58. This was not significantly different from the group of specialized esophageal surgeons (p = 0.26) but was significantly superior to that of the group of certified general gastrointestinal surgeons (p = 0.019). |
| Maktabi M et al. [22] | 2019 | 11 patients | Retrospective | Evaluation of intraoperative hyperspectral imaging for identifying the tumor margins intraoperatively | The support vector machines (SVM) algorithm has the best performance, with 63% sensitivity and 69% specificity for identifying cancerous tissue. |
| Furube T et al. [23] | 2024 | 94 videos | Retrospective | Development of an AI-based automated ESD phase recognition system based on video images | The model displayed an overall accuracy of 90%, an average precision of 91%, and a recall of 90%. |
| Takeuchi M et al. [24] | 2022 | 31 patients | Retrospective | Development of an AI-based automated surgical-phase recognition system for RAMIE | The model achieved an overall accuracy of 84%. |
| Jung JO et al. [25] | 2023 | 864 patients | Retrospective | Development of ML-based methods for predicting Clavien–Dindo grade IIIa or greater complications following esophagectomy | The neural network achieved an overall accuracy of 68.8% and accuracies of 69.2% for medical complications, and 66.7% for surgical complications. The AUC of neural network was 0.672 for Clavien–Dindo grade IIIa or higher, 0.695 for medical complications, and 0.653 for surgical complications. |
| van Kooten RT et al. [26] | 2022 | 4288 patients | Retrospective | Evaluation of the added value of ML methods for predicting postoperative complications following esophagectomy and development of a predictive model for anastomotic leakage and cardiopulmonary complications | The optimal model GLM displayed an AUC of 0.619 for anastomotic leakage and 0.644 for pulmonary complications. |
| Klontzas M et al. [27] | 2024 | 471 patients | Retrospective | Development of an ML model combining CT and clinical variables for predicting anastomotic leakage following esophagectomy | The XGboost model achieved an AUC of 0.792, 77.46% specificity, 69.22% sensitivity, 48.39% positive predictive value, and 87.3% negative predictive value. |
| Bolourani S et al. [28] | 2021 | 2037 patients | Retrospective | Development of ML-based prediction model for early readmissions within 30 days following esophagectomy | The ML model for clinical decision achieved a sensitivity of 71.7% and specificity of 51.4%, while the ML model for quality review achieved an accuracy of 84.8%, specificity of 98.7%, and sensitivity of 23% |
| Author | Year | Sample Size | Study Design | Treatment | Objective | Results |
|---|---|---|---|---|---|---|
| Hu Y et al. [29] | 2020 | 231 | Retrospective | nCRT and surgery | Development and validation of CT-based models to identify pCR using intratumoral and peritumoral features in ESCC patients | The model combining 7 intratumoral and 6 peritumoral features displayed superior discriminative performance, with an AUC of 0.852 (95% CI, 0.753–0.951), accuracy of 84.3%, sensitivity of 90.3% and specificity of 79.5% in the test set. |
| Hu Y et al. [30] | 2021 | 231 | Retrospective | nCRT and surgery | Evaluation and validation of a CT-based model using DL features for predicting pCR to nCRT in ESCC patients | The model integrating features from ResNet50 achieved an AUC of 0.805 (95% CI, 0.696–0.913) and an accuracy of 77.1%, compared with 0.725 (95% CI, 0.605- 0.846) and 67.1% for the radiomics model in the test set. |
| Wang J et al. [31] | 2023 | 112 | Retrospective | nCRT and surgery | Development and validation of a radiomics feature-based ML model for predicting pCR to nCRT in ESCC patients | For pCR prediction models, the model combining rad-score and clinical features achieved an AUC of 0.891 (95% CI, 0.823–0.950), compared with 0.817 (95% CI, 0.732–0.896) for the radiomics model in the test set. |
| Li X et al. [32] | 2021 | 306 | Prospective | nCRT | Development and validation of a 3-dimensional DL radiomics model for predicting response to nCRT in ESCC patients | The model achieved an AUC of 0.897 (95% CI, 0.840–0.959) in the training cohort and 0.833 (95% CI, 0.654–1.000) in the validation cohort |
| Zhu Y et al. [33] | 2021 | 64 | Retrospective | Chemotherapy and PD-1 inhibitor | Development and validation of a radiomics model for evaluating treatment response to chemotherapy and immunotherapy in ESCC patients | The 2-dimensional corrected model achieved an AUC of 0.843 (95% CI, 0.736–0.950) in the training cohort and 0.914 (95% CI, 0.775–1.000) in the validation cohort, compared to the 3-dimensional corrected model with an AUC 0.658 (95% CI, 0.502–0.813) and 0.670 (95% CI, 0.511–0.849) respectively |
