Meta-Analysis of Ocy-454 Showed Interrupted Osteocyte Maturation in Spaceflight Affects SOST Expression and Hypoxic Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Download and Preprocessing
2.2. Statistical Analysis (Differential Expression Analysis)
2.3. Data Visualization
3. Results
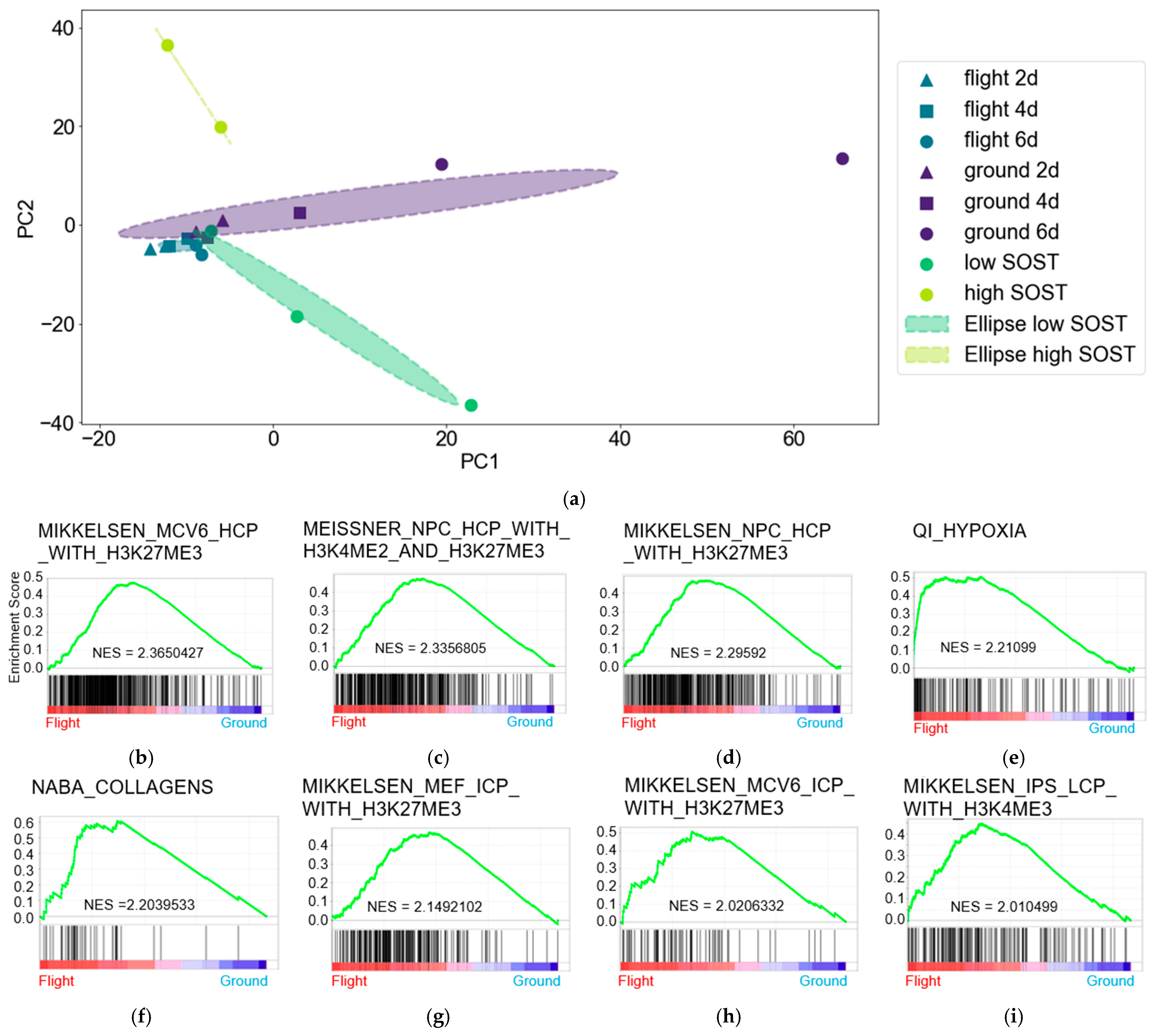
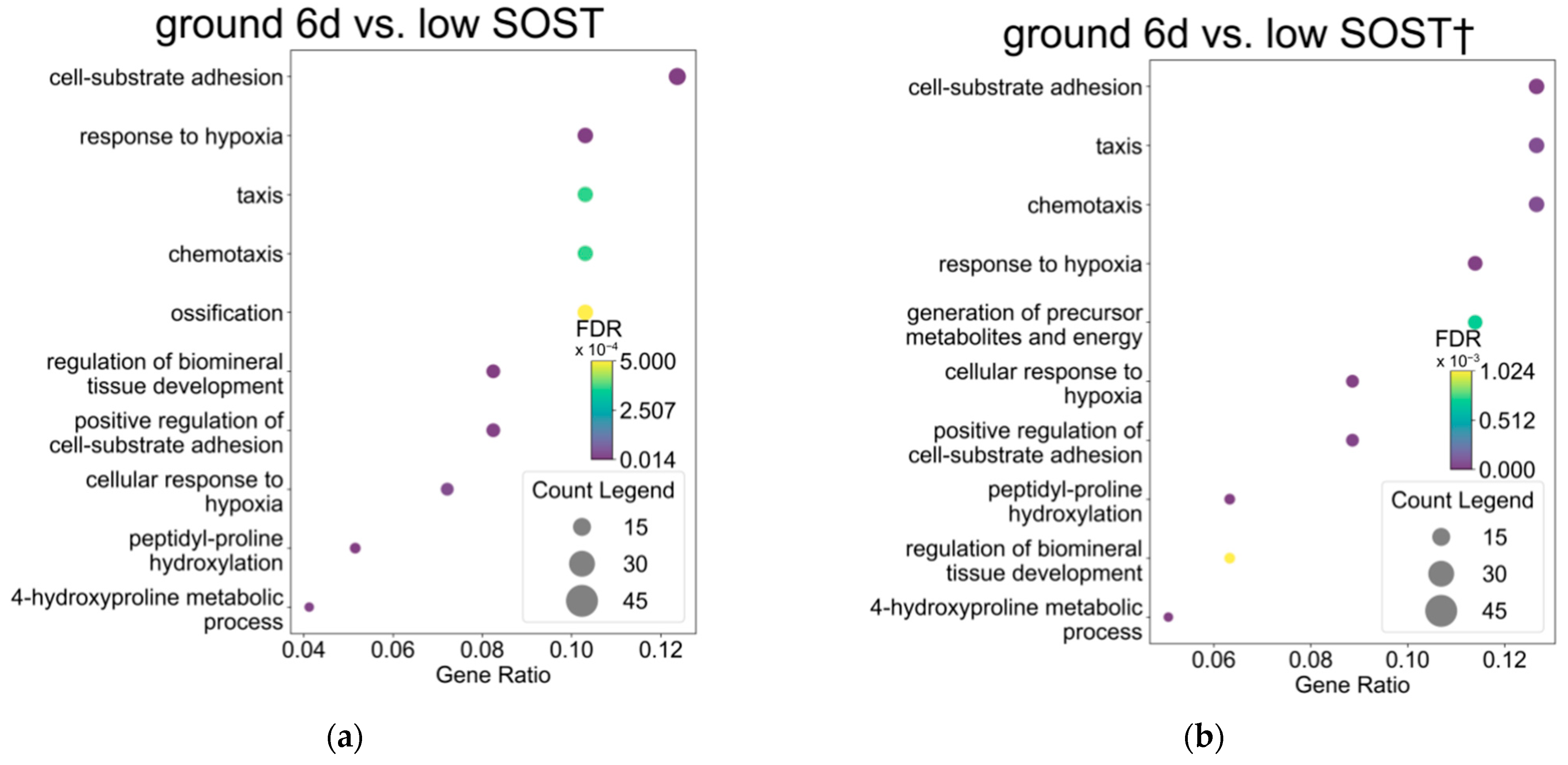
3.1. Gene Expression Profiles in Osteocytes Were Comparable After Six Days of Spaceflight and Low SOST Expression
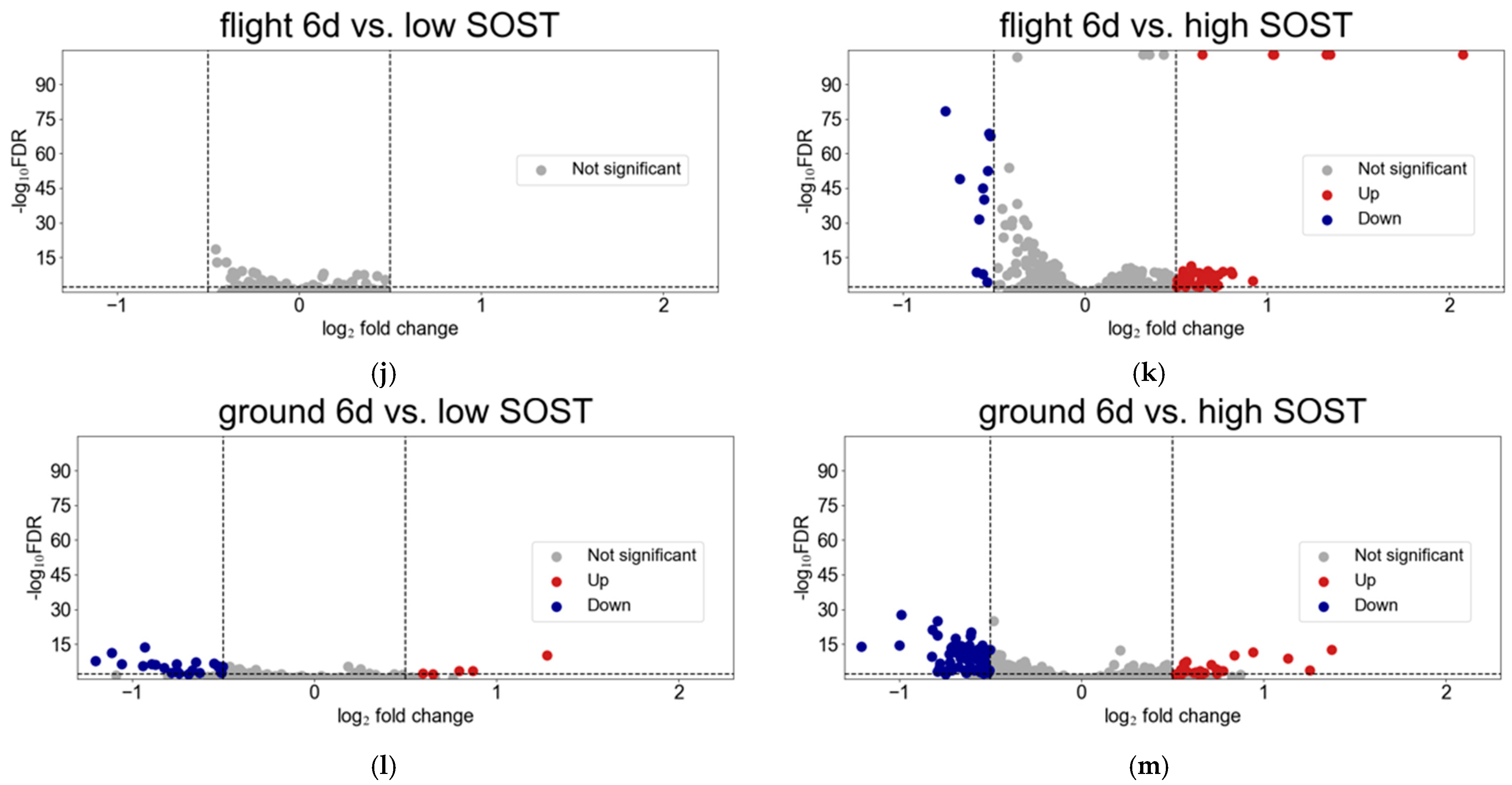
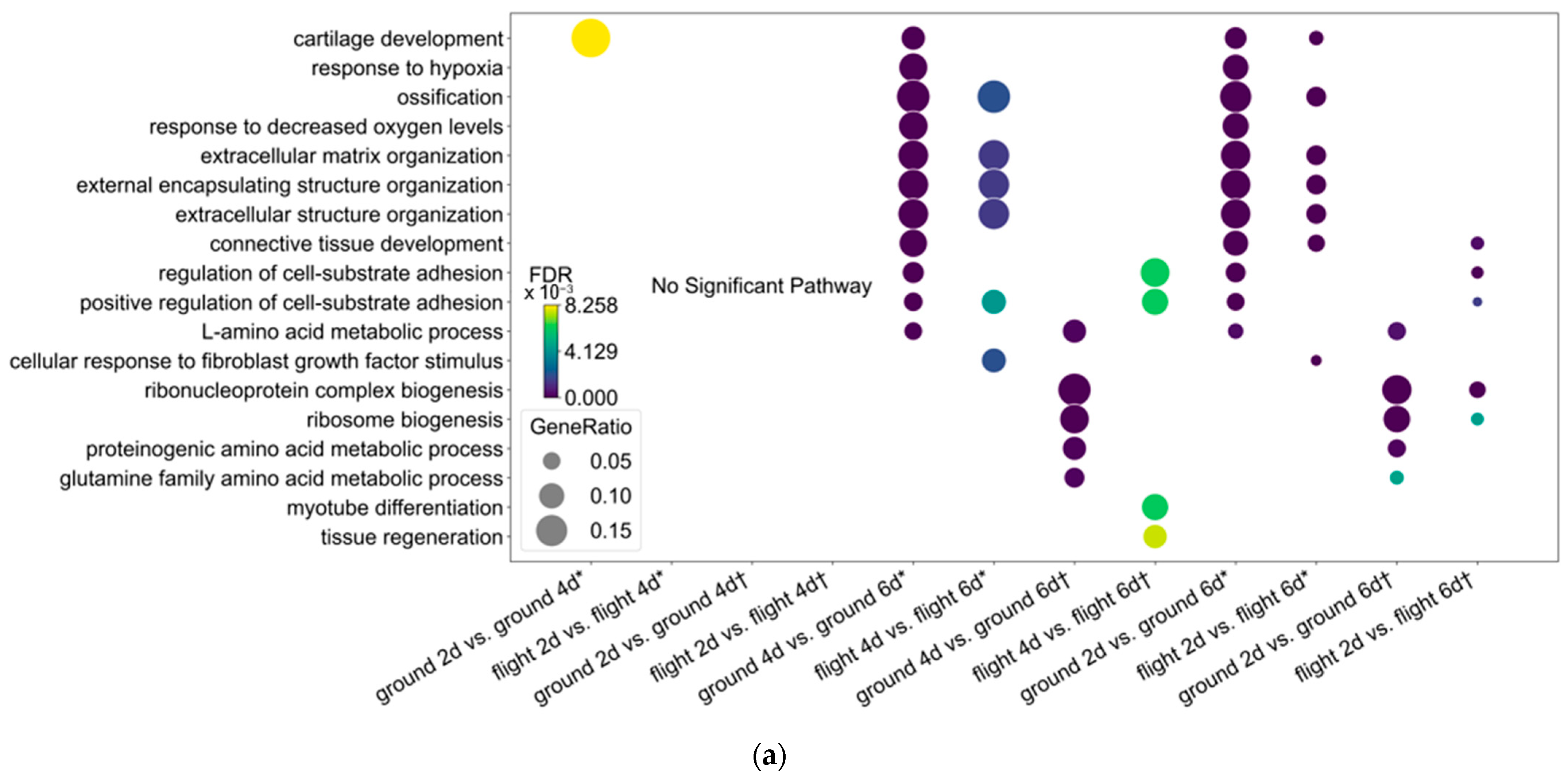
3.2. Ossification Is Downregulated in Osteocytes After Spaceflight in Contrast to Those with High SOST Expression
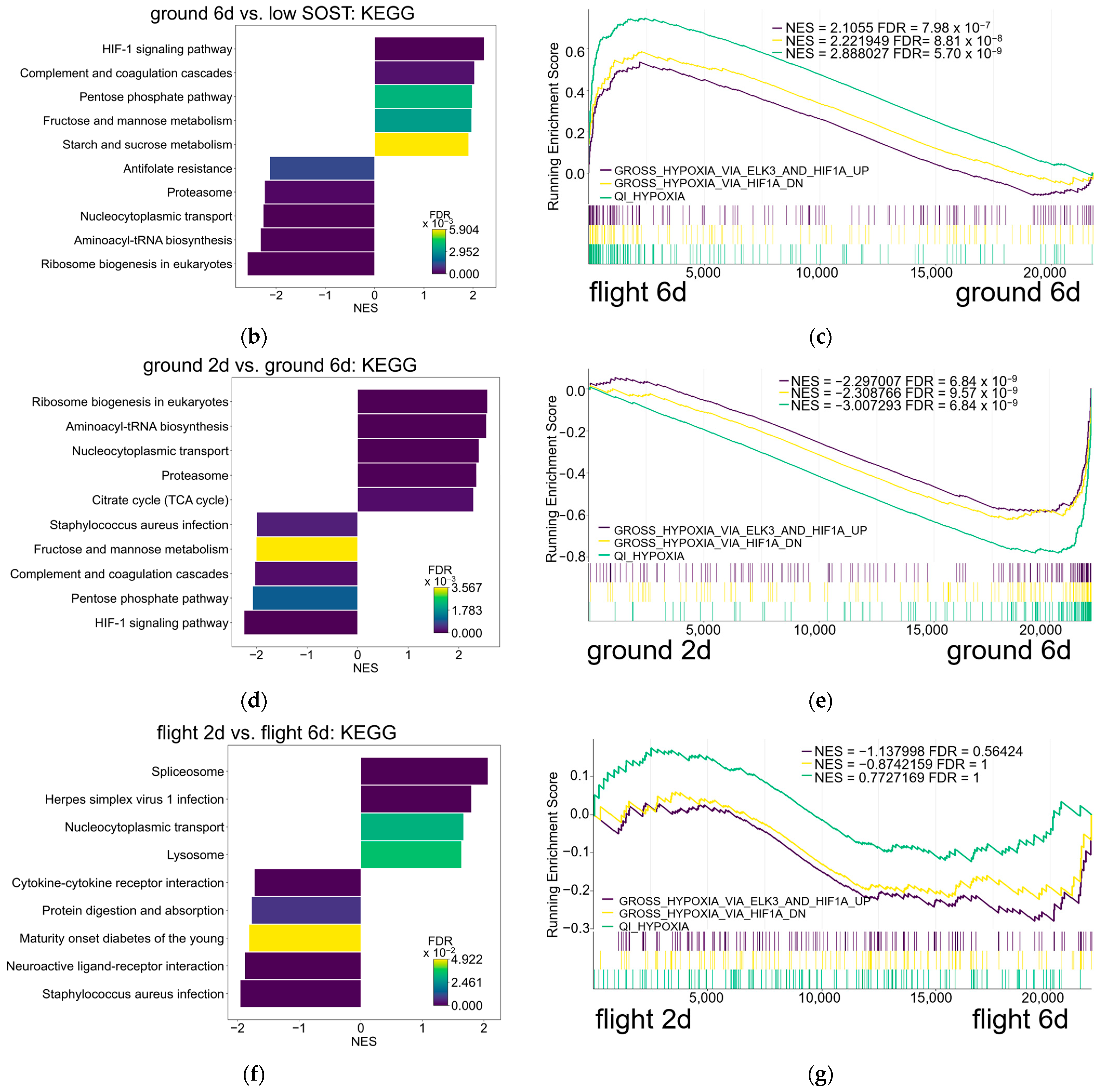
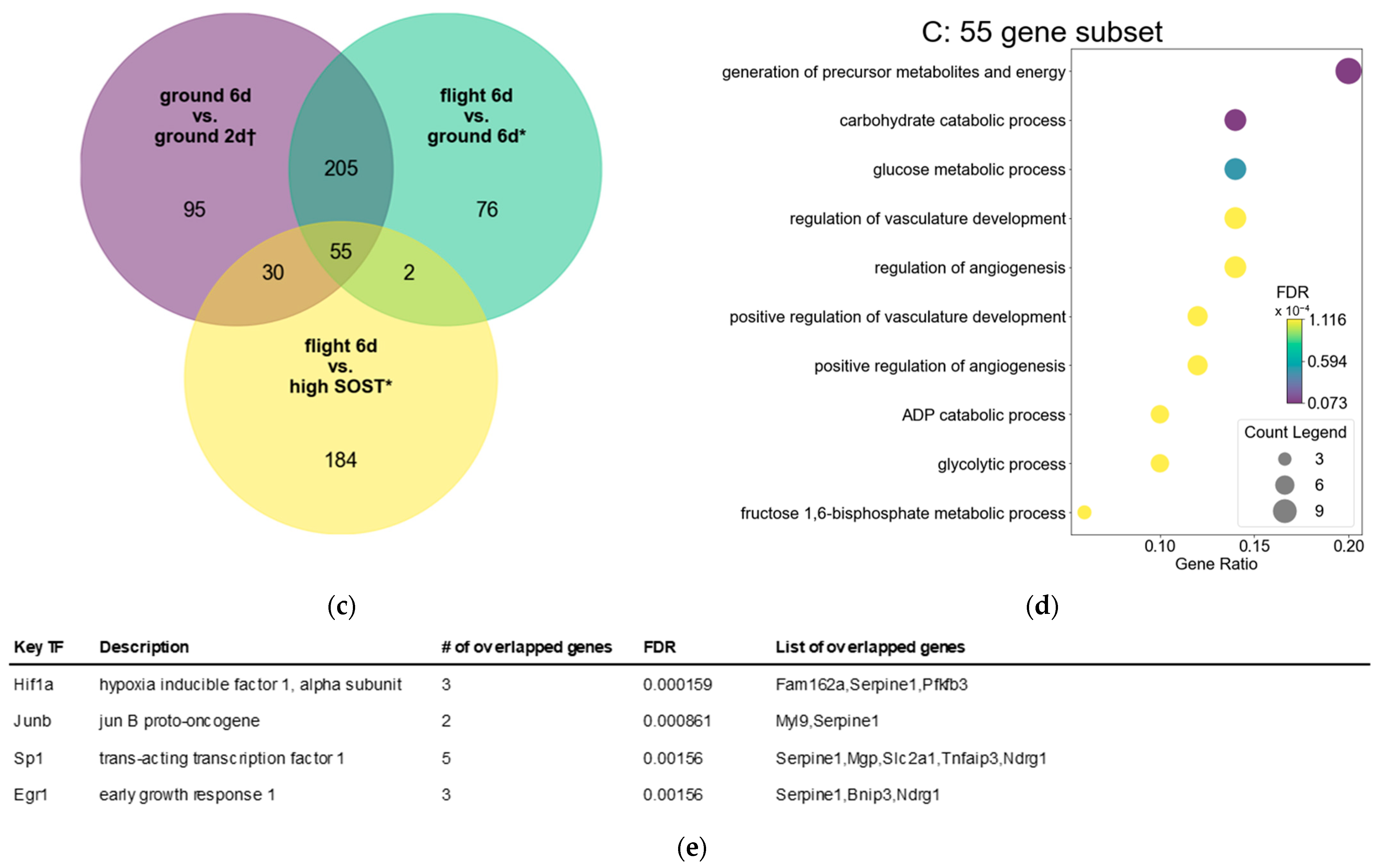
3.3. Hypoxic Pathway in Osteocytes Remained Enriched After Spaceflight Compared to Ground
3.4. Osteocytes After Spaceflight Maintain Energy Metabolism Pathway Downstream of Hif1a and Egr1 Compared to Osteocytes with High SOST Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OPG | Osteoprotegrin |
