Abstract
Background/Objectives: Accurate diagnosis, prognosis, and prediction of treatment response are essential in managing gynecologic cancers and maintaining patient quality of life. Computational pathology, powered by artificial intelligence (AI), offers a transformative opportunity for objective histopathological assessment. This review provides a comprehensive, user-oriented overview of existing AI tools for the characterization of gynecological cancers, critically evaluating their clinical applicability and identifying key challenges for future development. Methods: A systematic literature search was conducted in PubMed and Web of Science for studies published up to 2025. The search focused on AI tools developed for the diagnosis, prognosis, or treatment prediction of gynecologic cancers based on histopathological images. After applying selection criteria, 36 studies were included for in-depth analysis, covering ovarian, uterine, cervical, and other gynecological cancers. Studies on cytopathology and pure tumor detection were excluded. Results: Our analysis identified AI tools addressing critical clinical tasks, including histopathologic subtyping, grading, staging, molecular subtyping, and prediction of therapy response (e.g., to platinum-based chemotherapy or PARP inhibitors). The performance of these tools varied significantly. While some demonstrated high accuracy and promising results in internal validation, many were limited by a lack of external validation, potential biases from training data, and performance that is not yet sufficient for routine clinical use. Direct comparison between studies was often hindered by the use of non-standardized evaluation metrics and evolving disease classifications over the past decade. Conclusions: AI tools for gynecologic cancers represent a promising field with the potential to significantly support pathological practice. However, their current development is heterogeneous, and many tools lack the robustness and validation required for clinical integration. There is a pressing need to invest in the creation of clinically driven, interpretable, and accurate AI tools that are rigorously validated on large, multicenter cohorts. Future efforts should focus on standardizing evaluation metrics and addressing unmet diagnostic needs, such as the molecular subtyping of rare tumors, to ensure these technologies can reliably benefit patient care.
1. Introduction
Worldwide, millions of women of all ages are affected by gynecological cancer, which often leads to significant impairments in general health, quality of life, and, in many cases, early death. Over the recent decades, the classification of gynecological cancers, along with their respective treatment strategies, follow-up protocols, and prognosis for the vast majority of patients, has evolved significantly [1,2,3]. The incidence and mortality of cervical cancer have fallen considerably since human papillomavirus (HPV) was identified as its primary cause and screening and vaccination were introduced [4]. Currently, the greatest focus is on other prevalent cancer types, such as endometrial and ovarian cancers [5]. Despite the advancements in targeted treatments and innovative surgical strategies (including robotic surgery), the mortality rate for endometrial cancer has increased over the past 20 years and has plateaued (≥60%) for ovarian cancer [6].
Today, histopathological diagnostics remain the standard for the classification and therapeutic guidance of gynecological cancers [7]. The integration of computational pathology, supported by artificial intelligence (AI) tools, has the potential to enhance histopathological diagnostic accuracy and provide new prognostic and predictive insights [8,9,10]. Successful AI tools have already been developed for various cancer types (e.g., prostate, breast, and colon cancer) [11,12,13,14]. By utilizing such AI tools, pathologists can have an adjunct to tumor identification, histotyping, staging, and grading, as well as the evaluation of immunohistochemical (IHC) markers and the prediction of IHC results, molecular subtypes, prognosis, and sensitivity to chemotherapy/targeted therapy [15,16,17,18,19].
Over the past three decades, digital images have become increasingly common in medical practice. A major breakthrough in this field occurred in the 1990s with the advent of whole-slide images (WSIs), produced by scanning entire tissue sections on slides rather than focusing on specific regions of interest. Pathologists at different sites could then view these WSIs on a computer monitor, enhancing remote collaboration [20,21,22]. WSIs offer optimized navigation and precision measurement tools, facilitate the exchange of cases between pathologists, and allow the use of computational pathology tools [20,21,22,23,24]. The integration of computational pathology tools, such as those based on AI, into the digital workflow may affect the diagnostic accuracy and efficiency [11]. Moreover, AI outputs would become part of the pathologist’s portfolio, similar to immunohistochemistry, and the acceptance of their results, at this stage, should rely solely on the pathologists. Bringing AI results that are discordant with the pathologist’s observations to the Multidisciplinary Team (MDT) is still at an early stage, as the performance of algorithms still encompasses many errors and is limited to certain organs and pathologies. The discussion of prediction results provided by AI software should definitely take place in MDTs, but, for now, their use is not recommended [25]. In addition, we should mention that, while no FDA-approved digital pathology algorithms yet exist for gynecological diagnostics, future needs for precise molecular subtyping in endometrial and ovarian cancers will require such tools for clinical trial recruitment. Predicting genetic abnormalities directly from H&E slides would further streamline patient identification. Successful implementation will require standardized image acquisition, clear guidelines, and regulatory collaboration [26,27].
AI technologies are rapidly evolving, and the algorithms first based on deep learning’s simple convolutional neural networks (CNNs) are now based more on complex transformer-based algorithms [28]. In addition, taking into consideration all the new evidence on how the pre-scanning operation may influence the AI performance, detailed specifications for use should be a constant in every computational pathology product so that bias may be minimized [29].
This review outlines the current computational-pathology-based approaches to the histological diagnosis, prognosis, and treatment of gynecologic cancers, as well as the challenges and future directions in the field. Cytological diagnosis remains outside the scope of this review, due to the fact that digital cytology is currently an independent field, with commercial solutions and scientific developments so numerous and extensive that they require separate consideration. Furthermore, several comprehensive review publications are already dedicated to this topic [30,31,32,33].
Another limitation of this study is that, over the past 10 years of active development in digital pathology—during which most of the AI tools examined were developed—the approaches to female reproductive tract tumor classification have changed. This is particularly true for ovarian tumors, making it difficult to fully compare AI tools developed using previous classification systems with those designed according to current classification groups. In addition, as a limitation of the study, we recognize that there is an oversimplification of molecular subtyping: although well-known molecular classes (e.g., POLE-mutant and p53-abnormal) are mentioned, epigenetic features and methylation profiles are ignored despite their growing importance. This is because there are no dedicated algorithms to target these less frequent molecular alterations. Moreover, the utility of AI to prioritize variants of unknown significance (VUSs) for secondary review could be proposed as a novel frontier.
2. Methodology
A search query was developed in the PubMed and Web of Science databases for literature on this topic published up to and including 2025. The search string was as follows: (Pathology OR Histopathology OR Histopathological OR Whole slide image OR WSI OR Artificial Intelligence OR AI OR Neural Network OR NN or Computational Pathology OR Digital Pathology) AND (Gynecologic Pathology OR Reproductive System Pathology OR Endometrial cancer OR Endometrial carcinoma OR Ovarian cancer OR Ovarian carcinoma OR Fallopian tube cancer OR Fallopian tube carcinoma OR Fallopian tube tumors OR Cervical cancer OR Cervical carcinoma OR Vulvar cancer OR Vulvar carcinoma OR Vaginal cancer OR Vaginal carcinoma OR Uterine cancer OR Uterine carcinoma OR Uterine mesenchymal tumors OR trophoblastic disease OR hydatidiform mole). The selection criteria consisted of accepting only studies that involved the development of AI tools for the diagnosis, prognosis, or prediction of gynecologic cancers. The two databases provided a total of 4360 results, of which 72 were duplicates, and 4252 studies were not included as they did not meet the acceptance criteria.
The remaining 36 studies were analyzed in this review (17 related to ovarian and fallopian tube cancer, 16 to uterine cancer, 2 to cervical cancer, and 1 to lower reproductive tract cancer).
3. Computational Pathology Dedicated to Gynecological Cancers
In gynecologic pathology, the applications of AI tools have already been tested for their diagnostic, prognostic, and predictive functions, tailored to the location, molecular subtyping, and target therapy of each cancer. The most frequent AI tools for the characterization of gynecologic cancers are summarized in Figure 1.
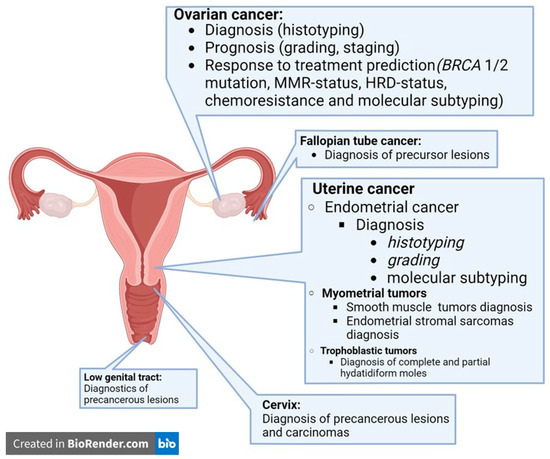
Figure 1.
Artificial intelligence tools for characterization of gynecological cancers reported in the literature up to today.
4. Ovarian Cancer
4.1. Diagnosis
There are three main groups of ovarian tumors, classified according to the World Health Organization (WHO) 5th Edition Classification (2020): epithelial, stromal, and germ cell tumors. The most frequent and deadliest are epithelial tumors, which include high-grade serous cancer (HGSC); low-grade serous cancer (LGSC); mucinous, endometrioid, clear cell, and seromucinous cancer; malignant Brenner tumors; and other rare types. Most AI tools for the diagnosis of ovarian cancer are designed to identify the five most common types of epithelial cancer (HGSC, LGSC, and endometrioid, mucinous, and clear cell cancer) (Table 1). Both convolutional neural network (CNN) and support vector machine (SVM) models have been used in the development of these tools. Most of these models use hematoxylin and eosin (H&E)-stained whole-slide images (WSIs) divided into patches, with the most popular sizes being 500 × 500 pixels [34,35] or 256 × 256 pixels [36,37]. However, the final algorithms more frequently operate on WSIs [34,35,38,39] rather than patches [36,37,40]. The metrics used to evaluate the precision of these models vary, which poses challenges for direct comparisons. Nevertheless, all algorithms included in this study achieve an area under the curve (AUC) greater than 0.92 or an accuracy exceeding 90%, although some did not use independent cohorts [34,35,37,40]. With refined training, the inclusion of additional histotypes, and the optimal performance in generalization tests, these AI tools are strong candidates for clinical practice. While some ovarian cancer histotypes have high inter-observer reproducibility, others have a moderate or even low agreement [41,42].
