Computer Vision-Assisted Spatial Analysis of Mitoses and Vasculature in Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Preparation
- DHMC (Dartmouth-Hitchcock Medical Center), a collection of 143 WSIs of lung adenocarcinoma from the Dartmouth-Hitchcock Pathology Center [15].
- NLST (National Lung Screening Trial), a large-scale clinical trial involving 451 WSIs from lung cancer patients enrolled in the NLST screening study (total cohort: 1225 WSIs) [16].
- TCGA-LUAD (The Cancer Genome Atlas Lung Adenocarcinoma), a collection containing 150 WSIs of lung adenocarcinoma, each paired with clinical, genomic, and radiologic metadata from the TCGA consortium [16].
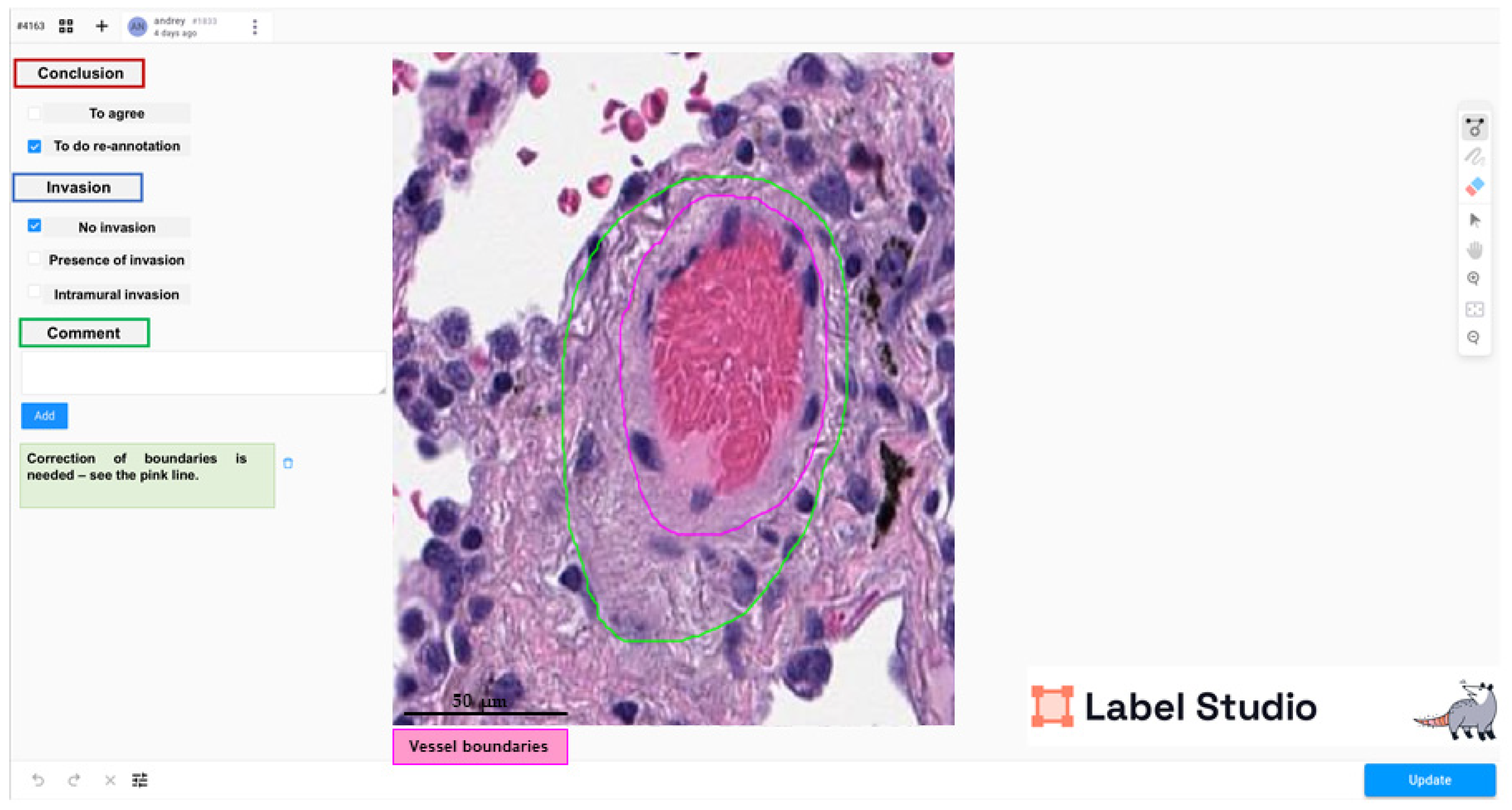
2.2. Annotation
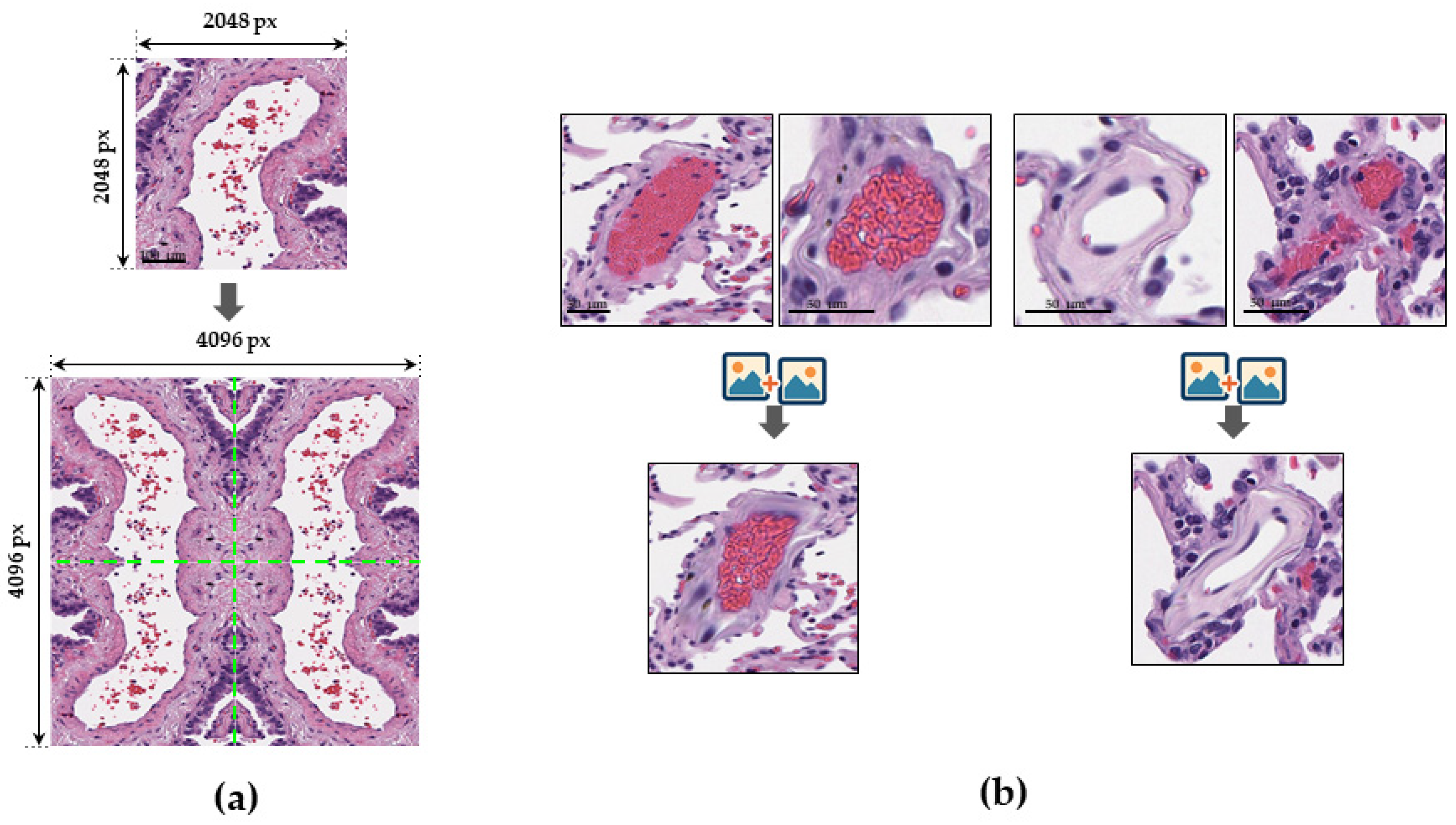
2.3. Training, Test, and Validation
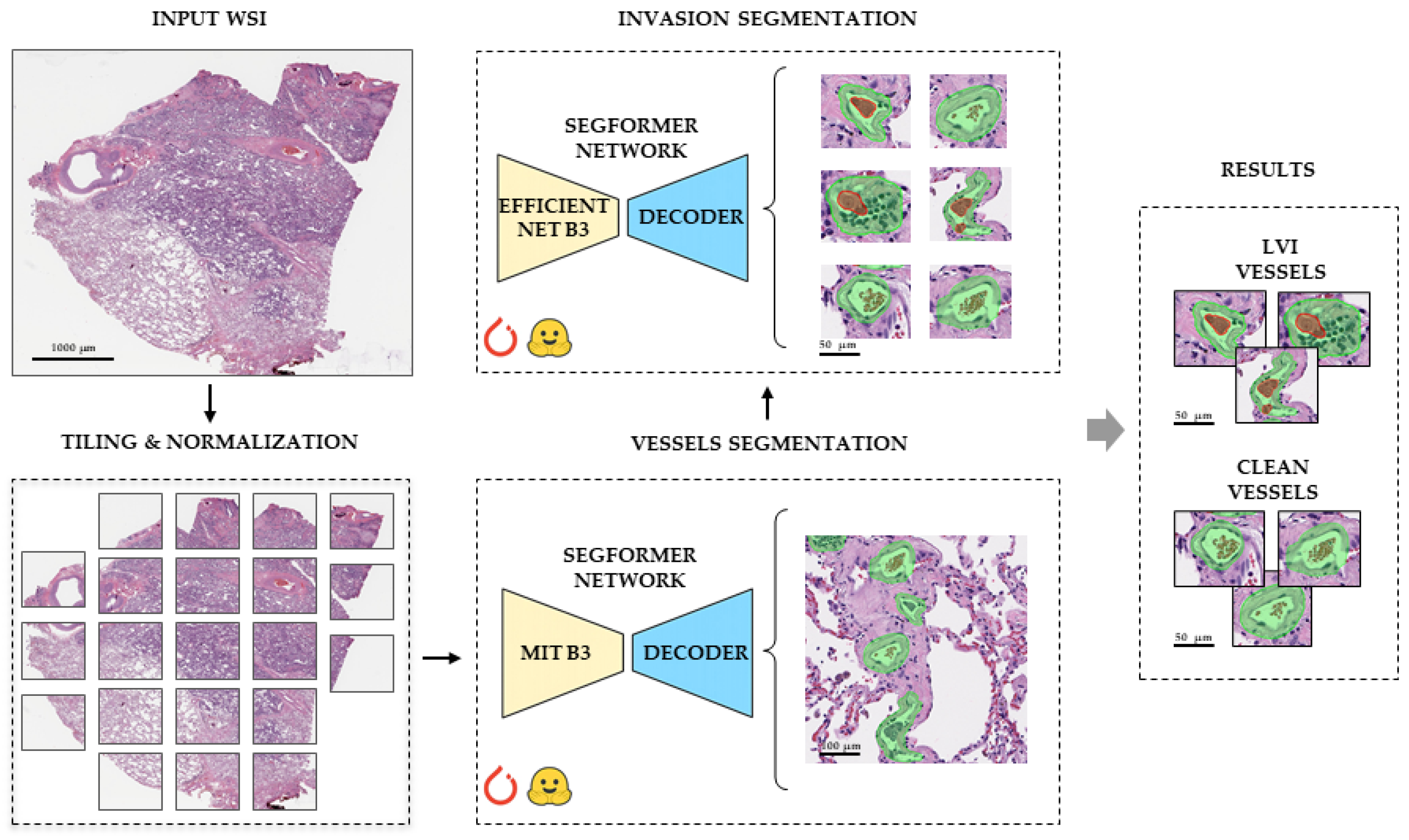
2.3.1. Vessel Segmentation
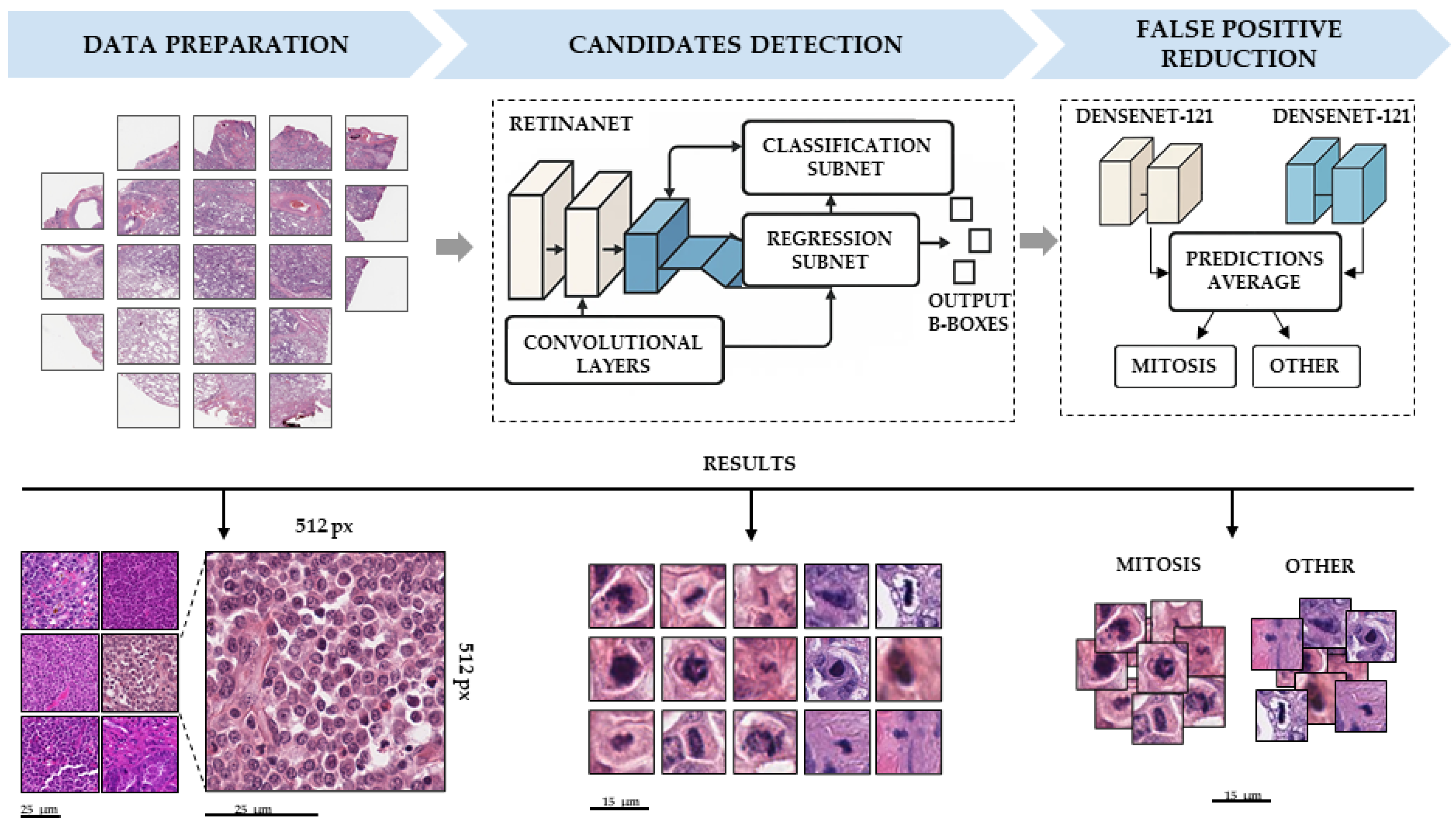
2.3.2. Mitosis Detection
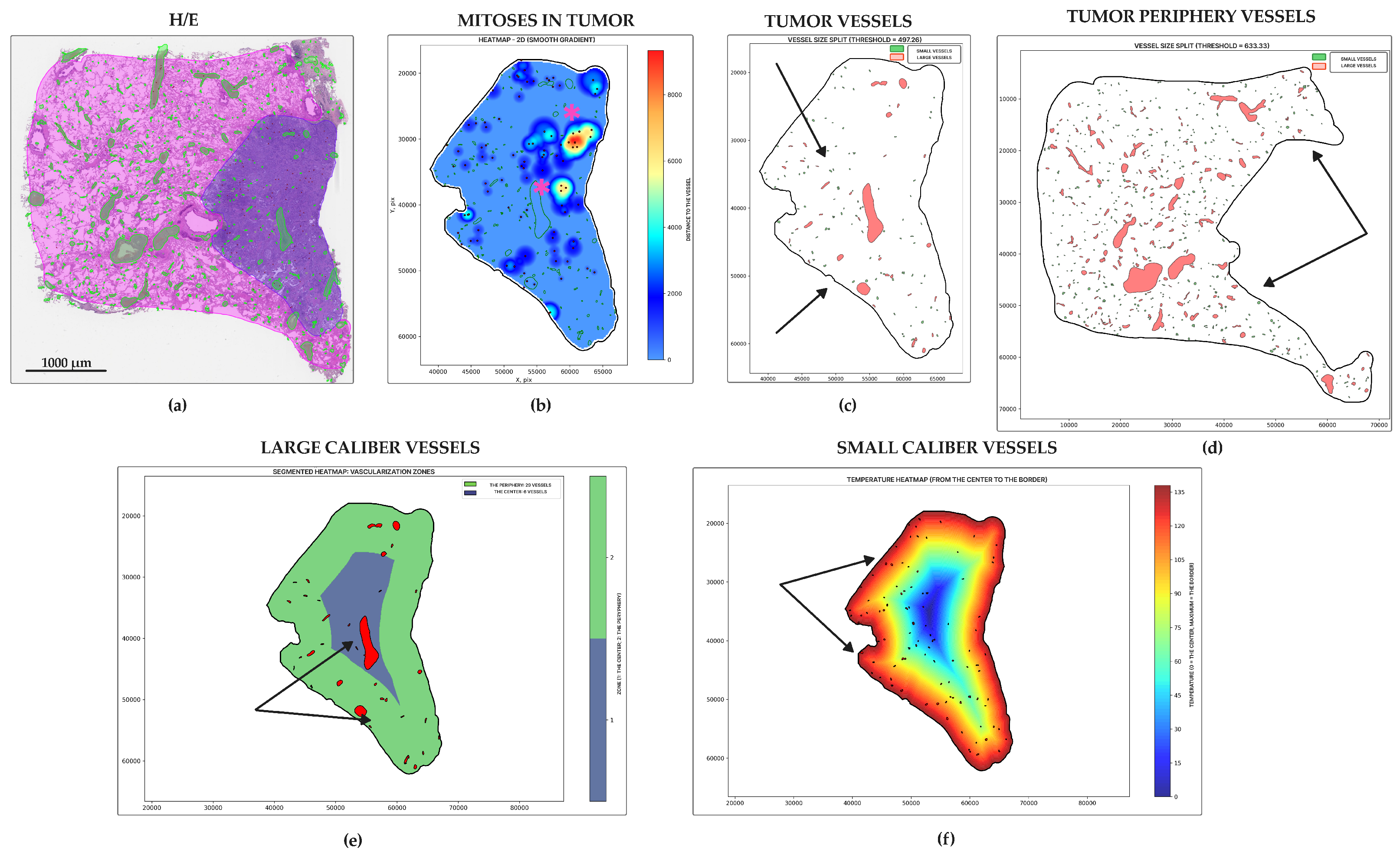
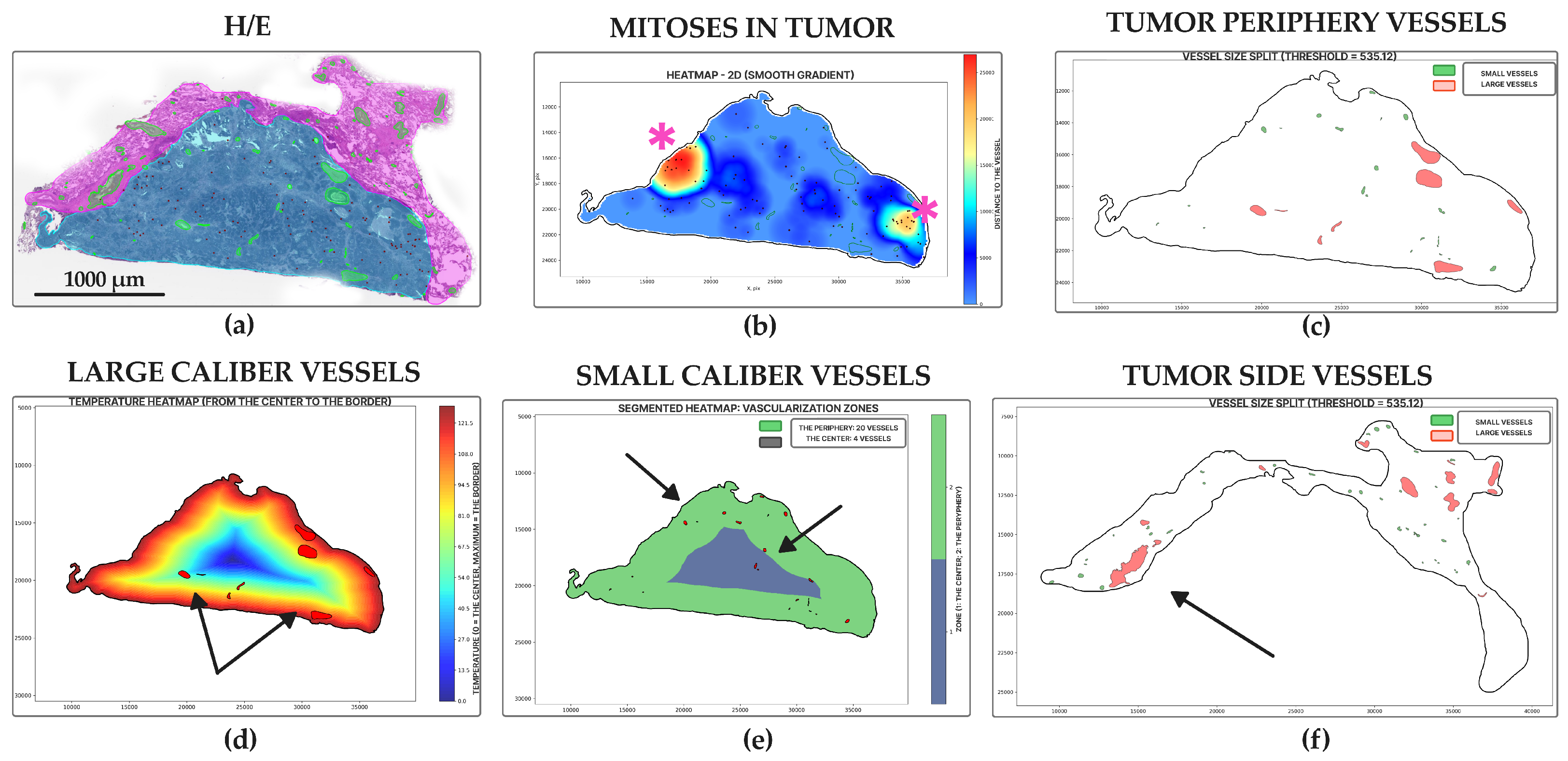
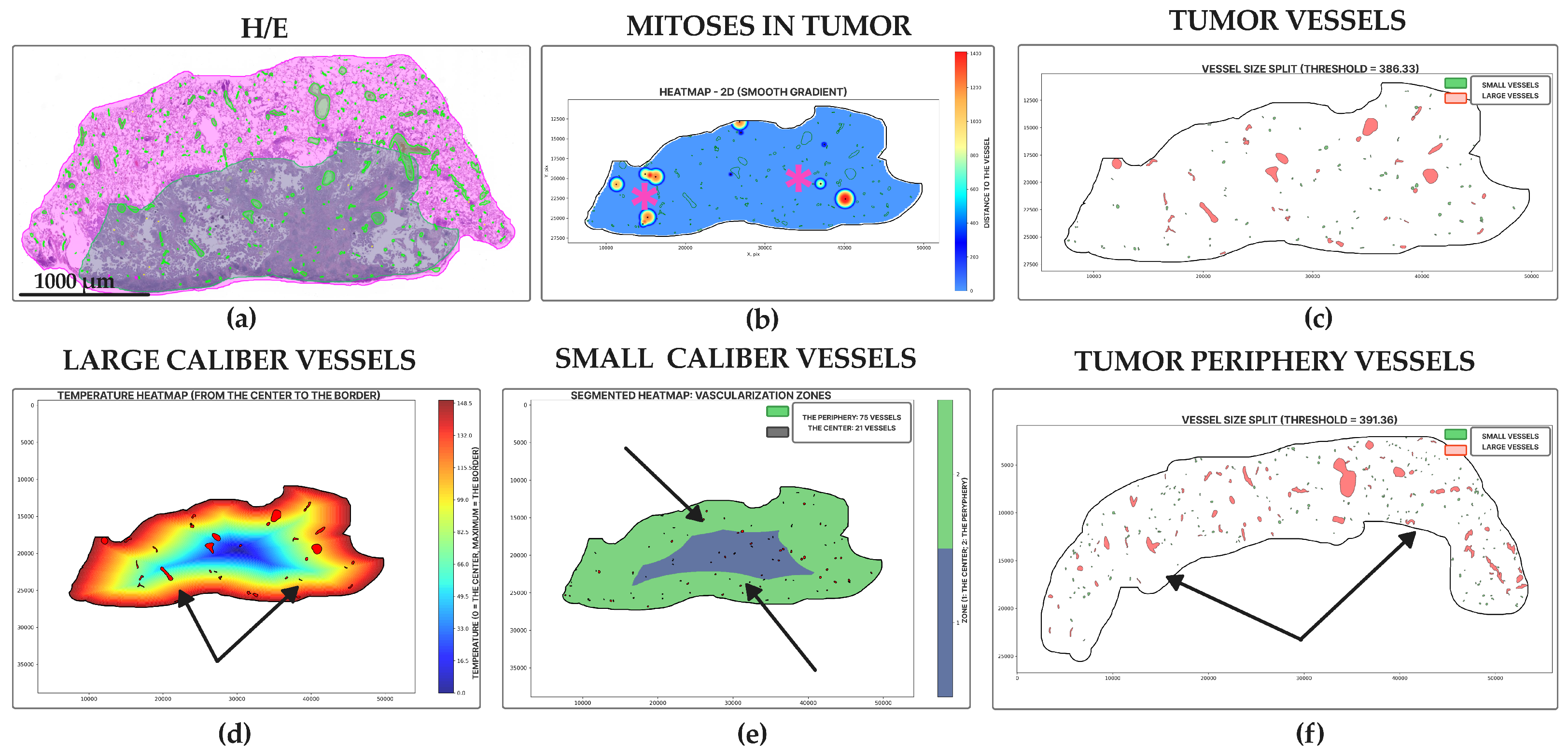
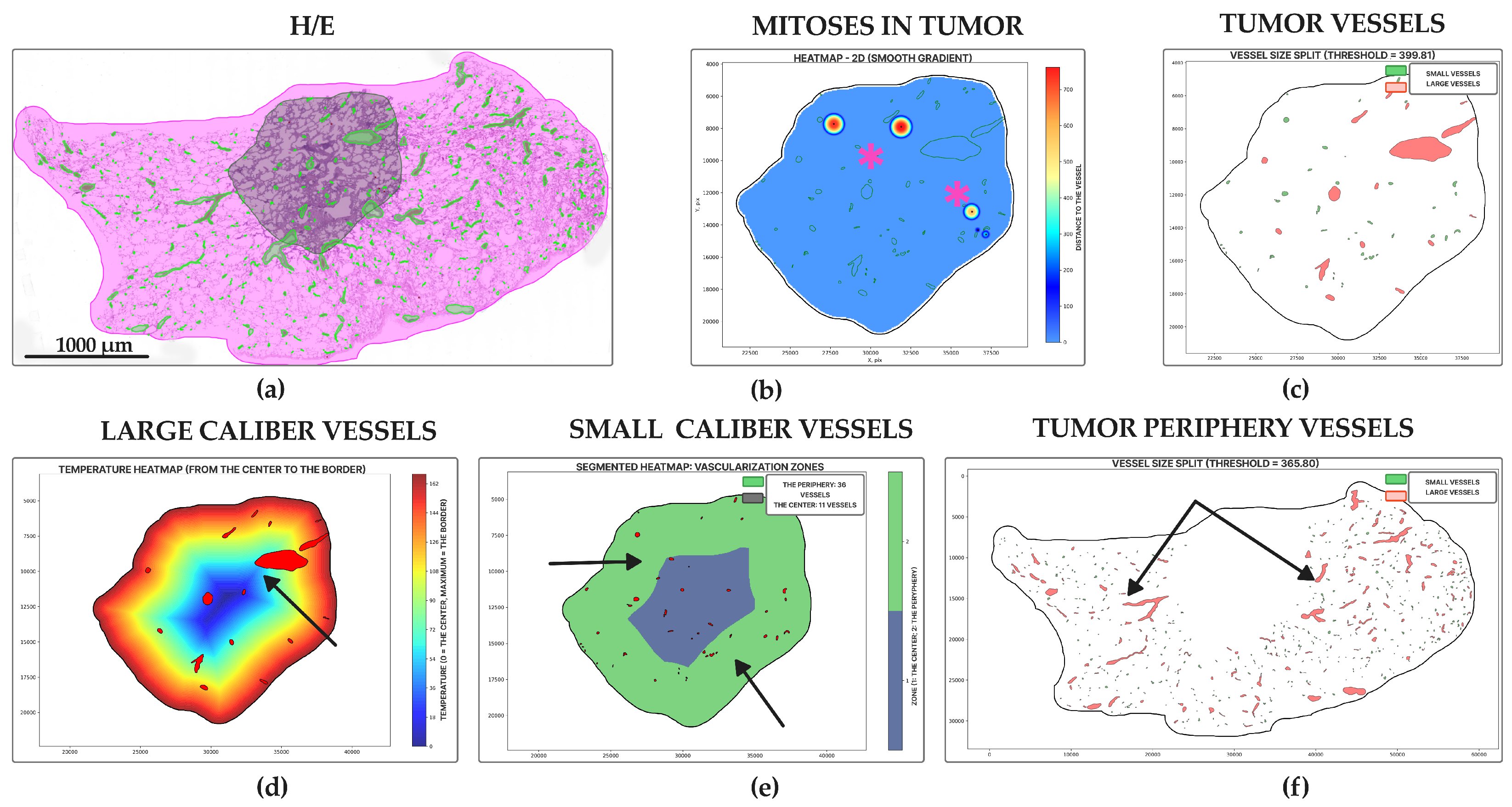
2.4. Morphology, Parameters, and Metrics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| DHMC | Dartmouth-Hitchcock Medical Center |
| EGFR | Epidermal growth factor receptor |
| FP | False positive |
| HIF | Hypoxia-inducible factor |
| NLST | National Lung Screening Trial |
| OWN_D | Own dataset |
| PD-L1 | Programmed death-ligand 1 |
| TCGA | The Cancer Genome Atlas Lung Adenocarcinoma |
| TP | True positive |
| VEGF | Vascular endothelial growth factor—VEGF |
| WSI | Whole-slide image |
Appendix A
| Name of the Parameter | Symbol | Dimension | Role of the Parameter Value |
|---|---|---|---|
| Area of tumor tissue | TA | µm2 | Tumor tissue area on WSI. Helps to evaluate the stage |