| RANKL | Nuclear factor kappa beta ligand |
| M-CSF | Macrophage colony-stimulating factor |
| IL-6 | Interleukin 6 |
| TNFα | Tumor necrosis factor-alpha |
| Dkk1 | Dickkopf WNT signaling pathway inhibitor 1 |
| NCBI | National Center for Biotechnology Information |
| NASA | National Aeronautics and Space Administration |
| GEO | Gene Expression Omnibus |
| LPE | Local Pooled Error |
| BH | Benjamini–Hochberg |
| PCA | Principal component analysis |
| CGP | Chemical and genetic perturbations |
| MSigDB | Molecular Signatures Database |
| GSEA | Gene Set Enrichment Analysis |
| NES | Normalized enrichment score |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| FC | Fold change |
| FDR | False discovery rate |
| DEGs | Differentially expressed genes |
| GO | Gene Ontology |
| BP | Biological Process |
| CC | Cellular Component |
| MF | Molecular Function |
| TRRUST | Transcriptional regulatory relationships unraveled by sentence-based text-mining |
| H3K27me3 | The trimethylation of H3K27 |
| Utx | X chromosome |
References
- Afshinnekoo, E.; Scott, R.T.; MacKay, M.J.; Pariset, E.; Cekanaviciute, E.; Barker, R.; Gilroy, S.; Hassane, D.; Smith, S.M.; Zwart, S.R.; et al. Fundamental Biological Features of Spaceflight: Advancing the Field to Enable Deep-Space Exploration. Cell 2020, 183, 1162–1184. [Google Scholar] [CrossRef] [PubMed]
- Coulombe, J.C.; Senwar, B.; Ferguson, V.L. Spaceflight-Induced Bone Tissue Changes that Affect Bone Quality and Increase Fracture Risk. Curr. Osteoporos. Rep. 2020, 18, 1–12. [Google Scholar] [CrossRef]
- Vico, L.; Hargens, A. Skeletal changes during and after spaceflight. Nat. Rev. Rheumatol. 2018, 14, 229–245. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kordsmeier, J.; Xiong, J. New Advances in Osteocyte Mechanotransduction. Curr. Osteoporos. Rep. 2021, 19, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Robling, A.G.; Bonewald, L.F. The Osteocyte: New Insights. Annu. Rev. Physiol. 2020, 82, 485–506. [Google Scholar] [CrossRef]