4.2. Prognosis
AI-based grading and staging on H&E-stained slides have been proposed to improve the diagnostic accuracy of pathologist evaluations (Table 1). The most relevant grading evaluation for ovarian cancer is the differentiation between HGSC and LGSC, as these tumors are considered different histotypes. Nonetheless, endometrioid and mucinous cancers also require accurate grading. Staging ovarian cancer remains one of the most controversial issues, as it is sometimes difficult to identify the original source of the tumor. Moreover, distinguishing between invasive and non-invasive implants in LGSC can significantly influence the final stage, though the reproducibility of their evaluation is rather low.
The study by Yu et al. was based on The Cancer Genome Atlas (TCGA) cohort, included only serous cancer [43], and focused on histotyping (HGSC and LGSC). The AUC of the AI tool reported in this study was 0.812, which is lower than other models and needs to be improved to be comparable with those achieving AUCs of 0.95–0.972 and also including independent cohorts [38,39]. The staging algorithm proposed by Ghoniem et al. demonstrated a very high accuracy (around 99%) [44]. This study also used the TCGA cohort, in which the majority of tumors were HGSC, whereas pathologists encounter more difficulties with staging LGSC, and endometrioid and mucinous cancers, which could be underrepresented in this study [45,46].
In ovarian cancer, the prediction of the prognosis is crucial for stratification in clinical trials, as most cases are diagnosed at an advanced stage [46,47]. Besides grading and staging, other factors can be used for prognostication, including IHC markers, and various -omics data related to histopathologic image analysis [48,49] (Table 1). The inclusion of different histotypes within the same study may lead to poor results due to the presence of many confounding factors that can affect the final outcome and reduce the accuracy of the prognosis prediction. The study by Poruthoor et al. used three ovarian cancer datasets (genomic, proteomic, and imaging) retrieved from The Cancer Genome Atlas (TCGA) to determine if the prediction of the ovarian cancer grade or patient survival rate can be predicted with AI tools. It was conducted over a decade ago, when CNNs were not as advanced as they are today [50].
A recent study by Yang et al. [51] presents more conclusive results, showing significant statistical differences between two ovarian cancer clusters subdivided according to the Ovarian Cancer Digital Pathology Index (OCDPI). The construction of the OCDPI involves two steps: a histopathological feature extractor and a graph-based deep-learning aggregation module, which integrates embeddings from all patches of the WSI. The use of diverse cohorts (TCGA, the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial, and the Harbin Medical University Cancer Hospital), well-organized external validation, and contemporary neural network approaches with a transformer deep-learning architecture has led to much more impressive outcomes [51].
4.3. Response to Treatment Prediction
Predicting the treatment response in ovarian cancer is crucial, given the low overall survival and progression-free survival rates of patients with this disease. The accurate identification of chemotherapy-resistant tumors, the evaluation of the potential efficacy of immunotherapy, and enrolling patients in suitable clinical trials with targeted or experimental agents are essential. In ovarian cancer patients, the mutations in BRCA1/2 and mismatch repair (MMR) genes, as well as homologous recombination deficiency (HRD) status and platinum resistance of the tumor, should be evaluated to predict the treatment response. It is well-known that patients with BRCA1/2 mutations can benefit from poly(ADP-ribose) polymerase (PARP) inhibitors [52,53]. Although only a small proportion of ovarian cancers are MMR-deficient (about 16%), these patients may benefit from checkpoint inhibition therapy. The HRD status can predict the response to platinum-based therapy and PARP inhibitors [54,55,56]. Several clinical studies have focused on accurately predicting the response to platinum-based therapy in patients with HGSC, as this is the first-line treatment for the majority of ovarian cancer patients [57]. Therefore, screening for these genetic disorders at the time of diagnosis using AI tools based on H&E-stained slides could be practical for the initial stratification and for further molecular genetic studies to confirm the specific tumor biology (Table 2).
Despite the long history of BRCA1/2 testing and the definition of reliable morphological characteristics [58], the currently developed AI tools for predicting their mutational status are not yet accurate enough for routine practice. Only the study by Zeng et al. demonstrated a high accuracy (0.912) [17], differing from others by using a random forest model and a patient-based predictive approach, while other studies used more traditional models and WSI-based predictions, despite recruiting more patients [18,59,60]. Zeng et al.’s study was not solely dedicated to BRCA1/2 mutation tumors but also included a prediction of the MMR status; however, the study focused only on HGSC and did not provide information on the number of MMR-deficient (dMMR) tumors. An MSI-high status is rare in HGSC and LGSC, and even endometrioid ovarian cancer shows dMMR in fewer than 20% of cases [19]. Nonetheless, the AUC results for the prediction of dMMR were also very high in this study [17], warranting further verification by others. This study is the only one assessing the MSI status in ovarian cancer using AI tools. The pan-cancer investigation by Arslan et al. on the deep-learning-based prediction of multi-omic biomarkers included ovarian cancer but focused on MMR gene mutations in gastrointestinal cancer [61]. Other cross-cancer surveys on the MSI status in solid tumors have only addressed endometrial cancer among gynecological malignancies [62,63].
The HRD status is predominantly associated with BRCA1/2 mutations in ovarian cancer, though some studies consider this parameter separately for the development of AI tools. The pan-cancer study by Loeffler et al. included ovarian cancer cohorts, achieving a reasonable accuracy only for endometrial cancer among gynecological malignancies [64]. Another study by Frenel et al. on HRD status prediction based on whole-slide imaging focused solely on ovarian cancer, achieving an AUC of 0.74 in the proprietary “Discovery cohort” and 0.67 in the TCGA cohort [65]. However, this study is not fully published yet (currently only an abstract is available), making it difficult to assess the strengths and weaknesses of this algorithm. At the same time, the most recent results related to HRD status prediction showed a 72% accuracy for internal cohorts and 57% for external cohorts in the study by Marmé et al. [66]; Bregstrom et al. achieved an AUC of 0.81 for the internal cohort [67], and Zhang et al. reported an AUC of 0.769 [68].
Several AI tools have been developed to predict the response to chemotherapy by combining clinical, molecular, and genetic data. Most proposed algorithms related to platinum-based therapy effectiveness have demonstrated a high accuracy (above 90%) and/or AUC (above 0.95) [43,69,70]. Yu et al.’s studies aimed not only to predict the chemotherapy response but also to distinguish the molecular subtypes of high-grade ovarian cancer [43]. Gilley et al. demonstrated how to predict the effectiveness of bevacizumab in ovarian cancer treatment using a pathomics biomarker; however, the final results were not particularly impactful (AUC 0.82–0.83) [71]. Meanwhile, the prediction of PARP-inhibitors’ efficacy was less successful: Marmé et al. demonstrated an only 72% accuracy for internal cohorts and 57% for external cohorts [66].
Molecular subtyping is critical for both the prognosis and treatment stratification of patients with HGSC. Previous approaches have been developed to distinguish these subtypes (immunoreactive, mesenchymal, differentiated, and proliferative) based on molecular and genetic data, morphological features, and IHC characteristics [15,72]. However, the results of this study were moderate, with a Spearman correlation with true molecular subtypes ranging from 0.111 to 0.576 [43]. The development of dedicated AI tools for subtyping LGSC could improve the outcomes of targeted treatment with mitogen-activated protein kinase (MEK) inhibitors and enhance prognostication.

Table 1.
AI tools for ovarian cancer diagnosis, prognosis, and response to treatment.
Table 1.
AI tools for ovarian cancer diagnosis, prognosis, and response to treatment.
| Patients/ Original Images (n) | Original Image Type | Image for AI Training Size (Pixels) | Features to Be Assessed/Final Model | AI Tool Input | AI Tool Output | Internal Results Metrics | Internal Results | |
|---|---|---|---|---|---|---|---|---|
| Histotyping | ||||||||
| BenTaieb et al., 2016 [34] | 80/80 | WSI | 500 × 500 | Color, texture, cellular morphology, cytology/SVM | WSI | 5 classes * | Accuracy | 95% |
| BenTaieb et al., 2017 [35] | 133/133 | WSI | 500 × 500 | CNN features novel K-means/SVM | WSI | 5 classes * | Accuracy | 90% |
| Levine et al., 2020 [36] | 406/406 | WSI | 256 × 256 | CNN VGG19 | patch | 5 classes * | Accuracy | 70.87% |
| AUC | 0.92 | |||||||
| Kasture et al., 2020 [37] | ≤500/500 | patch | N/D | CNN novel KK Net | patch | 5 classes * | Accuracy | 91% |
| AUC | 0.95 | |||||||
| Boschman et al., 2022 [38] | 160/308 | WSI | 256 × 256 | CNN ResNet 18 | WSI | 5 classes * | AUC | 0.97 |
| Farahani et al., 2022 [39] | 485/948 | WSI | 512 × 512 | CNN VGG19 | WSI | 5 classes * | AUC | 0.95 |