| Area of tumor side tissue | TS | µm2 | The area of adjacent non-tumor tissue. The size of the tumor microenvironment on WSI is important to assess the effect of a tumor on side tissues |
| Number of vessels | N_vessels | units | Number of tumor and tumor side vessels. Basic angiogenesis score |
| Tumor area small vessels | tumor_area_vessels_small_cnt | units | A total number of tumor small vessels in WSI |
| Tumor area big vessels | tumor_area_vessels_big_cnt | units | A total number of tumor big vessels on WSI |
| Tissue small vessels | tissue_area_vessels_small_cnt | units | A total number of tissue small vessels on WSI |
| Tissue big vessels | tissue_area_vessels_big_cnt | units | A total number of tissue big vessels on WSI |
| Area of vessels (in tumor) | A_vessels | µm2 | Total area of tumor vessels provides information about general vascularization |
| Number of invasion vessels | N_invasive | units | Number of vessels with LVI features. A prognostic marker of future metastases |
| Number of mitotic figures (in tumor) | N_mitosis | units | Shows the activity of cell proliferation. The higher the number, the higher the rate of tumor growth |
| Feret diameter of vessels | µm | Describes the geometric shape of the vessel |
| Name of the Metric | Abbreviation | Formula/Dimension | Role of the Metric Value |
| Vessels number density (in tumor) | VND | , vessels/µm2 | The number of vessels per unit area of the tumor. Assessment of perfusion level |
| Vessels relative area density (in tumor) | VAD_TUMOR | , % (×100) | The proportion of the tumor area with blood vessels. Assessment of relative vascularization |
| Vessels relative area density (in tissue) | VAD_TISSUE | , % (×100) | The proportion of the tumor side with blood vessels. Assessment of relative vascularization |