- Robling, A.G.; Niziolek, P.J.; Baldridge, L.A.; Condon, K.W.; Allen, M.R.; Alam, I.; Mantila, S.M.; Gluhak-Heinrich, J.; Bellido, T.M.; Harris, S.E.; et al. Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J. Biol. Chem. 2008, 283, 5866–5875. [Google Scholar] [CrossRef]
- Spatz, J.M.; Wein, M.N.; Gooi, J.H.; Qu, Y.; Garr, J.L.; Liu, S.; Barry, K.J.; Uda, Y.; Lai, F.; Dedic, C.; et al. The Wnt Inhibitor Sclerostin Is Up-regulated by Mechanical Unloading in Osteocytes in Vitro. J. Biol. Chem. 2015, 290, 16744–16758. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, P.; Wu, J.; Feng, X.; Cai, J.; Zhai, M.; Li, J.; Liu, X.; Jiang, M.; Luo, E.; et al. Fluid shear stress improves morphology, cytoskeleton architecture, viability, and regulates cytokine expression in a time-dependent manner in MLO-Y4 cells. Cell Biol. Int. 2018, 42, 1410–1422. [Google Scholar] [CrossRef]
- Spatz, J.M.; Ellman, R.; Cloutier, A.M.; Louis, L.; van Vliet, M.; Suva, L.J.; Dwyer, D.; Stolina, M.; Ke, H.Z.; Bouxsein, M.L. Sclerostin antibody inhibits skeletal deterioration due to reduced mechanical loading. J. Bone Miner. Res. 2013, 28, 865–874. [Google Scholar] [CrossRef]
