Type I Interferon-Related Gene Expression and Laboratory Abnormalities in Acute Infection Are Associated with Long COVID Symptom Burden
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Data Collection
2.3. Statistical Analysis
2.4. Ethical Considerations
3. Results
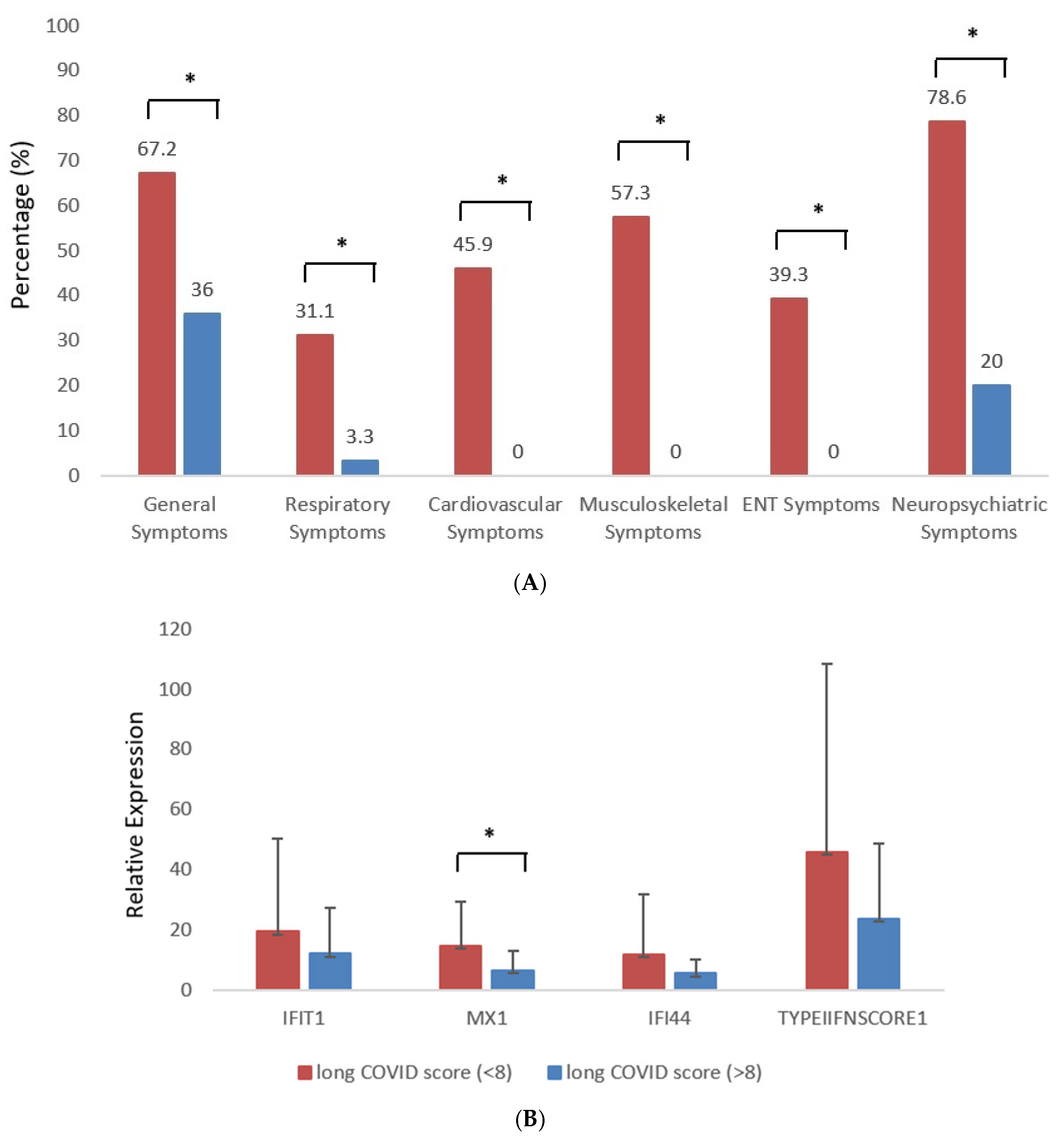
3.1. Demographics and Distribution of Long COVID Symptoms According to Organ Involvement in Study Participants and Controls
3.2. Association of Long COVID Symptom Burden with Clinical, Hematological, and Serological Variables at Baseline
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615. [Google Scholar] [CrossRef]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine 2021, 38, 101019. [Google Scholar] [CrossRef]
- Peluso, M.J.; Deeks, S.G. Mechanisms of long COVID and the path toward therapeutics. Cell 2024, 187, 5500–5529. [Google Scholar] [CrossRef]
- Su, H.C.; Jing, H.; Zhang, Y.; Members of the COVID Human Genetic Effort; Casanova, J.L. Interfering with Interferons: A Critical Mechanism for Critical COVID-19 Pneumonia. Annu. Rev. Immunol. 2023, 41, 561–585. [Google Scholar] [CrossRef] [PubMed]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulou, V.E.; Lembessis, P.; Skarlis, C.; Gkoufa, A.; Sipsas, N.V.; Mavragani, C.P. Hematological Abnormalities in COVID-19 Disease: Association with Type I Interferon Pathway Activation and Disease Outcomes. Front. Med. 2022, 9, 850472. [Google Scholar] [CrossRef]
- Penny’s Hill Practice. Post-COVID Newcastle Screening Tool; Penny’s Hill Practice: Ferndown, UK, 2021; Available online: https://www.pennyshillpractice.co.uk/wp-content/uploads/2021/02/Post-COVID-Newcastle-Screening-Tool.docx (accessed on 20 September 2025).
- Tsilingiris, D.; Vallianou, N.G.; Karampela, I.; Christodoulatos, G.S.; Papavasileiou, G.; Petropoulou, D.; Magkos, F.; Dalamaga, M. Laboratory Findings and Biomarkers in Long COVID: What Do We Know So Far? Insights into Epidemiology, Pathogenesis, Therapeutic Perspectives and Challenges. Int. J. Mol. Sci. 2023, 24, 10458. [Google Scholar] [CrossRef]
- Streiner, D.; Norman, G.; Cairney, J. Health Measurement Scales: A Practical Guide to Their Development and Use, 5th ed.; Oxford University Press: Oxford, UK, 2015; 399p, ISBN 0199685215, 9780199685219. [Google Scholar]
- Bencze, D.; Fekete, T.; Pázmándi, K. Correlation between Type I Interferon Associated Factors and COVID-19 Severity. Int. J. Mol. Sci. 2022, 23, 10968. [Google Scholar] [CrossRef]