| Peripheral/central ratio for small vessels | tumor_area_vessels_small_supply_ratio | tissue_area_vessels_small_cnt/tumor_area_vessels_small_cnt | The predominance of vessels on the tumor side may indicate active angiogenesis at the edges, which is typical for aggressive tumors |
| Peripheral/central ratio for big vessels | tumor_area_vessels_big_supply_ratio | tissue_area_vessels_big_cnt/tumor_area_vessels_big_cnt | The predominance of vessels on the tumor side may indicate active angiogenesis at the edges, which is typical for aggressive tumors |
| Vessels invasion index | VII | , % (x100) | The proportion of vessels with LVI features among all vessels. Indicates the risk of tumor metastasis |
| Mitoses number (in tumor) | MD N_mitoses/A_Tumor) | , mitosis/µm2 | The number of mitoses per unit area of the tumor. Indicates tumor growth points |
| Shannon entropy (in tumor) | SE | where -K—number of spatial segments (for example, grid cells or clusters), -—the relative frequency of mitosis in the I-th segment, dits | It shows how evenly or focused the mitotic foci are. Identification of “hot spots” (clusters) of aggressive growth |
| Fractal size of mitosis (in tumor) | FS | Method box-counting | Spatial estimation of the distribution of mitotic figures. High fractality indicates active branched tumor growth and allows us to evaluate this proliferation metric on a small amount of material. |
| Mitosis clusters | mitosis_dbscan_clusters | The presence of mitotic clusters may indicate local foci of high proliferative activity | |
| «Center-to-edge» symmetry coefficient | Sym_C | The ratio of vascular densities in the tumor center and on the tumor side. An indicator of asymmetric growth |
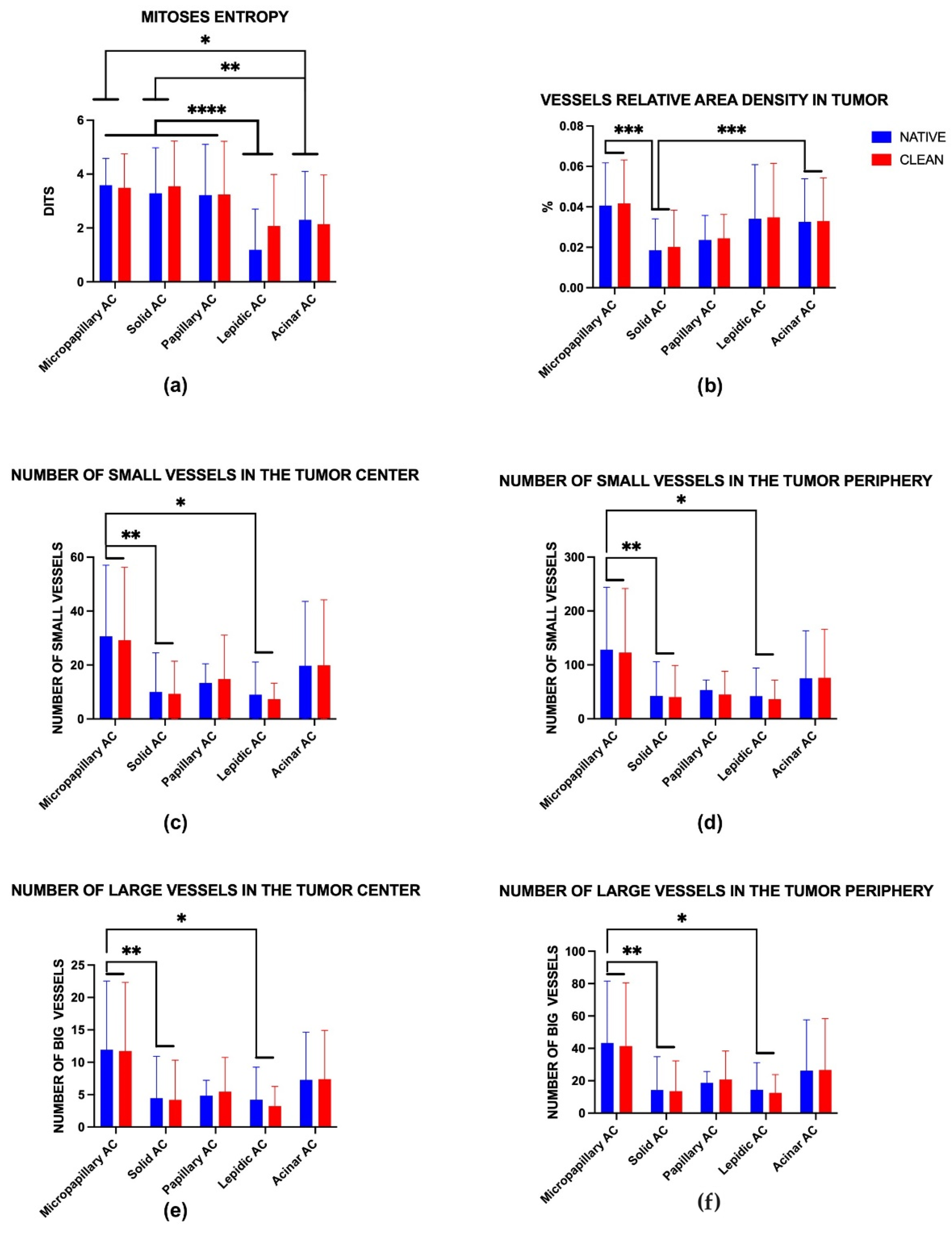