| Xie Y et al. [35] | 2023 | 248 | Retrospective | nCRT | Establishment and validation of an ANN-based radiomics model for predicting radiotherapy response in patients with stage III ESCC | The pretrained network ResNet50 displayed superior performance, with AUCs of 0.876, 0.802, and 0.732 in the training, internal validation, and external validation cohorts respectively |
| Jin X et al. [36] | 2019 | 94 | Retrospective | CRT | Evaluation of a model combining CT radiomics and dosimetric features for predicting response to CRT in esophageal cancer patients | The XGBoost plus principal component analysis model combining radiomics features with dosimetric parameters achieved a prediction accuracy of 70.8% and an AUC of 0.689 compared to the radiomics model with an accuracy of 62.5% and AUC of 0.412 |
| Yap WK et al. [37] | 2023 | 80 | Retrospective | nCRT and surgery | Development of radiotherapy dose map-guided DL model for predicting ypCR to nCRT in ESCC patients | The HRNetV2p model with dose contextual representations achieved superior performance, with an AUC of 0.928 (95% CI, 0.884–0.972) |
| Ypsilantis PP et al. [40] | 2015 | 107 | Prospective | nCT | Evaluation of the predictive ability of an ML algorithm (3S-CNN) for representing esophageal cancer’s metabolic profile using 18F-FDG PET imaging | The 3S-CNN achieved superior performance with an average 80.7% sensitivity, 81.6% specificity, and 73.4% accuracy in predicting non-responders |
| Rishi A et al. [41] | 2020 | 68 | Retrospective | nCRT and surgery | Development and validation of a combined CT and PET/CT radiomics model for predicting pCR to nCRT in patients with advanced esophageal cancer | The combined PET/CT radiomics model achieved improved predictive performance with an AUC of 0.87 compared to CT- or PET-only models with AUCs of 0.73 and 0.66 respectively |
| Qi WX et al. [42] | 2024 | 126 | Prospective | nCRT, anti-PD1 inhibitors and surgery | Evaluation of an integrated multimodal radiomics with ML model for predicting pCR to nCRT and anti-PD1 in ESCC patients | Support vector machine ML trained on CT, PET, and clinical features achieved superior performance compared to CT and PET-only models with an AUC of 0.997 in the training set and 0.852 in the testing set |
| Kawahara D et al. [43] | 2022 | 98 | Retrospective | nCRT and surgery | Proposal of a DL-based model using endoscopic images for predicting pCR to nCRT in ESCC patients | The model achieved 64% accuracy, 80.3% sensitivity, 36.3% specificity, and 0.58 AUC in the testing set compared to 80.6%, 79.8%, 80.6%, and 0.83 with wavelet filter, respectively |
| Matsuda S et al. [44] | 2023 | 123 | Retrospective | nCRT and surgery | Development of a deep neural network using endoscopic images for identifying pCR to nCT preoperatively in ESCC patients | In 20 models, the median sensitivity, specificity, positive predictive value, negative predictive value, and accuracy for endoscopic response evaluation were 60%, 81%, 77%, 67%, and 70% compared to 43%, 90%, 85%, 65%, and 66% for endoscopists respectively |
| Matsuda S et al. [45] | 2023 | 193 | Retrospective | nCRT and surgery | Development of a deep neural network for evaluating endoscopic response to nCT in ESCC patient development of a deep neural network for evaluating endoscopic response to nCT in ESCC patients | In 10 models the median sensitivity, specificity, positive predictive value, and negative predictive value were 60%, 100%, 100%, and 71% compared to 80%, 80%, 81%, and 81% for endoscopists respectively |
| Author | Year | Sample Size | Study Design | Objective | Results |
|---|---|---|---|---|---|
| Sato F et al. [47] | 2005 | 418 | Retrospective | Development of an ANN model for predicting 1- and 5-year survival for esophageal cancer patients | The optimal ANN models for predicting 1- and 5-year survival consisted of 65 variables (AUR = 0.883) and 60 variables (AUR = 0.884), respectively, and outperformed the TNM solely based model (p < 0.0001). |
| Modifi R et al. [48] | 2006 | 216 | Retrospective | Evaluation of the performance of an ANN model for predicting 1- and 3-year disease-free survival for esophageal and EGJ carcinoma patients | The accuracy, sensitivity, and specificity of the ANN for predicting survival at 1 year was 88%, 92.3%, and 84.5%, and at 3 years was 91.5%, 94.6%, and 88%, and was statistically significantly superior to TNM classification system. |