- Montenegro, A.F.L.; Clementino, M.A.F.; Yaochite, J.N.U. Type I interferon pathway genetic variants in severe COVID-19. Virus Res. 2024, 342, 199339. [Google Scholar] [CrossRef] [PubMed]
- Lechuga, G.C.; Morel, C.M.; De-Simone, S.G. Hematological alterations associated with long COVID-19. Front. Physiol. 2023, 14, 1203472. [Google Scholar] [CrossRef]
- Mohiuddin Chowdhury, A.T.M.; Karim, M.R.; Ali, M.A.; Islam, J.; Li, Y.; He, S. Clinical Characteristics and the Long-Term Post-recovery Manifestations of the COVID-19 Patients—A Prospective Multicenter Cross-Sectional Study. Front. Med. 2021, 8, 663670. [Google Scholar] [CrossRef] [PubMed]
- Radkhah, H.; Omidali, M.; Hejrati, A.; Bahri, R.A.; Arefi, S.; Behzadi, A.; Eslami, M.; Khadembashiri, M.; Khadembashiri, M.; Najafirashed, M.; et al. Correlations of Long COVID Symptoms and Inflammatory Markers of Complete Blood Count (CBC): A Cross-sectional Study. J. Community Hosp. Intern. Med. Perspect. 2023, 13, 112–119. [Google Scholar] [CrossRef]
- Hong, S.G.; Sato, N.; Legrand, F.; Gadkari, M.; Makiya, M.; Stokes, K.; Howe, K.N.; Yu, S.J.; Linde, N.S.; Clevenger, R.R.; et al. Glucocorticoid-Induced Eosinopenia Results from CXCR4-Dependent Bone Marrow Migration. Blood 2020, 136, 2667–2678. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-Cell Landscape of Bronchoalveolar Immune Cells in Patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Silvin, A.; Chapuis, N.; Dunsmore, G.; Goubet, A.G.; Dubuisson, A.; Derosa, L.; Almire, C.; Hénon, C.; Kosmider, O.; Droin, N.; et al. Elevated Calprotectin and Abnormal Myeloid Cell Subsets Discriminate Severe from Mild COVID-19. Cell 2020, 182, 1401–1418.e18. [Google Scholar] [CrossRef]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440.e23. [Google Scholar] [CrossRef]
- Fialek, B.; Pruc, M.; Smereka, J.; Jas, R.; Rahnama-Hezavah, M.; Denegri, A.; Szarpak, A.; Jaguszewski, M.J.; Peacock, F.W.; Szarpak, L. Diagnostic value of lactate dehydrogenase in COVID-19: A systematic review and meta-analysis. Cardiol. J. 2022, 29, 751–758. [Google Scholar] [CrossRef]
- Udeh, R.; Utrero-Rico, A.; Dolja-Gore, X.; Rahmati, M.; McEVoy, M.; Kenna, T. Lactate dehydrogenase contribution to symptom persistence in long COVID: A pooled analysis. Rev. Med. Virol. 2023, 33, e2477. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulou, V.E.; Bali, T.; Adamantou, M.; Asimakopoulou, S.; Makrodimitri, S.; Samara, S.; Triantafyllou, M.; Voutsinas, P.M.; Eliadi, I.; Karamanakos, G.; et al. Acute hepatitis and liver injury in hospitalized patients with COVID-19 infection. Exp. Ther. Med. 2022, 24, 691. [Google Scholar] [CrossRef]