| Idlahcen et al., 2025 [40] | 500/500 | WSI | 224 × 224 | Autoencoder + CNN (DenseNet-201) | patch | 5 classes * | Accuracy | 94.88% |
| Staging and grading | ||||||||
| Yu et al., 2020 [43] | 80/80 | WSI | N/D | CNN VGG16 | WSI | 2 grades (low/moderate and high) | AUC | 0.812 |
| Ghoniem et al., 2021 [44] | 160/308 | WSI | 256 × 256 | CNN altered VGG16 | WSI | 5 FIGO stages (I–IV) and N/D | Accuracy | 98.87% |
| Prognosis | ||||||||
| Poruthoor et al., 2013 [50] | 382/≤382 | WSI | 512 × 512 | CNN features/novel SVM | WSI | 2 classes of survival rate (<5 years/ ≥5 years | Accuracy | 55% |
| Yang et al., 2024 [51] | 874/1826 | WSI | 224 × 224 | Transformer network/graph deep-learning analysis | WSI | 2 classes of survival rate (high and low OCDPI) | Comparison of survival rate | <0.001 |
| BRCA1/2 mutation status | ||||||||
| Zeng et al., 2021 [17] | 229/≥229 | WSI | 256 × 256 | CNN features VGG19/ random forest | WSI | 2 classes of BRCA mutations (BRCAmut and BRCAwt) | AUC | 0.912 |
| Nero et al., 2022 [59] | 664/664 | N/D | 256 × 256 | CNN Features ResNet50/ CNN (CLAM) | WSI | 2 classes of BRCA mutations (BRCAmut and BRCAwt) | AUC | 0.59 |
| Ho et al., 2023 [60] | 609/609 | WSI | 224 × 224 | CNN features novel KK Net/CNNResNet 182 | WSI | 2 classes of BRCA mutations (BRCAmut and BRCAwt) | AUC | 0.43 |
| Borgade et al., 2023 [18] | 867/867 | WSI | 512 × 512 | PyTorch 3.7, Deepflash2 U-Net, DeepLabv3, UNet++, LinkNet, ResNet, Inception, EfficientNet, ResNeSt | WSI | 2 classes of BRCA mutations (BRCAmut and BRCAwt) | AUC | 0.681 |
| MMR mutation status | ||||||||
| Zeng et al., 2021 [17] | 229/≥229 | WSI | 1000 × 1000 | texture, cellular, and nuclear morphology/random forest | WSI | 3 classes of MMR status (MSI high/ MSI stable/N/A | AUC dMMR | 0.919 |
| AUC pMMR | 0.924 | |||||||
| HRD status | ||||||||
| Loeffler et al., 2023 [64] | 520/520 | WSI | 224 × 224 | ResNet50 (pretrained) + attMIL | WSI | 2 classes: HRD-high/low and HRD prediction score | AUROC | 0.61 |
| Frenel et al., 2024 [65] | 244/≥244 | WSI | N/D | Fusion-like DNN | WSI | 2 classes: HRD+/− and HRD prediction score | AUC (internal) | 0.74 |
| AUC (external) | 0.67 | |||||||
| Marmé et al., 2025 [66] | 669/675 | WSI | 224 × 224 | ResNet18&Transformer | WSI | 2 classes HRD status positive/negative and HRD prediction score | AUROC (internal) | 72% |
| AUROC (external) | 57% | |||||||
| Bergstrom et al., 2024 [67] | 600/≥1356 | WSI | 256 × 256 | Multiresolution MIL-ResNet18 | WSI | classes: HRD+/− and HRD prediction score | AUC (internal) | 0.81 |
| AUC (external) | N/D | |||||||
| Zhang et al., 2025 [68] | 205/205 | WSI | 512 × 512 | UNet++ and Hover-Net | patch | HRD status (deficient/proficient) at WSI level | AUC | 0.769 |
| F1-score | 0.762 | |||||||
| Chemotherapy response 2prediction | ||||||||
| Wang et al., 2023 [65] | <180/180 | WSI | 512 × 512 | CNN features novel/SVM | TMA | 2 classes of CT efficacy (effective/ invalid) | Accuracy | 90% |
| Wang et al., 2022 [69] | 78/288 | WSI | 256 × 256 | CNN (Inception V3) | WSI | 2 classes of CT efficacy (effective/ invalid) | AUC | 0.99 |
| Yu et al., 2020 [43] | 570 ≤ 1358 | WSI | N/D | CNN features VGG16 | WSI | 2 classes of relapse (early/late relapse) | AUC | 0.95 |
| Accuracy | 91% | |||||||
| Gilley et al., 2024 [71] | 78/288 | WSI | 1000 × 1000 | SVM | patch | 2 classes of relapse (responders/nonresponders) | AUC (linear SVM) | 0.83 |
| AUC (Gaussian SVM | 0.82 | |||||||
* 5 classes include HGSC, LGSC, and endometrioid, mucinous, and clear cell cancer. WSI—whole-slide image; CNN—convolution neural network; SVM—support vector machine; AUC—area under the curve; N/D—no data; OCDPI—Ovarian Cancer Digital Pathology Index; CT—chemotherapy; MMR—mismatch repair; MSI—microsatellite instability; attMIL—Attention-based Multiple Instance Learning.

Table 2.
AI tools for endometrial cancer histotyping, grading, and molecular subtyping.
Table 2.
AI tools for endometrial cancer histotyping, grading, and molecular subtyping.
| Patients/ Original Images (n) | Original Image Type | Image for AI Training Size (Pixels) | Features to Be Assessed/ Final Model | AI Tool Input | AI Tool Output | Internal Results Metrics | Internal Results | |
|---|---|---|---|---|---|---|---|---|
| Histotyping | ||||||||
| Hong et al., 2021 [73] | 456/ 20,000 | WSI | 299 × 299 | Inception Resnet-based/CNN | WSI, patch | 2 histotypes (endometrioid/ serous) | AUROC patient | 0.969 |
| AUROC patch | 0.870 | |||||||
| Song et al., 2022 [74] | 109 | WSI | 360 × 360 | Inception-v3 | WSI | 2 histotypes (endometrioid/ serous) | AUROC | 0.944 |
| Grading | ||||||||
| Goyal et al., 2024 [75] | 929/ N/D | WSI | N/D | EndoNet | WSI | 2 grades: low/high | F1-score | 0.91 |
| AUC | 0.9 | |||||||
| Immunohistochemical markers automatic assessment | ||||||||
| Kildal et al., 2024 [76] | 1228/ 2456 | WSI | 800 × 800 | YOLOv5 for nuclear model | patch | 2 classes of nuclei as (positive/negative); fraction of positive tumor cells | CCR (PMS2) | 95.3% |
| CCR (MSH6) | 90.0% | |||||||
| CCR (MSI, combined PMS2 and MSH6) | 90.7% | |||||||
| Ji et al., 2024 [77] | 57/114 | WSI | 256 × 256 | U-Net and DenseNet-121 | patch | Digitally generated H-DAB IHC-stained images | Rintraslide validation: | 0.98 |
| Rcross-case validation | 0.66 | |||||||
| Molecular subtypes | ||||||||
| Hong, 2021 [73] | 456/ 20,000 | WSI * | 299 × 299 | Inception Resnet-based CNN | WSI, patch | 4 molecular subtypes | AUROC patient CNV-L | 0.889 |
| AUROC patch CNV-L | 0.710 | |||||||
| AUROC patient CNV-H | 0.873 | |||||||
| AUROC patch CNV-H | 0.713 | |||||||
| AUROC patient MSI-H | 0.827 | |||||||
| AUROC patch MSI-H | 0.638 | |||||||
| Fremond, 2023 [78] | 2028/ 1,170,931 | WSI | 224 × 224 | ResNet 50, MoCo-v2 | WSI | POLE | AUROC | 0.849 |
| dMMR | AUROC | 0.844 | ||||||
| NSMP | AUROC | 0.883 | ||||||
| p53 abn | AUROC | 0.928 | ||||||
| Goyal, 2024 [79] | 2072/ 3,702,447 | WSI | 224 × 224 | HECTOR | WSI | 2: low/high grade | F1-score | 0.91 |
| AUC | 0.95 | |||||||
| MMR status | ||||||||
| Zhang, 2018 [75] | N/A | N/A | 1000 × 1000 | Inception-V3 | WSI | 2 (MSI, MSS) | Accuracy | 84.2% |
| Kather, 2019 [80] | 81/94 | WSI | N/D | ResNet18 | Patch | 2 (MSI, MSS) | AUC | 0.75 |
| Wang, 2020 [81] | N/A | N/A | 512 × 512 | ResNet18 | WSI | 2 (MSI, MSS) | AUC | 0.73 |
| Zhang, 2023 [75] | 95/ 22,044 | WSI | 256 × 256 | ResNet34 VGG16 | Patch | 2 (MSI, MSS) | AUC | 0799 |
| F1-score | 0786 | |||||||
| Wang, 2024 [82] | 344/ N/A | WSI | 512× 512 | Inception-V3 | WSI | 2 (MSI, MSS) | AUC | 87% |
| F1-score | 84% | |||||||
| Arslan, 2024 [61] | 61/ 12,093 (totally) | WSI | 256 × 256 | ResNet34 | WSI | 2 (MSI, MSS) | AUC | 0.771 |
| Whangbo et al., 2024 [83] | 325/1168 | WSI | N/D | EfficientNetB2 | patch | 2 (MSI, MSS) on WSI level | AUC | 0.821 |
| Accuracy | 0.778 | |||||||
| Umemoto et al., 2024 [84] | 114 | WSI | 512 × 512 | ResNet50 | patch | 2 (MSI, MSS) on WSI level | AUC | 0.91 |
| Accuracy | 0.80 | |||||||
| Liu et al., 2025 [85] | 1027/1678 | WSI | 224 × 224 | ResNet18 and EfficientNet | patch | 2 (MSI, MSS) on WSI level via patch-level probability averaging. | AUC (internal) | 0.897 |
| AUC (external) | 0.790–0.863 | |||||||
* H/E and IHC WSI were used; N/A—not available; H/E—hematoxylin and eosin; WSI—whole-slide image; CNN—convolution neural network; POLE—Polymerase ɛ; dMMR—Deficient DNA Mismatch Repair; NSMP—no specific molecular profile; AUROC—area under the receiver operating characteristic; AUC—area under the curve; CCR—Correct Classification Rate.
5. Fallopian Tube Cancer
The most common fallopian tube malignancy is high-grade serous carcinoma; however, tumors confined to the fallopian tube are rare, and tubal HGSC is predominantly diagnosed at an advanced stage after spreading to the ovaries and peritoneum [86,87]. Consequently, some authors recommend the term “pelvic HGSC” because it is often challenging to determine the original source of such cancer [86,88]. A putative precursor of tubal HGSC, known as serous tubal intraepithelial carcinoma (STIC), has been described and is located in the endosalpinx of the fallopian tube, predominantly in the fimbrial part [89]. Detecting STIC or early-stage tumors in fallopian tubes removed by prophylactic or opportunistic salpingectomy can prevent the development of HGSC [90,91]. A two-step procedure, involving morphological and IHC determination with an assessment of p53 and Ki-67 expression, has been proposed for accurate and reproducible STIC diagnosis [92]. Bogaerts et al. explored this procedure using a neural-network-based model with U-net and ResNet50. The final algorithm was developed for the WSI-based verification of two classes (serous tubal intraepithelial lesion (STIL) and STIC), but the F1-score was rather low (0.35) [93]. This is likely only a pilot model, but the unsatisfactory result could be partly explained by the very small size of the lesions (<30 cells) and the difficulties in slide mapping.
6. Uterine Cancer
6.1. Endometrial Cancer
Endometrial cancer has undergone changes in its WHO classification [7], molecular subtyping, and risk stratification [3]. A new International Federation of Gynecology and Obstetrics (FIGO) staging system (2023) has been published [94], and new treatment strategies have been proposed, including the prominent role of immunotherapy [95], providing several opportunities for the application of AI tools [96]. Currently, most AI tools focus on detecting only two histotypes (endometrioid and serous), while other histotypes (such as clear cell cancer or carcinosarcoma) are overlooked [73,74]. Additionally, there is a lack of AI tools for detecting the polymerase epsilon (POLE)-mutant molecular subtype, even though these patients could benefit from de-escalated therapy due to the extremely benign nature of this subtype [73,78].