- Shi, C.; Uda, Y.; Dedic, C.; Azab, E.; Sun, N.; Hussein, A.I.; Petty, C.A.; Fulzele, K.; Mitterberger-Vogt, M.C.; Zwerschke, W.; et al. Carbonic anhydrase III protects osteocytes from oxidative stress. FASEB J. 2018, 32, 440–452. [Google Scholar] [CrossRef]
- Yamamoto, T.; Ikegame, M.; Hirayama, J.; Kitamura, K.-I.; Tabuchi, Y.; Furusawa, Y.; Sekiguchi, T.; Endo, M.; Mishima, H.; Seki, A.; et al. Expression of sclerostin in the regenerating scales of goldfish and its increase under microgravity during space flight. Biomed. Res. 2020, 41, 279–288. [Google Scholar] [CrossRef]
- Zhang, B.; Cory, E.; Bhattacharya, R.; Sah, R.; Hargens, A.R. Fifteen days of microgravity causes growth in calvaria of mice. Bone 2013, 56, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, T.R.; Siamwala, J.H.; Hargens, A.R.; Macias, B.R. Thirty days of spaceflight does not alter murine calvariae structure despite increased Sost expression. Bone Rep. 2017, 7, 57–62. [Google Scholar] [CrossRef]
- Vico, L.; Van Rietbergen, B.; Vilayphiou, N.; Linossier, M.T.; Locrelle, H.; Normand, M.; Zouch, M.; Gerbaix, M.; Bonnet, N.; Novikov, V.; et al. Cortical and Trabecular Bone Microstructure Did Not Recover at Weight-Bearing Skeletal Sites and Progressively Deteriorated at Non-Weight-Bearing Sites During the Year Following International Space Station Missions. J. Bone Miner. Res. 2017, 32, 2010–2021. [Google Scholar] [CrossRef]
- Uda, Y.; Spatz, J.M.; Hussein, A.; Garcia, J.H.; Lai, F.; Dedic, C.; Fulzele, K.; Dougherty, S.; Eberle, M.; Adamson, C.; et al. Global transcriptomic analysis of a murine osteocytic cell line subjected to spaceflight. FASEB J. 2021, 35, e21578. [Google Scholar] [CrossRef]
- Ray, S.; Gebre, S.; Fogle, H.; Berrios, D.C.; Tran, P.B.; Galazka, J.M.; Costes, S.V. GeneLab: Omics database for spaceflight experiments. Bioinformatics 2019, 35, 1753–1759. [Google Scholar] [CrossRef]