| Metric/Parameter | Predicted Mean of Native | Predicted Mean of Clean | Difference Between Predicted Means | Standard Error of Difference | 95% CI of Difference |
| Mitoses entropy | 2.76 | 2.96 | −0.20 | 0.17 | −0.53 to 0.13 |
| Vessels relative area density in tumor | 0.03 | 0.03 | 0.00 | 0.00 | 0.00 |
| Number of small vessels in the tumor center | 16.54 | 16.10 | 0.44 | 2.47 | −4.41 to 5.28 |
| Number of small vessels in the tumor periphery | 68.20 | 64.18 | 4.02 | 9.15 | −13.96 to 21.99 |
| Number of large vessels in the tumor center | 6.54 | 6.40 | 0.14 | 0.89 | −1.62 to 1.91 |
| Number of large vessels in the tumor periphery | 23.41 | 22.98 | 0.44 | 3.37 | −6.19 to 7.06 |
References
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The trends projection analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- Ramos, R.; Moura, C.S.; Costa, M.; Lamas, N.J.; Castro, L.P.e.; Correia, R.; Garcez, D.; Pereira, J.M.; Sousa, C.; Vale, N. Heterogeneity of lung cancer: The histopathological diversity and tumour classification in the Artificial Intelligence era. Pathobiology 2025, 92, 239–250. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, Q.; Zhang, T.; Li, L.; Xu, C. Minor histological components predict the recurrence of patients with resected stage I acinar-or papillary-predominant lung adenocarcinoma. Front. Oncol. 2022, 12, 1090544. [Google Scholar] [CrossRef]
- Yan, T.; Shi, J. Angiogenesis and EMT regulators in the tumor microenvironment in lung cancer and immunotherapy. Front. Immunol. 2024, 15, 1509195. [Google Scholar] [CrossRef]
- Brierley, J.D.; Van Eycken, E.; Rous, B.; Giuliani, M. TNM Classification of Malignant Tumours; John Wiley & Sons: Hoboken, NJ, USA, 2025. [Google Scholar]