AI tools for histotyping, grading, and staging endometrial cancer are summarized in Table 2. All these algorithms have demonstrated a high predictive value, although the histotyping models only considered the two most common cancer types (endometrioid and serous), which may lead to the misinterpretation of rarer tumors (e.g., clear cell cancer, carcinosarcomas, and mesonephric cancer). There is limited research on predictive algorithms for grading, but Volinsky-Fremond et al. included this evaluation in their enriched cohort, which was also used for predicting the MSI status, showing a cumulative AUC of 0.844 with the Histopathology-based Endometrial Cancer Tailored Outcome Risk (HECTOR) model [97]. Some models have been developed to evaluate the endometrial immunophenotype, as this is an important part for diagnosis and prognosis.
These algorithms were constructed either as a digital IHC algorithm (for digital Ki-67 expression, as demonstrated by Ji et al. [77]) or for the automatic assessment of physically stained slides (for PMS2/MSH6 expression evaluation, as described by Kildal et al. [76]). Both algorithms demonstrated a high internal accuracy (>90%). Nevertheless, the digital IHC algorithm still requires improvement for cross-case validation, as the proposed model showed a Pearson correlation of only 0.66 between digital and physical Ki-67 labelling indices.
Since molecular subtypes were proposed in 2013 [98] and surrogate markers have been developed to distinguish between these subtypes in pathological practice [99], several AI tools have been proposed to evaluate them. Among the molecular subtyping models, the algorithm by Fremond et al. [78] is the most impressive, with a high accuracy for all subtypes, developed using data from more than 2000 patients and over 1 million images. Other models for predicting the MSI status showed reasonably good, but not excellent, accuracy/AUC, though none examined such a large number of patients [61,75,80,81,83,84,100]. The AI tool developed by Fremond et al. could benefit from further training with an independent cohort to achieve high-level performance for clinical use. In pan-cancer studies, endometrial cancers have also been included in prediction models. For instance, Arslan et al. reported an AUC of 0.771 for endometrial cancers, higher than the average AUC of 0.653 [61]. Another pan-cancer study including endometrial cancer cohorts did not analyze the prediction of the MSI status for these tumors, focusing instead on the prediction of PTEN, TP53, and APC mutations dependent of the MSI status [62]. The most recent investigation by Liu et al. [85] demonstrated impactful results, analyzing more than 1000 patients and achieving an AUC of approximately 0.9. In addition, their algorithm showed a high AUC for external cohorts (0.790–0.863), which have not been reported previously.
6.2. Uterine Mesenchymal Tumors
6.2.1. Uterine Smooth Muscle Tumors
The morphological criteria—nuclear atypia, necrosis, and mitotic figures—remain the most important features for diagnosing uterine smooth muscle tumors [7,101,102]. Although the classification of atypia is not easily addressed by AI tools due to challenges in establishing the ground truth, the AI-based evaluation of mitotic activity has been widely explored in other tumor models [103,104,105]. In uterine mesenchymal tumors, simple machine-learning-based AI tools have been developed to assess the mitotic index and generally achieve a high predictive value (over 90%).
It has also been demonstrated that uterine leiomyosarcoma can be subdivided into molecular subtypes with different prognoses and potentially different therapeutic targets [106,107,108]. However, we could not find AI tools specifically designed to detect such molecular subtypes.
6.2.2. Uterine Stromal Sarcoma
Some AI tools have been proposed to distinguish uterine stromal sarcomas from other tumors and tumor-like conditions. Yang et al. reported a comparison of different models to differentiate low-grade stromal sarcomas (LGSSs) from leiomyomas [108]. The authors concluded that the neural network PCA (Principal Component Analysis) with an SVM is the most effective of the basic techniques for training their model, achieving a final test accuracy of 0.8535 for LGSS. These technical comparisons are useful for designing the experiments; nevertheless, the overall accuracy of AI tools depends not only on the neural network design but also on the characteristics of the training sets. In the study by Yang et al., there was no information on the size or origin of the cohort, so further research is needed to substantiate this result.
According to the WHO 2020 classification [7], endometrial stromal sarcoma includes many entities with different molecular subtypes that cannot be distinguished based on morphological features alone. This is an area that could benefit from a decision support AI tool.
6.3. Trophoblastic Tumors
Trophoblastic tumors are among the most difficult categories to diagnose, as there are few reliable morphological and IHC features to support the differential diagnosis. The fragility of the morphological criteria makes this cancer type less attractive for developing AI tools, since the ground truth is difficult to establish. One of the differential diagnostic challenges in trophoblastic tumors is the correct labelling of hydatidiform moles, as this can significantly affect the follow-up strategy. We found only one AI tool for the diagnosis of trophoblastic tumors. Palee et al. proposed a model for the differential diagnosis of complete and partial hydatidiform moles [109], based on a neural network classifier and machine-learning-based classifiers, with an accuracy of 81.2%, trained on more than 900 images captured from WSI. The accuracy achieved by the Palee et al. model is quite good; however, the differential diagnosis between complete and partial hydatidiform moles is not a significant problem for pathologists, as the p57 IHC expression is an accurate marker that is differentially expressed in these two conditions [110]. Currently, the most important problem in the pathological evaluation of villi is the differentiation between the hydropic abortus (non-hydatidiform mole) and partial hydatidiform mole, as this is not possible without a genetic analysis of short tandem repeats (STRs) from the paraffin-embedded material [111,112]. Although this method has been developed in detail and has been proven to be excellent, many pathology and genetics departments are unable to perform this analysis regularly. An initial screening with an AI tool would provide an effective triage approach for the verification of partial hydatidiform moles.
7. Lower Genital Tract
7.1. Cervical Cancer
There are not many AI tools for histological cervical cancer diagnosis, partly because the incidence of cervical cancer has decreased dramatically in recent decades due to the introduction of screening and vaccination programs [1,4]. Screening options expanded by digital cytology are now enhanced by dedicated AI tools [30,113,114,115].
The main drawback of AI tools for the histopathological diagnosis of cervical cancer is the patch-based analysis used in some models, which prevents the adoption into routine practice and requires further improvement. Nevertheless, rather accurate AI tools were proposed by Habtemariam et al. [114] and Li et al. [116]. The AI tool developed by Habtemariam et al. was trained on 915 patients (WSI) and demonstrated a 94.5% accuracy for four classes (normal, precancerous (squamous intraepithelial lesions), adenocarcinoma, and squamous cell carcinoma (SCC)) [114]. Li et al. developed an AI tool for distinguishing between only two classes (adenocarcinoma and SCC) with an AUC of 0.966 [116]. Since the 2020 WHO classification focuses on the distinction between HPV-associated and HPV-independent cervical cancers [7], the HPV status should instead be the fundamental feature to target in future AI training. The same applies to the key role of p16, which is a single marker recommended by the Lower Anogenital Squamous Terminology Project for a high-grade squamous lesion diagnosis to improve the sensitivity and specificity of the histopathological diagnosis [117]. An AI-based quantification and spatial assessment of p16 could be very helpful for pathologists, as it is sometimes difficult to distinguish between patchy and block-type p16 staining. An et al. demonstrated an AI tool that can accurately identify HSIL among p16-positive areas with a high accuracy, sensitivity, and specificity (0.845, 0.922, and 0.829, respectively) [118].
7.2. Vulvar and Vaginal Cancers
Precancerous lesions of the vulva and vagina are even more difficult to diagnose than cervical lesions, as there is still no uniform classification for HPV-negative p53 wild-type lesions [3]. These lesions have a very similar phenotype, and IHC markers such as p16, p53, and CK17 cannot help to differentiate them [119,120]. HPV-dependent lesions (vulvar intraepithelial neoplasia of the usual type, uVIN) and HPV-independent p53 mutant lesions (differentiated VIN, dVIN) cannot be differentiated in more than 20% of cases based on morphological features alone [121,122]. Currently, the already developed AI tools can only contribute to the distinction between VINI/II/III. Choschzick et al. presented an AI tool for Ki-67 expression assessment in precancerous vulvar lesions. However, the reproducibility between pathologists and the AI tool was rather low, a result that could be partially explained by the mild atypia and marked keratinization in VINs, which is difficult to assess [123].
In summarizing the presented algorithms across different tumor localizations, the key strengths and limitations of this review must be highlighted. This review provides a comprehensive and critical synthesis of a substantial body of literature (125 references) on AI in gynecological pathology. It maintains a clear, clinically oriented focus on the diagnostic, prognostic, and predictive applications relevant to pathologists. The work not only describes existing tools but also critically evaluates their methodologies, identifying common pitfalls like the lack of external validation. It concludes with practical recommendations for future standardization and the safe integration of AI into clinical workflows. Nevertheless, the scope is intentionally limited to histopathology, excluding the vast field of digital cytology. The direct comparison of AI tools is challenging due to the evolving tumor classifications over the past decade, rendering some models outdated. The analysis focuses on common molecular classes, omitting emerging aspects like epigenetics due to a lack of dedicated algorithms. Furthermore, the review does not address the critical impact of pre-analytical variables on AI classifier robustness.
8. Conclusions
In general, AI-based-tools for assessing gynecologic cancers should be considered a promising area of computational pathology, already targeting clinically relevant diagnostic, prognostic, and predictive goals. In this review, many accurate models with potential application in clinical routine are identified as supportive tools that remain largely dependent on the initial diagnostic framing by the pathologist (narrow AI). Some studies lack information on their training sets, such as the diseases included, the number of patients, the number of samples or images used, whether patients or images were excluded, and the methods by which tissue findings were reported. Many studies do not report the external validation results and lack cross-validation. In addition, some studies used inappropriate datasets, including frozen tissue sections, datasets without sufficient classes or histotypes, or unbalanced frequencies of cancers that do not reflect reality, and/or do not include challenging or rare cancer types. These observations highlight the clear need for a standardized algorithm specification for each AI tool.