- Berrios, D.C.; Galazka, J.; Grigorev, K.; Gebre, S.; Costes, S.V. NASA GeneLab: Interfaces for the exploration of space omics data. Nucleic Acids Res. 2021, 49, D1515–D1522. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Affymetrix. [MTA-1_0] Affymetrix Mouse Transcriptome Array 1.0 [Transcript (Gene) CDF Version]. 2018. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL20258 (accessed on 17 September 2025).
- Affymetrix. [MoGene-2_0-st] Affymetrix Mouse Gene 2.0 ST Array [mogene20st_Mm_ENTREZG_17.1.0]. 2015. Available online: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL17791 (accessed on 17 September 2025).
- MacDonald, J. mta10transcriptcluster.db: Affymetrix mta10 Annotation Data (Chip mta10transcriptcluster), R Package Version 8.7.0; R Core Team: Vienna, Austria, 2017. [Google Scholar]
- MacDonald, J. mogene20sttranscriptcluster.db: Affymetrix mogene20 Annotation Data (Chip mogene20sttranscriptcluster), R Package Version 8.7.0; R Core Team: Vienna, Austria, 2017. [Google Scholar]
- Microsoft Corporation. Microsoft Excel. 2018. Available online: https://office.microsoft.com/excel (accessed on 17 September 2025).