- Cholongitas, E.; Bali, T.; Georgakopoulou, V.E.; Kamiliou, A.; Vergos, I.; Makrodimitri, S.; Samara, S.; Triantafylou, M.; Basoulis, D.; Eliadi, I.; et al. Comparison of liver function test- and inflammation-based prognostic scores for coronavirus disease 2019: A single center study. Eur. J. Gastroenterol. Hepatol. 2022, 34, 1165–1171. [Google Scholar] [CrossRef]
- Cholongitas, E.; Bali, T.; Georgakopoulou, V.E.; Giannakodimos, A.; Gyftopoulos, A.; Georgilaki, V.; Gerogiannis, D.; Basoulis, D.; Eliadi, I.; Karamanakos, G.; et al. Prevalence of abnormal liver biochemistry and its impact on COVID-19 patients’ outcomes: A single-center Greek study. Ann. Gastroenterol. 2022, 35, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Bali, T.; Georgakopoulou, V.E.; Kamiliou, A.; Vergos, I.; Adamantou, M.; Vlachos, S.; Ermidis, G.; Sipsas, N.V.; Samarkos, M.; Cholongitas, E. Abnormal liver function tests and coronavirus disease 2019: A close relationship. J. Viral. Hepat. 2023, 30, 79–80. [Google Scholar] [CrossRef] [PubMed]
- de Lima, I.C.; de Menezes, D.C.; Uesugi, J.H.E.; Bichara, C.N.C.; da Costa Vasconcelos, P.F.; Quaresma, J.A.S.; Falcão, L.F.M. Liver Function in Patients with Long-Term Coronavirus Disease 2019 of up to 20 Months: A Cross-Sectional Study. Int. J. Environ. Res. Public Health 2023, 20, 5281. [Google Scholar] [CrossRef]
- Kim, Y.M.; Shin, E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021, 53, 750–760. [Google Scholar] [CrossRef]
- Mavragani, C.P.; Skarlis, C.; Kostopoulos, I.V.; Maratou, E.; Moutsatsou, P.; Terpos, E.; Tsitsilonis, O.E.; Dimopoulos, M.A.; Sfikakis, P.P. Distinct type I interferon responses between younger women and older men contribute to the variability of COVID-19 outcomes: Hypothesis generating insights from COVID-19 convalescent individuals. Cytokine 2022, 157, 155964. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- van der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; van den Heuvel, G.; Mantere, T.; Kersten, S.; van Deuren, R.C.; Steehouwer, M.; van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants Among Young Men with Severe COVID-19. JAMA 2020, 324, 663–673. [Google Scholar] [CrossRef]