The works included in this review reported different metrics to evaluate the effectiveness of the proposed AI tools (accuracy, AUC, F1-score, kappa, etc.), which makes it difficult to compare the models. Establishing standardized metrics would make the evaluation and comparison clearer for users. Some researchers also chose technology that is now outdated for the development of their models, so their results could be partially improved by using updated technology. However, the most important problem in computational gynecologic pathology is the lack of investment by researchers in issues that are essential for pathologists in routine practice. Consequently, AI tools for clinically relevant subtyping (e.g., the molecular subtyping of LGSC, leiomyosarcomas, and high-grade stromal sarcomas of the uterus; hydatidiform moles and vulvar precancerous lesions) represent areas where accurate, explainable, and generalizable AI tools would have a positive impact on clinical practice for the benefit of patients. At the same time, the already developed algorithms that have demonstrated convincing results in tasks such as the classification of histological subtypes, tumor grading, prognosis determination, and potential sensitivity to chemotherapy and targeted therapy must be validated and tested on larger patient cohorts using scanners from different manufacturers and slides prepared in various laboratories in order to be fully standardized before being implemented into clinical practice. Finally, future research should not only examine patients’ psychological responses and clinicians’ readiness to use AI but also adopt structured evaluation frameworks to ensure AI systems are integrated into oncology workflows safely, transparently, and equitably. As proposed by Erul et al. [124], implementing a quality assurance framework with standardized reporting, explainability mechanisms, and external validation can enhance trust among both patients and physicians. Such frameworks are essential in low-resource settings, where deploying unvalidated or non-transparent AI tools could exacerbate existing healthcare disparities.
Author Contributions
Conceptualization, A.A. and C.E.; formal analysis, A.A.; data curation, A.A. and E.K.; writing—original draft preparation, A.A.; writing—review and editing, C.E., A.P., J.P., E.K. and X.M.-G.; supervision, C.E. and X.M.-G.; funding acquisition, E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant provided by the Ministry of Economic Development of the Russian Federation in accordance with the subsidy agreement (agreement identifier 000000C313925P4G0002) and the agreement with the Ivannikov Institute for System Programming of the Russian Academy of Sciences, dated 20 June 2025, No. 139-15-2025-011.
Informed Consent Statement
Compliance with Ethical Standards: Formal ethical approval was not required for this study, as it did not involve any patient data collection or impact on patient care.
Data Availability Statement
No data was generated.
Conflicts of Interest
C.E. consulted in the last two years for MSD, Mindpeak, Sakura, Diapath, Diaceutics, and Leica. A.P. consulted in the last two years for Indica Labs. The other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Abbreviations
The following abbreviations are used in this manuscript:
| AI | Artificial intelligence |
| AUC | Area under the curve |
| CNN | Convolutional neural network |
| CTransPat | Transformer-based unsupervised contrastive learning for histopathological image classification |
| dMMR | MMR deficient mismatch repair |
| dVIN | Differentiated VIN |
| FDA | Food and Drug Administration |
| FIGO | The International Federation of Gynecology and Obstetrics |
| H&E | Hematoxylin and eosin |
| HECTOR | Histopathology-based Endometrial Cancer Tailored Outcome Risk |
| HGSC | High-grade serous cancer |
| HPV | Human papillomavirus |
| IHC | Immunohistochemical |
| LGSC | Low-grade serous cancer |
| LGSS | Low-grade stromal sarcoma |
| MDT | Multidisciplinary Team |
| MEK | Mitogen-activated protein kinase |
| ML | Machine learning |
| PAIP | Pathology Artificial Intelligence Platform |
| PARP | Poly(ADP-ribose) polymerase |
| PCA | Principal component analysis |
| POLE | Polymerase epsilon |
| RetCCL | Retrieval with Clustering-guided Contrastive Learning |
| SCC | Squamous cell carcinoma |
| STIC | Inhibitors serous tubal intraepithelial carcinoma |
| STIL | Serous tubal intraepithelial lesion |
| STRs | Short tandem repeats |
| SVM | Support vector machine |
| TCGA | The Cancer Genome Atlas |
| uVIN | Vulvar intraepithelial neoplasia of usual type |
| WHO | World Health Organization |
| WSI | Whole-slide imaging |
References
- Marth, C.; Landoni, F.; Mahner, S.; McCormack, M.; Gonzalez-Martin, A.; Colombo, N.; ESMO Guidelines Committee. Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2017, 28, iv72–iv83. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.A.; Matias-Guiu, X.; Amant, F.; Concin, N.; Davidson, B.; Fotopoulou, C.; González-Martin, A.; Gourley, C.; Leary, A.; Lorusso, D.; et al. ESGO-ESMO-ESP consensus conference recommendations on ovarian cancer: Pathology and molecular biology and early, advanced and recurrent disease. Ann. Oncol. 2024, 35, 248–266. [Google Scholar] [CrossRef]
- Oaknin, A.; Bosse, T.J.; Creutzberg, C.L.; Giornelli, G.; Harter, P.; Joly, F.; Lorusso, D.; Marth, C.; Makker, V.; Mirza, M.R.; et al. Endometrial cancer: ESMO Clinical Practice Guideline for diagnosis, treatment and follow-up. Ann. Oncol. 2022, 33, 860–877. [Google Scholar] [CrossRef] [PubMed]
- Fokom-Defo, V.; Dille, I.; Fokom-Domgue, J. Single Dose HPV Vaccine in Achieving Global Cervical Cancer Elimination. Lancet Glob. Health 2024, 12, e360–e361. [Google Scholar] [CrossRef]
- Silva, E. Cancer Statistics. Mixed Results in Gynecologic Oncology. Int. J. Gynecol. Cancer 2024, 34, 964. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Vang, R.; Davidson, B.; Kong, C.S.; Longacre, T.A.; Malpica, A. WHO Classification of Female Genital Tumors; International Agency for Research on Cancer: Lyon, France, 2021; pp. 43–44. [Google Scholar]
- Kiehl, T.R. Digital and Computational Pathology: A Specialty Reimagined. In The Future Circle of Healthcare. Future of Business and Finance; Ehsani, S., Glauner, P., Plugmann, P., Thieringer, F.M., Eds.; Springer: Cham, Switzerland, 2021; pp. 227–250. [Google Scholar] [CrossRef]
- Kaplan, K.J.; Rao, L.K.F. Digital Pathology: Historical Perspectives, Current Concepts and Future Applications; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 1–116. [Google Scholar] [CrossRef]
- Parwani, A.V. Whole Slide Imaging: Current Applications and Future Directions; Springer International Publishing: Berlin/Heidelberg, Germany, 2021; pp. 1–242. [Google Scholar]
- Eloy, C.; Marques, A.; Pinto, J.; Pinheiro, J.; Campelos, S.; Curado, M.; Vale, J.; Polónia, A. Artificial intelligence–assisted cancer diagnosis improves the efficiency of pathologists in prostatic biopsies. Virchows Arch. 2023, 82, 595–604. [Google Scholar] [CrossRef]
- Viswanathan, V.S.; Toro, P.; Corredor, G.; Mukhopadhyay, S.; Madabhushi, A. The state of the art for artificial intelligence in lung digital pathology. J. Pathol. 2022, 257, 413–429. [Google Scholar] [CrossRef]
- Zhu, J.; Liu, M.; Li, X. Progress on deep learning in digital pathology of breast cancer: A narrative review. Gland. Surg. 2022, 11, 751–766. [Google Scholar] [CrossRef]
- Griem, J.; Eich, M.L.; Schallenberg, S.; Pryalukhin, A.; Bychkov, A.; Fukuoka, J.; Zayats, V.; Hulla, W.; Munkhdelger, J.; Seper, A.; et al. Artificial Intelligence-Based Tool for Tumor Detection and Quantitative Tissue Analysis in Colorectal Specimens. Artificial Intelligence-Based Tool for Tumor Detection and Quantitative Tissue Analysis in Colorectal Specimens. Mod. Pathol. 2023, 36, 100327. [Google Scholar] [CrossRef]
- Chen, G.M.; Kannan, L.; Geistlinger, L.; Kofia, V.; Safikhani, Z.; Gendoo, D.M.A.; Parmigiani, G.; Birrer, M.; Haibe-Kains, B.; Waldron, L. Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma. Clin. Cancer Res. 2018, 24, 5037–5047. [Google Scholar] [CrossRef]
- Ahn, B.; Moon, D.; Kim, H.S.; Lee, C.; Cho, N.H.; Choi, H.K.; Kim, D.; Lee, J.Y.; Nam, E.J.; Won, D.; et al. Histopathologic image-based deep learning classifier for predicting platinum-based treatment responses in high-grade serous ovarian cancer. Nat. Commun. 2024, 15, 4253. [Google Scholar] [CrossRef]
- Zeng, H.; Chen, L.; Zhang, M.; Luo, Y.; Ma, X. Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol. Oncol. 2021, 163, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Bourgade, R.; Rabilloud, N.; Perennec, T.; Pécot, T.; Garrec, C.; Guédon, A.F.; Delnatte, C.; Bézieau, S.; Lespagnol, A.; de Tayrac, M.; et al. Deep Learning for Detecting BRCA Mutations in High-Grade Ovarian Cancer Based on an Innovative Tumor Segmentation Method From Whole Slide Images. Mod. Pathol. 2023, 36, 100304. [Google Scholar] [CrossRef] [PubMed]