- Cancer Protocol Templates|College of American Pathologists. Available online: https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/cancer-protocol-templates (accessed on 17 October 2025).
- Qiu, Z.; Wu, J.; Pang, G.; Xu, X.; Lin, J.; Wang, P. CD34 evaluation of microvasculature in lung adenocarcinoma and its microvascular density predicts postoperative tumor recurrence. Pathol. Oncol. Res. 2025, 31, 1611985. [Google Scholar] [CrossRef] [PubMed]
- Anai, M.; Saruwatari, K.; Imamura, K.; Fujino, K.; Jodai, T.; Sakata, S.; Iyama, S.; Tomita, Y.; Saeki, S.; Ichiyasu, H.; et al. Negative impact of ratio of the microvascular area to tumor area on the response to EGFR-TKI in non-small cell lung cancer with an EGFR mutation. J. Thorac. Dis. 2024, 16, 1151. [Google Scholar] [CrossRef]
- Fu, X.; Sahai, E.; Wilkins, A. Application of digital pathology-based advanced analytics of tumour microenvironment organisation to predict prognosis and therapeutic response. J. Pathol. 2023, 260, 578–591. [Google Scholar] [CrossRef]
- Timakova, A.; Ananev, V.; Fayzullin, A.; Makarov, V.; Ivanova, E.; Shekhter, A.; Timashev, P. Artificial intelligence assists in the detection of blood vessels in whole slide images: Practical benefits for oncological pathology. Biomolecules 2023, 13, 1327. [Google Scholar] [CrossRef] [PubMed]
- Cree, I.A. Counting mitoses: SI (ze) matters. Pathology 2022, 54, S2–S3. [Google Scholar] [CrossRef]
- Stathonikos, N.; Aubreville, M.; de Vries, S.; Wilm, F.; Bertram, C.A.; Veta, M.; van Diest, P.J. Breast cancer survival prediction using an automated mitosis detection pipeline. J. Pathol. Clin. Res. 2024, 10, e70008. [Google Scholar] [CrossRef]