- Ghorra, N.; Popotas, A.; Besse-Hammer, T.; Rogiers, A.; Corazza, F.; Nagant, C. Cytokine Profile in Patients with Postacute Sequelae of COVID-19. Viral Immunol. 2024, 37, 346–354. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, L.; Cui, D.; Zhang, H.; Zhang, Q.; Ren, L.; Wang, G.; Zhang, X.; Huang, T.; Chen, L.; et al. Immune Responses in Discharged COVID-19 Patients with and Without Long COVID Symptoms. Open Forum. Infect. Dis. 2024, 11, ofae137. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Carballa, A.; Pischedda, S.; Pardo-Seco, J.; Gómez-Rial, J.; Martinón-Torres, F.; Salas, A. Interferon gene expression declines over time post-COVID infection and in long COVID patients. Infect. Dis. 2024, 57, 35–48. [Google Scholar] [CrossRef]
- Vavougios, G.D.; Tseriotis, V.S.; Liampas, A.; Mavridis, T.; de Erausquin, G.A.; Hadjigeorgiou, G. Type I interferon signaling, cognition and neurodegeneration following COVID-19: Update on a mechanistic pathogenetic model with implications for Alzheimer’s disease. Front. Hum. Neurosci. 2024, 18, 1352118. [Google Scholar] [CrossRef] [PubMed]
- Fracella, M.; Mancino, E.; Nenna, R.; Virgillito, C.; Frasca, F.; D’Auria, A.; Sorrentino, L.; Petrarca, L.; La Regina, D.; Matera, L.; et al. Age-related transcript changes in type I interferon signaling in children and adolescents with long COVID. Eur. J. Immunol. 2024, 54, e2350682. [Google Scholar] [CrossRef] [PubMed]
- Gross, R.S.; Thaweethai, T.; Kleinman, L.C.; Snowden, J.N.; Rosenzweig, E.B.; Milner, J.D.; Tantisira, K.G.; Rhee, K.E.; Jernigan, T.L.; Kinser, P.A.; et al. Characterizing Long COVID in Children and Adolescents. JAMA 2024, 21, e2412747. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, M.A.F.; Brito, W.R.D.S.; Pereira, K.A.S.; Pereira, L.M.S.; Amoras, E.D.S.G.; Lima, S.S.; Santos, E.F.D.; Costa, F.P.D.; Sarges, K.M.L.; Cantanhede, M.H.D.; et al. Severe COVID-19 and long COVID are associated with high expression of STING, cGAS and IFN-α. Sci. Rep. 2024, 14, 4974. [Google Scholar] [CrossRef]
| Parameter | Long COVID (Score ≤ 8) | Long COVID (Score > 8) | p-Value |
|---|---|---|---|
| Age | 58.5 ± 12.6 | 56.8 ± 15.3 | 0.96 |
| Female Gender (%) | 69.2 | 30.8 | 0.06 |
| Comorbidities—Diabetes | 2 (4.2%) | 2 (15.4%) | 0.196 |
| Comorbidities—Cardiovascular Disease | 24 (50.0%) | 3 (23.1%) | 0.118 |
| Comorbidities—Autoimmune Disease/Hypothyroidism | 7 (14.6%) | 5 (38.5%) | 0.108 |
| Comorbidities—Chronic Respiratory Disease | 8 (16.7%) | 5 (38.5%) | 0.126 |
| Intubation (%) | 25 | 75 | 0.004 * |
| High Disease Severity (%) | 22.7 | 77.3 | 0.33 |
| Hemoglobin (g/L) | 13.5 ± 1.7 | 13.1 ± 1.4 | 0.35 |
| Platelet Count (×103/μL) | 221 ± 97 | 377 ± 531 | 0.97 |
| WBC (×103/μL) (mean ± SD) | 5.9 ± 2.4 | 7.4 ± 2.7 | 0.04 * |