- Fraune, C.; Rosebrock, J.; Simon, R.; Hube-Magg, C.; Makrypidi-Fraune, G.; Kluth, M.; Büscheck, F.; Höflmayer, D.; Schmalfeldt, B.; Müller, V.; et al. High homogeneity of MMR deficiency in ovarian cancer. Gynecol. Oncol. 2020, 156, 669–675. [Google Scholar] [CrossRef]
- Jain, E.; Patel, A.; Parwani, A.V.; Shafi, S.; Brar, Z.; Sharma, S.; Mohanty, S.K. Whole Slide Imaging Technology and Its Applications: Current and Emerging Perspectives. Int. J. Surg. Pathol. 2024, 32, 433–448. [Google Scholar] [CrossRef] [PubMed]
- Mastrosimini, M.G.; Eccher, A.; Nottegar, A.; Montin, U.; Scarpa, A.; Pantanowitz, L.; Girolami, I. WSI validation studies in breast and gynecological pathology. Pathol. Res. Pract. 2022, 240, 154191. [Google Scholar] [CrossRef]
- Prewitt, J.M.; Mendelsohn, M.L. The analysis of cell images. Ann. N. Y. Acad. Sci. 1966, 128, 1035–1053. [Google Scholar] [CrossRef] [PubMed]
- Snead, D.R.J.; Tsang, Y.-W.; Meskiri, A.; Kimani, P.; Crossman, R.J.; Rajpoot, N.M.; Blessing, E.; Chen, K.; Gopalakrishnan, K.; Matthews, P.; et al. Validation of digital pathology imaging for primary histopathological diagnosis. Histopathology 2016, 68, 1063–1072. [Google Scholar] [CrossRef] [PubMed]
- Aloqaily, A.; Polonia, A.; Campelos, S.; Alrefae, N.; Vale, J.; Caramelo, A.; Eloy, C. Digital Versus Optical Diagnosis of Follicular Patterned Thyroid Lesions. Head. Neck Pathol. 2021, 15, 537–543. [Google Scholar] [CrossRef]
- Marra, A.; Morganti, S.; Pareja, F.; Campanella, G.; Bibeau, F.; Fuchs, T.; Loda, M.; Parwani, A.; Scarpa, A.; Reis-Filho, J.S.; et al. Artificial intelligence entering the pathology arena in oncology: Current applications and future perspectives. Ann. Oncol. 2025, 36, 712–725. [Google Scholar] [CrossRef] [PubMed]
- Pell, R.; Oien, K.; Robinson, M.; Pitman, H.; Rajpoot, N.; Rittscher, J.; Snead, D.; Verrill, C.; UK National Cancer Research Institute (NCRI) Cellular-Molecular Pathology (CM-Path) Quality Assurance Working Group. The use of digital pathology and im-age analysis in clinical trials. J. Pathol. Clin. Res. 2019, 5, 81–90. [Google Scholar] [CrossRef]
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 2022, 35, 23–32. [Google Scholar] [CrossRef]
- Ersavas, T.; Smith, M.A.; Mattick, J.S. Novel applications of Convolutional Neural Networks in the age of Transformers. Sci. Rep. 2024, 14, 10000. [Google Scholar] [CrossRef]
- Shah, M.; Polónia, A.; Curado, M.; Vale, J.; Janowczyk, A.; Eloy, C. Impact of Tissue Thickness on Computational Quantification of Features in Whole Slide Images for Diagnostic Pathology. Endocr. Pathol. 2025, 36, 10, Erratum in Endocr. Pathol. 2025, 36, 26. https://doi.org/10.1007/s12022-025-09872-1. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Li, X.; Shen, H.; Chen, Y.; Wang, L.; Chen, H.; Feng, J.; Liu, J. A systematic review of deep learning-based cervical cytology screening: From cell identification to whole slide image analysis. Artif. Intell. Rev. 2023, 56, 2687–2758. [Google Scholar] [CrossRef]
- Kim, D.; Sundling, K.; Virk, R.; Thrall, M.; Alperstein, S.; Bui, M.; Chen-Yost, H.; Donnelly, A.; Lin, O.; Liu, X.; et al. Digital cytology part 2: Artificial intelligence in cytology: A concept paper with review and recommendations from the American Society of Cytopathology Digital Cytology Task Force. J. Am. Soc. Cytopathol. 2024, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Sundling, K.; Virk, R.; Thrall, M.; Alperstein, S.; Bui, M.; Chen-Yost, H.; Donnelly, A.; Lin, O.; Liu, X.; et al. Digital cytology part 1: Digital cytology implementation for practice: A concept paper with review and recommendations from the American Society of Cytopathology Digital Cytology Task Force. J. Am. Soc. Cytopathol. 2024, 13, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yu, Y.; Tan, Y.; Wan, H.; Zheng, N.; He, Z.; Mao, L.; Ren, W.; Chen, K.; Lin, Z.; et al. Artificial intelligence enables precision diagnosis of cervical cytology grades and cervical cancer. Nat. Commun. 2024, 15, 4369. [Google Scholar] [CrossRef] [PubMed]
- BenTaieb, A.; Nosrati, M.; Li-Chang, H.; Huntsman, D.; Hamarneh, G. Clinically-inspired automatic classification of ovarian carcinoma subtypes. J. Pathol. Inform. 2016, 7, 28. [Google Scholar] [CrossRef]
- BenTaieb, A.; Li-Chang, H.; Huntsman, D.; Hamarneh, G. A structured latent model for ovarian carcinoma subtyping from histopathology slides. Med. Image Anal. 2017, 39, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Peng, J.; Farnell, D.; Nursey, M.; Wang, Y.; Naso, J.; Ren, H.; Farahani, H.; Chen, C.; Chiu, D.; et al. Synthesis of diagnostic quality cancer pathology images by generative adversarial networks. J. Pathol. 2020, 252, 178–188. [Google Scholar] [CrossRef]
- Kasture, K.; Choudhari, D.; Matte, P. Prediction and Classification of Ovarian Cancer Using Enhanced Deep Convolutional Neural Network. IJETT 2022, 70, 310–318. [Google Scholar] [CrossRef]
- Boschman, J.; Farahani, H.; Darbandsari, A.; Ahmadvand, P.; Van Spankeren, A.; Farnell, D.; Levine, A.; Naso, J.; Churg, A.; Jones, S.; et al. The utility of color normalization for AI-based diagnosis of hematoxylin and eosin-stained pathology images. J. Pathol. 2022, 256, 15–24. [Google Scholar] [CrossRef]
- Farahani, H.; Boschman, J.; Farnell, D.; Darbandsari, A.; Zhang, A.; Ahmadvand, P.; Jones, S.; Huntsman, D.; Köbel, M.; Gilks, C.; et al. Deep learning-based histotype diagnosis of ovarian carcinoma whole-slide pathology images. Mod. Pathol. 2022, 35, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Idlahcen, F.; Mboukou, P.F.C.; Idri, A.; El Attar, H. PathoCoder: Rethinking the Flaws of Patch-Based Learning for Multi-Class Classification in Computational Pathology. Microsc. Res. Tech. 2025, 88, 1712–1726. [Google Scholar] [CrossRef] [PubMed]
- Genestie, C.; Auguste, A.; Al Battal, M.; Scoazec, J.Y.; Gouy, S.; Lacroix, L.; Morice, P.; Pautier, P.; Leary, A.; Devouassoux-Shisheboran, M. Histological classification of mucinous ovarian tumors: Inter-observer reproducibility, clinical relevance, and role of genetic biomarkers. Virchows Arch. 2021, 478, 885–891. [Google Scholar] [CrossRef]
- Handley, K.F.; Sims, T.T.; Bateman, N.W.; Glassman, D.; Foster, K.I.; Lee, S.; Yao, J.; Yao, H.; Fellman, B.M.; Liu, J.; et al. Classification of High-Grade Serous Ovarian Cancer Using Tumor Morphologic Characteristics. JAMA Netw. Open 2022, 5, e2236626. [Google Scholar] [CrossRef]
- Yu, K.; Hu, V.; Wang, F.; Matulonis, U.; Mutter, G.; Golden, J.; Kohane, I. Deciphering serous ovarian carcinoma histopathology and platinum response by convolutional neural networks. BMC Med. 2020, 18, 236. [Google Scholar] [CrossRef] [PubMed]
- Ghoniem, R.M.; Algarni, A.D.; Refky, B.; Ewees, A.A. Multi-Modal Evolutionary Deep Learning Model for Ovarian Cancer Diagnosis. Symmetry 2021, 13, 643. [Google Scholar] [CrossRef]
- Yu, K.; Hu, V.; Wang, F.; Matulonis, U.; Mutter, G.; Golden, J.; Kohane, I. Histological grading of ovarian mucinous carcinoma—An outcome-based analysis of traditional and novel systems. Histopathology 2020, 77, 26–34. [Google Scholar] [CrossRef]
- De Decker, K.; Wenzel, H.H.B.; Bart, J.; van der Aa, M.A.; Kruitwagen, R.F.P.M.; Nijman, H.W.; Kruse, A.J. Stage, treatment and survival of low-grade serous ovarian carcinoma in the Netherlands: A nationwide study. Acta Obstet. Gynecol. Scand. 2023, 102, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Shiradkar, R.; Liu, Z. Integrating pathomics with radiomics and genomics for cancer prognosis: A brief review. Chin. J. Cancer Res. 2021, 33, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Bülow, R.D.; Hölscher, D.L.; Costa, I.G.; Boor, P. Extending the landscape of omics technologies by pathomics. NPJ Syst. Biol. Appl. 2023, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Poruthoor, A.; Phan, J.H.; Kothari, S.; Wang, M.D. Exploration of Genomic, Proteomic, and Histopathological Image Data Integration Methods for Clinical Prediction. IEEE China Summit Int. Conf. Signal Inf. Process. 2013, 2013, 259–263. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, Y.; Zhuo, L.; Sun, K.; Meng, F.; Zhou, M.; Sun, J. Prediction of prognosis and treatment response in ovarian cancer patients from histopathology images using graph deep learning: A multicenter retrospective study. Eur. J. Cancer. 2024, 199, 113532. [Google Scholar] [CrossRef]
- Li, S.; Tao, L.; Dai, H.; Gong, X.; Zhuo, Y.; Xiang, H.; Zhao, Y.; Gao, Q.; Deng, L. BRCA1 Versus BRCA2 and PARP Inhibitors Efficacy in Solid Tumors:A Meta-Analysis of Randomized Controlled Trials. Front. Oncol. 2021, 11, 718871. [Google Scholar] [CrossRef] [PubMed]
- Bhamidipati, D.; Haro-Silerio, J.I.; Yap, T.A.; Ngoi, N. PARP inhibitors: Enhancing efficacy through rational combinations. Br. J. Cancer. 2023, 129, 904–916. [Google Scholar] [CrossRef]