- Bakoglu, N.; Cesmecioglu, E.; Sakamoto, H.; Yoshida, M.; Ohnishi, T.; Lee, S.-Y.; Smith, L.; Yagi, Y.J.P.; Research, O. Artificial intelligence-based automated determination in breast and colon cancer and distinction between atypical and typical mitosis using a cloud-based platform. Pathol. Oncol. Res. 2024, 30, 1611815. [Google Scholar] [CrossRef]
- Timakova, A.; Ananev, V.; Fayzullin, A.; Zemnuhov, E.; Rumyantsev, E.; Zharov, A.; Zharkov, N.; Zotova, V.; Shchelokova, E.; Demura, T.; et al. LVI-PathNet: Segmentation-classification pipeline for detection of lymphovascular invasion in whole slide images of lung adenocarcinoma. J. Pathol. Inform. 2024, 15, 100395. [Google Scholar] [CrossRef]
- Wei, J.W.; Tafe, L.J.; Linnik, Y.A.; Vaickus, L.J.; Tomita, N.; Hassanpour, S. Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks. Sci. Rep. 2019, 9, 3358. [Google Scholar] [CrossRef]
- National Lung Screening Trial Research Team; Aberle, D.R.; Berg, C.D.; Black, W.C.; Church, T.R.; Fagerstrom, R.M.; Galen, B.; Gareen, I.F.; Gatsonis, C.; Goldin, J.; et al. The national lung screening trial: Overview and study design. Radiology 2011, 258, 243–253. [Google Scholar] [CrossRef]
- Aubreville, M.; Wilm, F.; Stathonikos, N.; Breininger, K.; Donovan, T.A.; Jabari, S.; Veta, M.; Ganz, J.; Ammeling, J.; van Diest, P.J.; et al. A comprehensive multi-domain dataset for mitotic figure detection. Sci. Data 2023, 10, 484. [Google Scholar] [CrossRef] [PubMed]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and efficient design for semantic segmentation with transformers. Adv. Neural Inf. Process. Syst. 2021, 34, 12077–12090. [Google Scholar]
- Tekin, E.; Yazıcı, Ç.; Kusetogullari, H.; Tokat, F.; Yavariabdi, A.; Iheme, L.O.; Çayır, S.; Bozaba, E.; Solmaz, G.; Darbaz, B.; et al. Tubule-U-Net: A novel dataset and deep learning-based tubule segmentation framework in whole slide images of breast cancer. Sci. Rep. 2023, 13, 128. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Li, G.; Zhang, M.; Li, J.; Lv, F.; Tong, G. Efficient densely connected convolutional neural networks. Pattern Recognit. 2021, 109, 107610. [Google Scholar] [CrossRef]