| Absolute Neutrophil Count (×103/μL) | 6.90 ± 1.13 | 7.72 ± 1.13 | 0.02 * |
| Absolute Lymphocyte Count (×103/μL) | 2.24 ± 0.95 | 1.72 ± 0.98 | 0.08 |
| Absolute Monocyte Count (×103/μL) | 0.94 ± 0.15 | 0.51 ± 0.15 | 0.009 * |
| Absolute Eosinophil Count (×103/μL) | 0.74 ± 0.13 | 0.15 ± 0.31 | 0.03 * |
| Troponin (pg/mL) | 13.8 ± 25.8 | 9.5 ± 5.7 | 0.74 |
| Creatinine (mg/dL) | 1.2 ± 2.2 | 0.7 ± 0.1 | 0.14 |
| Urea (mg/dL) | 37.3 ± 28.7 | 30.5 ± 9.6 | 0.52 |
| SGOT (U/L) | 31.3 ± 17.2 | 47.2 ± 19.7 | 0.005 * |
| SGPT (U/L) | 32.4 ± 32.7 | 44.3 ± 22.9 | 0.35 |
| GGT (U/L) | 39.2 ± 33.1 | 47 ± 34.4 | 0.26 |
| Alkaline Phosphatase (U/L) | 68.2 ± 28.2 | 55.3 ± 17 | 0.07 |
| CK (U/L) | 143.8 ± 175 | 218.1 ± 231.8 | 0.18 |
| LDH (U/L) | 264.7 ± 94 | 397.2 ± 143.6 | 0.002 * |
| D-dimers (µg/mL) | 0.97 ± 0.78 | 0.74 ± 0.4 | 0.44 |
| ESR (mm/1 h) | 46 ± 33 | 80 ± 26 | 0.08 |
| CRP (mg/L) | 43.8 ± 50.7 | 91.3 ± 96.8 | 0.08 |
| Ferritin (ng/mL) | 517 ± 583 | 699 ± 499 | 0.10 |
| Fibrinogen (mg/dL) | 527 ± 158 | 573 ± 176 | 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emmanouil, M.; Georgakopoulou, V.E.; Drougkas, K.; Lembessis, P.; Skarlis, C.; Gkoufa, A.; Sipsas, N.V.; Mavragani, C.P. Type I Interferon-Related Gene Expression and Laboratory Abnormalities in Acute Infection Are Associated with Long COVID Symptom Burden. J. Clin. Med. 2025, 14, 7875. https://doi.org/10.3390/jcm14217875
Emmanouil M, Georgakopoulou VE, Drougkas K, Lembessis P, Skarlis C, Gkoufa A, Sipsas NV, Mavragani CP. Type I Interferon-Related Gene Expression and Laboratory Abnormalities in Acute Infection Are Associated with Long COVID Symptom Burden. Journal of Clinical Medicine. 2025; 14(21):7875. https://doi.org/10.3390/jcm14217875
Chicago/Turabian StyleEmmanouil, Mary, Vasiliki E. Georgakopoulou, Konstantinos Drougkas, Panagiotis Lembessis, Charalampos Skarlis, Aikaterini Gkoufa, Nikolaos V. Sipsas, and Clio P. Mavragani. 2025. "Type I Interferon-Related Gene Expression and Laboratory Abnormalities in Acute Infection Are Associated with Long COVID Symptom Burden" Journal of Clinical Medicine 14, no. 21: 7875. https://doi.org/10.3390/jcm14217875
APA StyleEmmanouil, M., Georgakopoulou, V. E., Drougkas, K., Lembessis, P., Skarlis, C., Gkoufa, A., Sipsas, N. V., & Mavragani, C. P. (2025). Type I Interferon-Related Gene Expression and Laboratory Abnormalities in Acute Infection Are Associated with Long COVID Symptom Burden. Journal of Clinical Medicine, 14(21), 7875. https://doi.org/10.3390/jcm14217875