- Romey, M.; Rodepeter, F.; Hattesohl, A.; Kaiser, K.; Teply-Szymanski, J.; Heitz, F.; Staebler, A.; Serra, V.; Grass, A.; Marmé, F.; et al. Systematic Analysis of Homologous Recombination Deficiency Testing in Ovarian Cancer-Development of Recommendations for Optimal Assay Performance. Mod. Pathol. 2024, 37, 100445. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Shao, D.; Cai, Y.; Bi, R.; Ju, X.; Chen, D.; Song, C.; Chen, X.; Li, J.; An, N.; et al. Homologous recombination deficiency status predicts response to platinum-based chemotherapy in Chinese patients with high-grade serous ovarian carcinoma. J. Ovarian Res. 2023, 16, 53. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Gu, Y.; Zhang, M.; Shi, X.; Li, Z.; Xu, X.; Sun, T.; Dong, Y.; Xue, C.; Zhu, X.; et al. HRD effects on first-line adjuvant chemotherapy and PARPi maintenance therapy in Chinese ovarian cancer patients. NPJ Precis. Oncol. 2023, 7, 51. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef] [PubMed]
- Soslow, R.A.; Han, G.; Park, K.J.; Garg, K.; Olvera, N.; Spriggs, D.R.; Kauff, N.D.; Levine, D.A. Morphologic patterns associated with BRCA1 and BRCA2 genotype in ovarian carcinoma. Mod. Pathol. 2012, 25, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Nero, C.; Boldrini, L.; Lenkowicz, J.; Giudice, M.T.; Piermattei, A.; Inzani, F.; Pasciuto, T.; Minucci, A.; Fagotti, A.; Zannoni, G.; et al. Learning to Predict BRCA Mutation and Survival from Digital H&E Slides of Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 11326. [Google Scholar] [CrossRef]
- Ho, D.J.; Chui, M.H.; Vanderbilt, C.M.; Jung, J.; Robson, M.E.; Park, C.S.; Roh, J.; Fuchs, T.J. Deep Interactive Learning-based ovarian cancer segmentation of H&E-stained whole slide images to study morphological patterns of BRCA mutation. J. Pathol. Inform. 2022, 14, 100160. [Google Scholar] [CrossRef] [PubMed]
- Arslan, S.; Schmidt, J.; Bass, C.; Mehrotra, D.; Geraldes, A.; Singhal, S.; Hense, J.; Li, X.; Raharja-Liu, P.; Maiques, O.; et al. A systematic pan-cancer study on deep learning-based prediction of multi-omic biomarkers from routine pathology images. Commun. Med. 2024, 4, 48. [Google Scholar] [CrossRef]
- Saldanha, O.L.; Loeffler, C.M.L.; Niehues, J.M.; van Treeck, M.; Seraphin, T.P.; Hewitt, K.J.; Cifci, D.; Veldhuizen, G.P.; Ramesh, S.; Pearson, A.T.; et al. Self-supervised attention-based deep learning for pan-cancer mutation prediction from histopathology. NPJ Precis. Oncol. 2023, 7, 35. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, E.Y.; Luchini, C.; Eccher, A.; Tizaoui, K.; Shin, J.I.; Lim, B.J. Artificial Intelligence for Predicting Microsatellite Instability Based on Tumor Histomorphology: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 2462. [Google Scholar] [CrossRef]
- Loeffler, C.M.L.; El Nahhas, O.S.M.; Muti, H.S.; Seibel, T.; Cifci, D.; van Treeck, M.; Gustav, M.; Carrero, Z.I.; Gaisa, N.T.; Lehmann, K.V.; et al. Direct prediction of Homologous Recombination Deficiency from routine histology in ten different tumor types with attention-based Multiple Instance Learning: A development and validation study. medRxiv, 2023; preprint. [Google Scholar] [CrossRef]
- Frenel, J.-S.; Bossard, C.; Rynkiewicz, J.; Thomas, F.; Salhi, Y.; Salhi, S.; Chetritt, J. Artificial intelligence to predict homologous recombination deficiency in ovarian cancer from whole-slide histopathological images. J. Clin. Oncol. 2024, 42, 5578. [Google Scholar] [CrossRef]
- Marmé, F.; Krieghoff-Henning, E.I.; Kiehl, L.; Wies, C.; Hauke, J.; Hahnen, E.; Harter, P.; Schouten, P.C.; Brodkorb, T.; Kayali, M.; et al. Predicting benefit from PARP inhibitors using deep learning on H&E-stained ovarian cancer slides. Eur. J. Cancer. 2025, 216, 115199. [Google Scholar] [CrossRef] [PubMed]
- Bergstrom, E.N.; Abbasi, A.; Díaz-Gay, M.; Galland, L.; Ladoire, S.; Lippman, S.M.; Alexandrov, L.B. Deep Learning Artificial Intelligence Predicts Homologous Recombination Deficiency and Platinum Response from Histologic Slides. J. Clin. Oncol. 2024, 42, 3550–3560. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Qiu, Y.; Feng, S.; Yin, H.; Liu, Q.; Zhu, Y.; Cui, H.; Wei, X.; Wang, G.; Wang, X.; et al. Development of model for identifying homologous recombination deficiency (HRD) status of ovarian cancer with deep learning on whole slide images. J. Transl. Med. 2025, 23, 267. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lee, Y.; Lin, Y.; Chang, C.; Sai, A.; Wang, C. Interpretable attention-based deep learning ensemble for personalized ovarian cancer treatment without manual annotations. Comput. Med. Imaging Graph. 2023, 107, 102233. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lee, Y.; Chang, C.; Lin, Y.; Liou, Y.; Hsu, P.; Chang, C.; Sai, A.; Wang, C.; Chao, T. A Weakly Supervised Deep Learning Method for Guiding Ovarian Cancer Treatment and Identifying an Effective Biomarker. Cancers 2022, 14, 1651. [Google Scholar] [CrossRef] [PubMed]
- Gilley, P.; Zhang, K.; Abdoli, N.; Sadri, Y.; Adhikari, L.; Fung, K.; Qiu, Y. Utilizing a Pathomics Biomarker to Predict the Effectiveness of Bevacizumab in Ovarian Cancer Treatment. Bioengineering 2024, 11, 678. [Google Scholar] [CrossRef]
- Miyagawa, C.; Nakai, H.; Otani, T.; Murakami, R.; Takamura, S.; Takaya, H.; Murakami, K.; Mandai, M.; Matsumura, N. Histopathological subtyping of high-grade serous ovarian cancer using whole slide imaging. J. Gynecol. Oncol. 2023, 34, e47. [Google Scholar] [CrossRef] [PubMed]
- Hong, R.; Liu, W.; DeLair, D.; Razavian, N.; Fenyö, D. Predicting endometrial cancer subtypes and molecular features from histopathology images using multi-resolution deep learning models. Cell Rep. Med. 2021, 2, 100400. [Google Scholar] [CrossRef]
- Song, J.; Im, S.; Lee, S.; Jang, H. Deep Learning-Based Classification of Uterine Cervical and Endometrial Cancer Subtypes from Whole-Slide Histopathology Images. Diagnostics 2022, 12, 2623. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Wang, Y.; Li, J.; Xu, K.; Chen, J.; Zhao, J. Deep learning-based methods for classification of microsatellite instability in endometrial cancer from HE-stained pathological images. J. Cancer Res. Clin. Oncol. 2023, 149, 8877–8888. [Google Scholar] [CrossRef]
- Kildal, W.; Cyll, K.; Kalsnes, J.; Islam, R.; Julbø, F.M.; Pradhan, M.; Ersvær, E.; Shepherd, N.; Vlatkovic, L.; OSBREAC—Oslo Breast Cancer Consortium; et al. Deep learning for automated scoring of immunohistochemically stained tumour tissue sections—Validation across tumour types based on patient outcomes. Heliyon. 2024, 10, e32529. [Google Scholar] [CrossRef]
- Ji, C.; Oshima, K.; Urata, T.; Kimura, F.; Ishii, K.; Uehara, T.; Suzuki, K.; Takeyama, S.; Yamaguchi, M. Transformation from hematoxylin-and-eosin staining to Ki-67 immunohistochemistry digital staining images using deep learning: Experimental validation on the labeling index. J. Med. Imaging 2024, 11, 047501. [Google Scholar] [CrossRef] [PubMed]
- Fremond, S.; Andani, S.; Barkey Wolf, J.; Dijkstra, J.; Melsbach, S.; Jobsen, J.; Brinkhuis, M.; Roothaan, S.; Jurgenliemk-Schulz, I.; Lutgens, L.; et al. Interpretable deep learning model to predict the molecular classification of endometrial cancer from haematoxylin and eosin-stained whole-slide images: A combined analysis of the PORTEC randomised trials and clinical cohorts. Lancet Digit. Health. 2023, 5, e71–e82. [Google Scholar] [CrossRef] [PubMed]
- Goyal, M.; Tafe, L.; Feng, J.; Muller, K.; Hondelink, L.; Bentz, J. Deep Learning for Grading Endometrial Cancer. Am. J. Pathol. 2024, 194, 1701–1711. [Google Scholar] [CrossRef]
- Kather, J.; Pearson, A.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.; et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef]
- Wang, T.; Lu, W.; Yang, F.; Liu, L. Microsatellite instability prediction of uterine corpus endometrial carcinoma based on H&E histology whole-slide imaging. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1289–1292. [Google Scholar] [CrossRef]
- Wang, C.; Muzakky, H.; Firdi, N.; Liu, T.; Lai, P.; Wang, Y.; Yu, M.; Chao, T. Deep learning to assess microsatellite instability directly from histopathological whole slide images in endometrial cancer. Digit. Med. 2024, 7, 143. [Google Scholar] [CrossRef] [PubMed]
- Whangbo, J.; Lee, Y.S.; Kim, Y.J.; Kim, J.; Kim, K.G. Predicting Mismatch Repair Deficiency Status in Endometrial Cancer through Multi-Resolution Ensemble Learning in Digital Pathology. J. Imaging Inform. Med. 2024, 37, 1674–1682. [Google Scholar] [CrossRef]
- Umemoto, M.; Mariya, T.; Nambu, Y.; Nagata, M.; Horimai, T.; Sugita, S.; Kanaseki, T.; Takenaka, Y.; Shinkai, S.; Matsuura, M.; et al. Prediction of Mismatch Repair Status in Endometrial Cancer from Histological Slide Images Using Various Deep Learning-Based Algorithms. Cancers 2024, 16, 1810. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Jing, B.; Liu, X.; Li, R.; Wan, Z.; Zhang, J.; Ouyang, X.; Kong, Q.; Kang, X.; Wang, D.; et al. MMRNet: Ensemble deep learning models for predicting mismatch repair deficiency in endometrial cancer from histopathological images. Cell Rep. Med. 2025, 6, 102099. [Google Scholar] [CrossRef]