- Shamos, M.I. Computational Geometry. Ph.D. Thesis, Yale University, New Haven, CT, USA, 1978. [Google Scholar]
- Ortmann, B.M. Hypoxia-inducible factor in cancer: From pathway regulation to therapeutic opportunity. BMJ Oncol. 2024, 3, e000154. [Google Scholar] [CrossRef]
- Er Özilhan, S.; Efil, S.C.; Çanakçı, D.; Ağaçkıran, Y.; Şener Dede, D.; Onak Kandemir, N.; Doğan, M.; Ünal, T.D.K.; Kıran, M.M.; Kayaçetin, S.; et al. Correlation of PD-L1 and HIF-1 Alpha Expression with KRAS Mutation and Clinicopathological Parameters in Non-Small Cell Lung Cancer. Curr. Issues Mol. Biol. 2025, 47, 121. [Google Scholar] [CrossRef]
- Li, Y.; Sharma, A.; Schmidt-Wolf, I.G. Evolving insights into the improvement of adoptive T-cell immunotherapy through PD-1/PD-L1 blockade in the clinical spectrum of lung cancer. Mol. Cancer 2024, 23, 80. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chan, L.; Zhu, D.; Pang, Y.; Jin, M.; Wang, Y.; Wang, W. Mitosis targeting in non-small lung cancer cells by inhibition of PAD4. Heliyon 2024, 10, e27313. [Google Scholar] [CrossRef] [PubMed]




| Dataset | Data Format | WSI Count |
|---|---|---|
| NLST | SVS | 57 |
| DHMC | TIFF | 77 |
| TCGA-LUAD | SVS | 9 |
| OWN_DB | SVS, TIFF | 56 |
| Quality Metrics of the DHMC Dataset Before and After Model Tuning | ||||||
|---|---|---|---|---|---|---|
| Before tuning | ||||||
| Negative class | TP | FP | TN | FN | Sensitivity | Specificity |
| Ink (4) | 193 | 5 | 294 | 15 | 0.92 | 0.98 |
| Muscle/fibroblast Nucleus (5) | 193 | 240 | 19 | 15 | 0.92 | 0.07 |