| Larue RTHM et al. [49] | 2018 | 239 | Retrospective | Training and validation of two models using radiomics or clinical variables for predicting 3-year overall survival for esophageal cancer patients following nCRT. | The radiomics-based RF model achieved superior prognostic performance for 3-year overall survival with an AUC of 0.69 and 0.61 in the training and validation dataset compared to 0.63 and 0.62 for the clinical feature-based model, respectively. |
| Luo HS et al. [50] | 2021 | 221 | Retrospective | Development and validation of a radiomics and clinical feature-based model for predicting local progression-free survival for ESCC patients following concurrent CRT | The C-index of the prediction model was 0.745 (95% CI, 0.770–0.790) in the training cohort and 0.723 (95% CI, 0.654–0.791) in the validation cohort. |
| Cui Y et al. [51] | 2022 | 204 | RCT | Development of ML models for predicting progression-free survival and overall survival for ESCC patients | Combined models using clinical and radiomics features achieved over 70% accuracy, an AUC of 0.833, and a C-index of 0.79 for predicting PFS and 0.768 and 0.71 for predicting OS in the test cohort. |
| Wang J et al. [52] | 2022 | 154 | Retrospective | Development and validation of a DL radiomics-based model for predicting 3-year overall survival for esophageal cancer patients | The deep-learning radiomics model achieved an AUC of 0.984 and a C-index of 0.76 in the training cohort and 0.942 and 0.784 in the validation cohort for predicting 3-year OS, respectively. |
| Zhang H et al. [53] | 2023 | 6020 | Retrospective | Development and validation of a DL model for predicting overall survival and a novel staging system for ESCC patients | The deep-learning model achieved superior performance than the traditional nomogram for predicting OS with C-index 0.732 [95% CI 0.714–0.750] and 0.671 [95% CI 0.647–0.695], respectively. The model in the test cohort displayed an AUC of 0.805 at 3-year OS and 0.825 at 5-year OS |
| Zhang K et al. [54] | 2023 | 2441 | Retrospective | Development of an ML survival prediction model using 6 different ML g approaches for ESCC patients | The ML-extended CoxPH model achieved superior discriminative performance, with AUCs for 1-, 3-, and 5-year overall survival of 0.760, 0.735, and 0.746 in the training cohort and 0.725, 0.720, and 0.752 in the validation cohort, respectively. |
| Chu F et al. [55] | 2022 | 434 | RCT | Development and validation of an MRI-based radiomics model for predicting disease-free survival and overall survival in ESCC patients | The combined model based on 7 radiomics features and clinical features achieved the best performance, with a C-index of 0.730 in the training cohort and 0.712 in the validation cohort for OS and 0.714 and 0.729 for DFS respectively. |
| Chen H et al. [58] | 2020 | 159 | Retrospective | Development and validation of a 7 immune gene model for predicting prognosis of esophageal cancer | The model achieved an AUC of 0.825 for 1 year and 0.596 for 3 years in the test set. |
| Li B et al. [59] | 2024 | 163 | Retrospective | Assessment of a DL-based pathomics signature for predicting clinical benefits of immunotherapy in ESCC patients | The ESCC-PS achieved an accuracy of 84.5% in the validation cohort and the combined model based on ESCC-PS and PD-L1 expression displayed an AUC of 0.904 and a C-index of 0.814 for a 6-month PFS. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mela, E.; Tsapralis, D.; Papaconstantinou, D.; Sakarellos, P.; Vergadis, C.; Klontzas, M.E.; Rouvelas, I.; Tzortzakakis, A.; Schizas, D. Current Role of Artificial Intelligence in the Management of Esophageal Cancer. J. Clin. Med. 2025, 14, 1845. https://doi.org/10.3390/jcm14061845
Mela E, Tsapralis D, Papaconstantinou D, Sakarellos P, Vergadis C, Klontzas ME, Rouvelas I, Tzortzakakis A, Schizas D. Current Role of Artificial Intelligence in the Management of Esophageal Cancer. Journal of Clinical Medicine. 2025; 14(6):1845. https://doi.org/10.3390/jcm14061845
Chicago/Turabian StyleMela, Evgenia, Dimitrios Tsapralis, Dimitrios Papaconstantinou, Panagiotis Sakarellos, Chrysovalantis Vergadis, Michail E. Klontzas, Ioannis Rouvelas, Antonios Tzortzakakis, and Dimitrios Schizas. 2025. "Current Role of Artificial Intelligence in the Management of Esophageal Cancer" Journal of Clinical Medicine 14, no. 6: 1845. https://doi.org/10.3390/jcm14061845
APA StyleMela, E., Tsapralis, D., Papaconstantinou, D., Sakarellos, P., Vergadis, C., Klontzas, M. E., Rouvelas, I., Tzortzakakis, A., & Schizas, D. (2025). Current Role of Artificial Intelligence in the Management of Esophageal Cancer. Journal of Clinical Medicine, 14(6), 1845. https://doi.org/10.3390/jcm14061845