- Wickham, H.; François, R.; Henry, L.; Müller, K.; Vaughan, D.; dplyr:A Grammar of Data Manipulation_. R Package Version 1.1.3. 2023. Available online: https://cran.r-project.org/package=dplyr (accessed on 17 September 2025).
- Posit Team. RStudio: Integrated Development Environment for R. 2024. Available online: https://posit.co/download/rstudio-desktop/ (accessed on 17 September 2025).
- R Core Team. R: A Language and Environment for Statistical Computing_. R Foundation for Statistical Computing. 2024. Available online: https://cran.r-project.org/mirrors.html (accessed on 17 September 2025).
- Jain, N.; O’Connell, M.; Lee, J.K.; Cho, H. LPE: Methods for Analyzing Microarray Data Using Local Pooled Error (LPE) Method. 2024. Available online: https://doi.org/10.18129/B9.bioc.LPE (accessed on 17 September 2025).
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- Team, T. Pandas Development. Pandas-dev/pandas: Pandas. 2020. Available online: https://doi.org/10.5281/zenodo.3509134 (accessed on 17 September 2025).
- Python Software Foundation. Python Language Reference, Version 3.12.5. 2023. Available online: https://docs.python.org/3/reference/index.html (accessed on 17 September 2025).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Castanza, A.S.; Recla, J.M.; Eby, D.; Thorvaldsdóttir, H.; Bult, C.J.; Mesirov, J.P. Extending support for mouse data in the Molecular Signatures Database (MSigDB). Nat. Methods 2023, 20, 1619–1620. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef]
- Carlson, M. GO.db: A set of Annotation Maps Describing the Entire Gene Ontology; R Package Version 3.8.2.; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Han, H.; Cho, J.-W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Yang, D.; Yu, B.; Sun, H.; Qiu, L. The Roles of Histone Demethylase Jmjd3 in Osteoblast Differentiation and Apoptosis. J. Clin. Med. 2017, 6, 24. [Google Scholar] [CrossRef]
- Xia, Y.; Ikedo, A.; Lee, J.-W.; Iimura, T.; Inoue, K.; Imai, Y. Histone H3K27 demethylase, Utx, regulates osteoblast-to-osteocyte differentiation. Biochem. Biophys. Res. Commun. 2022, 590, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Kamath, T.; Mazur, C.M.; Mirzamohammadi, F.; Rotter, D.; Hojo, H.; Castro, C.D.; Tokavanich, N.; Patel, R.; Govea, N.; et al. Control of osteocyte dendrite formation by Sp7 and its target gene osteocrin. Nat. Commun. 2021, 12, 6271. [Google Scholar] [CrossRef] [PubMed]
- Fournier, R.; Harrison, R.E. Methods for studying MLO-Y4 osteocytes in collagen-hydroxyapatite scaffolds in the rotary cell culture system. Connect. Tissue Res. 2021, 62, 436–453. [Google Scholar] [CrossRef]
- Wang, J.S.; Wein, M.N. Pathways Controlling Formation and Maintenance of the Osteocyte Dendrite Network. Curr. Osteoporos. Rep. 2022, 20, 493–504. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Honjo, M.; Hasegawa, T.; Muratani, M.; Bochimoto, H. Meta-Analysis of Ocy-454 Showed Interrupted Osteocyte Maturation in Spaceflight Affects SOST Expression and Hypoxic Response. J. Clin. Med. 2025, 14, 8100. https://doi.org/10.3390/jcm14228100
Honjo M, Hasegawa T, Muratani M, Bochimoto H. Meta-Analysis of Ocy-454 Showed Interrupted Osteocyte Maturation in Spaceflight Affects SOST Expression and Hypoxic Response. Journal of Clinical Medicine. 2025; 14(22):8100. https://doi.org/10.3390/jcm14228100
Chicago/Turabian StyleHonjo, Mayuka, Takanori Hasegawa, Masafumi Muratani, and Hiroki Bochimoto. 2025. "Meta-Analysis of Ocy-454 Showed Interrupted Osteocyte Maturation in Spaceflight Affects SOST Expression and Hypoxic Response" Journal of Clinical Medicine 14, no. 22: 8100. https://doi.org/10.3390/jcm14228100
APA StyleHonjo, M., Hasegawa, T., Muratani, M., & Bochimoto, H. (2025). Meta-Analysis of Ocy-454 Showed Interrupted Osteocyte Maturation in Spaceflight Affects SOST Expression and Hypoxic Response. Journal of Clinical Medicine, 14(22), 8100. https://doi.org/10.3390/jcm14228100