| Chromatin (6) | 193 | 80 | 6 | 15 | 0.92 | 0.06 |
| 4 + 5 + 6 | 193 | 325 | 319 | 15 | 0.92 | 0.49 |
| After tuning | ||||||
| Negative class | TP | FP | TN | FN | Sensitivity | Specificity |
| Ink (4) | 182 | 2 | 297 | 26 | 0.97 | 0.99 |
| Muscle/fibroblast Nucleus (5) | 182 | 1 | 258 | 26 | 0.97 | 0.99 |
| Chromatin (6) | 182 | 21 | 65 | 26 | 0.97 | 0.76 |
| 4 + 5 + 6 | 182 | 24 | 620 | 26 | 0.97 | 0.96 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timakova, A.; Fayzullin, A.; Ananev, V.; Zemnuhov, E.; Alfimov, V.; Baranov, A.; Smirnova, Y.; Shatalov, V.; Konukhova, N.; Karpulevich, E.; et al. Computer Vision-Assisted Spatial Analysis of Mitoses and Vasculature in Lung Cancer. J. Clin. Med. 2025, 14, 7526. https://doi.org/10.3390/jcm14217526
Timakova A, Fayzullin A, Ananev V, Zemnuhov E, Alfimov V, Baranov A, Smirnova Y, Shatalov V, Konukhova N, Karpulevich E, et al. Computer Vision-Assisted Spatial Analysis of Mitoses and Vasculature in Lung Cancer. Journal of Clinical Medicine. 2025; 14(21):7526. https://doi.org/10.3390/jcm14217526
Chicago/Turabian StyleTimakova, Anna, Alexey Fayzullin, Vladislav Ananev, Egor Zemnuhov, Vadim Alfimov, Alexey Baranov, Yulia Smirnova, Vitaly Shatalov, Natalia Konukhova, Evgeny Karpulevich, and et al. 2025. "Computer Vision-Assisted Spatial Analysis of Mitoses and Vasculature in Lung Cancer" Journal of Clinical Medicine 14, no. 21: 7526. https://doi.org/10.3390/jcm14217526
APA StyleTimakova, A., Fayzullin, A., Ananev, V., Zemnuhov, E., Alfimov, V., Baranov, A., Smirnova, Y., Shatalov, V., Konukhova, N., Karpulevich, E., Timashev, P., & Makarov, V. (2025). Computer Vision-Assisted Spatial Analysis of Mitoses and Vasculature in Lung Cancer. Journal of Clinical Medicine, 14(21), 7526. https://doi.org/10.3390/jcm14217526