- Linz, V.C.; Löwe, A.; van der Ven, J.; Hasenburg, A.; Battista, M.J. Incidence of pelvic high-grade serous carcinoma after isolated STIC diagnosis: A systematic review of the literature. Front. Oncol. 2022, 12, 951292. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.A.; Fu, L.; Goyeneche, A.; Gao, Z.H.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- Nik, N.N.; Vang, R.; Shih, I.; Kurman, R.J. Origin and pathogenesis of pelvic (ovarian, tubal, and primary peritoneal) serous carcinoma. Annu. Rev. Pathol. 2014, 9, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, J.M.A.; Steenbeek, M.P.; van Bommel, M.H.D.; Bulten, J.; van der Laak, J.A.W.M.; de Hullu, J.A.; Simons, M. Recommendations for diagnosing STIC: A systematic review and meta-analysis. Virchows Arch. 2022, 480, 725–737. [Google Scholar] [CrossRef]
- Ruel-Laliberté, J.; Kasasni, S.M.; Oprea, D.; Viau, M. Outcome and Management of Serous Tubal Intraepithelial Carcinoma Following Opportunistic Salpingectomy: Systematic Review and Meta-Analysis. J. Obstet. Gynaecol. Can. 2022, 44, 1174–1180. [Google Scholar] [CrossRef]
- Leblanc, E.; Narducci, F.; Ferron, G.; Mailliez, A.; Charvolin, J.; Houssein, E.; Guyon, F.; Fourchotte, V.; Lambaudie, E.; Crouzet, A.; et al. Prophylactic Radical Fimbriectomy with Delayed Oophorectomy in Women with a High Risk of Developing an Ovarian Carcinoma: Results of a Prospective National Pilot Study. Cancers 2023, 15, 1141. [Google Scholar] [CrossRef]
- Vang, R.; Visvanathan, K.; Gross, A.; Maambo, E.; Gupta, M.; Kuhn, E.; Li, R.; Ronnett, B.; Seidman, J.; Yemelyanova, A.; et al. Validation of an algorithm for the diagnosis of serous tubal intraepithelial carcinoma. Int. J. Gynecol. Pathol. 2012, 31, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, J.; Bokhorst, J.; Simons, M.; Bommel, M.; Steenbeek, M.; Hullu, J.; Linmans, J.; Bart, J.; Bentz, J.; Bosse, T.; et al. Deep learning detects premalignant lesions in the Fallopian tube. J. Womens Health 2024, 2, 11. [Google Scholar] [CrossRef]
- Berek, J.; Matias-Guiu, X.; Creutzberg, C.; Fotopoulou, C.; Gaffney, D.; Kehoe, S.; Lindemann, K.; Mutch, D.; Concin, N. Endometrial Cancer Staging Subcommittee, FIGO Women’s Cancer Committee. FIGO staging of endometrial cancer: 2023. Int. J. Gynaecol. Obstet. 2023, 162, 383–394. [Google Scholar] [CrossRef]
- Baker-Rand, H.; Kitson, S.J. Recent Advances in Endometrial Cancer Prevention, Early Diagnosis and Treatment. Cancers 2024, 16, 1028. [Google Scholar] [CrossRef]
- Wang, Y.; Gao, S.; Xiao, Q.; Li, C.; Grzegorzek, M.; Zhang, Y.; Li, X.; Kang, Y.; Liu, F.; Huang, D.; et al. Role of artificial intelligence in digital pathology for gynecological cancers. Comput. Struct. Biotechnol. J. 2024, 24, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Volinsky-Fremond, S.; Horeweg, N.; Andani, S.; Barkey Wolf, J.; Lafarge, M.; Kroon, C.; Ørtoft, G.; Høgdall, E.; Dijkstra, J.; Jobsen, J.; et al. Prediction of recurrence risk in endometrial cancer with multimodal deep learning. Nat. Med. 2024, 30, 1962–1973. [Google Scholar] [CrossRef]
- Levine, D.; The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.; Leung, S.; Li-Chang, H.; Kwon, J.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer. 2015, 113, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Osinski, B.L.; Taxter, T.J.; Perera, J.; Lau, D.J.; Khan, A.A. Adversarial deep learning for microsatellite instability prediction from histopathology slides. In Proceedings of the 1st Conference on Medical Imaging with Deep Learning (MIDL 2018), Amsterdam, The Netherlands, 4–6 July 2018; pp. 4–6. [Google Scholar]
- Rubisz, P.; Ciebiera, M.; Hirnle, L.; Zgliczyńska, M.; Łoziński, T.; Dzięgiel, P.; Kobierzycki, C. The Usefulness of Immunohistochemistry in the Differential Diagnosis of Lesions Originating from the Myometrium. Int. J. Mol. Sci. 2019, 20, 1136. [Google Scholar] [CrossRef] [PubMed]
- Ubago, J.; Zhang, Q.; Kim, J.; Kong, B.; Wei, J. Two Subtypes of Atypical Leiomyoma: Clinical, Histologic, and Molecular Analysis. Am. J. Surg. Pathol. 2016, 40, 923–933. [Google Scholar] [CrossRef]
- Balkenhol, M.; Tellez, D.; Vreuls, W.; Clahsen, P.; Pinckaers, H.; Ciompi, F.; Bult, P.; Laak, J. Deep learning assisted mitotic counting for breast cancer. Lab. Invest. 2019, 99, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Du, D.; Zheng, T.; Liu, L.; Wang, Z.; Du, J.; Yi, H.; Cui, Y.; Liu, D.; Fang, Y. Deep learning and radiomics to predict the mitotic index of gastrointestinal stromal tumors based on multiparametric MRI. Front. Oncol. 2022, 12, 948557. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, J.; Yang, S.; Xiang, J.; Luo, F.; Wang, M.; Zhang, J.; Yang, W.; Huang, J.; Han, X. A generalizable and robust deep learning algorithm for mitosis detection in multicenter breast histopathological images. Med. Image Anal. 2023, 84, 102703. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Jo, V.; Mills, A.; Zhu, S.; Lee, C.; Espinosa, I.; Nucci, M.; Varma, S.; Forgó, E.; Hastie, T.; et al. Clinically Relevant Molecular Subtypes in Leiomyosarcoma. Clin. Cancer Res. 2015, 21, 3501–3511. [Google Scholar] [CrossRef]
- Beck, A.H.; Lee, C.H.; Witten, D.M.; Gleason, B.C.; Edris, B.; Espinosa, I.; Zhu, S.; Li, R.; Montgomery, K.D.; Marinelli, R.J.; et al. Discovery of molecular subtypes in leiomyosarcoma through integrative molecular profiling. Oncogene 2010, 29, 845–854. [Google Scholar] [CrossRef]
- Yang, X.; Stamp, M. Computer-aided diagnosis of low grade endometrial stromal sarcoma (LGESS). Comput. Biol. Med. 2021, 138, 104874. [Google Scholar] [CrossRef] [PubMed]
- Palee, P.; Sharp, B.; Noriega, L.; Sebire, N.; Platt, C. Heuristic neural network approach in histological sections detection of hydatidiform mole. J. Med. Imaging 2019, 6, 044501. [Google Scholar] [CrossRef] [PubMed]
- Xing, D.; Adams, E.; Huang, J.; Ronnett, B. Refined diagnosis of hydatidiform moles with p57 immunohistochemistry and molecular genotyping: Updated analysis of a prospective series of 2217 cases. Mod Pathol. 2021, 34, 961–982. [Google Scholar] [CrossRef]
- Ronnett, B.M. Hydatidiform Moles: Ancillary Techniques to Refine Diagnosis. Arch. Pathol. Lab. Med. 2018, 142, 1485–1502. [Google Scholar] [CrossRef]
- Zhao, Y.; Cai, L.; Zhang, X.; Zhang, H.; Cai, L.; Zhou, L.; Huang, B.; Qian, J. Hematoxylin and Eosin Staining Helps Reduce Maternal Contamination in Short Tandem Repeat Genotyping for Hydatidiform Mole Diagnosis. Int. J. Gynecol. Pathol. 2024, 43, 253–263. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, A.; Valls, J.; Baena, A.; Rojas, F.; Ramírez, K.; Álvarez, R.; Cristaldo, C.; Henríquez, O.; Moreno, A.; Reynaga, D.; et al. Performance of cervical cytology and HPV testing for primary cervical cancer screening in Latin America: An analysis within the ESTAMPA study. Lancet Reg. Health Am. 2023, 26, 100593. [Google Scholar] [CrossRef] [PubMed]
- Habtemariam, L.W.; Zewde, E.T.; Simegn, G.L. Cervix Type and Cervical Cancer Classification System Using Deep Learning Techniques. Med. Devices 2022, 16, 163–176. [Google Scholar] [CrossRef]
- Youneszade, N.; Marjani, M.; Pei, C.P. Deep Learning in Cervical Cancer Diagnosis: Architecture, Opportunities, and Open Research Challenges. IEEE Access. Inst. Electr. Electron. Eng. Inc. 2023, 11, 6133–6149. [Google Scholar] [CrossRef]
- Li, Y.X.; Chen, F.; Shi, J.J.; Huang, Y.L.; Wang, M. Convolutional Neural Networks for Classifying Cervical Cancer Types Using Histological Images. J. Digit. Imaging 2023, 36, 441–449. [Google Scholar] [CrossRef]
- Nuño, T.; García, F. The Lower Anogenital Squamous Terminology Project and its implications for clinical care. Obs. Gynecol. Clin. North. Am. 2013, 40, 225–233. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Ding, L.; Ma, M.; Huang, A.; Gan, Y.; Sheng, D.; Jiang, Z.; Zhang, X. Deep Learning-Based Recognition of Cervical Squamous Interepithelial Lesions. Diagnostics 2023, 13, 1720. [Google Scholar] [CrossRef]
- Jeffus, S.K.; Quick, C.M. Coexpression of p53 and p16 in Vulvar Squamous Neoplasia. Mod. Pathol. 2023, 36, 100319. [Google Scholar] [CrossRef]
- Podoll, M.B.; Singh, N.; Gilks, C.B.; Moghadamfalahi, M.; Sanders, M.A. Assessment of CK17 as a Marker for the Diagnosis of Differentiated Vulvar Intraepithelial Neoplasia. Int. J. Gynecol. Pathol. 2017, 36, 273–280. [Google Scholar] [CrossRef]
- Thompson, E.F.; Chen, J.; Huvila, J.; Pors, J.; Ren, H.; Ho, J.; Chow, C.; Ta, M.; Proctor, L.; McAlpine, J.N.; et al. p53 Immunohistochemical patterns in HPV-related neoplasms of the female lower genital tract can be mistaken for TP53 null or missense mutational patterns. Mod. Pathol. 2020, 33, 1649–1659. [Google Scholar] [CrossRef]
- Carreras-Dieguez, N.; Saco, A.; Del Pino, M.; Marimon, L.; López Del Campo, R.; Manzotti, C.; Fusté, P.; Pumarola, C.; Torné, A.; Garcia, A.; et al. Human papillomavirus and p53 status define three types of vulvar squamous cell carcinomas with distinct clinical, pathological, and prognostic features. Histopathology 2023, 83, 17–30. [Google Scholar] [CrossRef]
- Choschzick, M.; Alyahiaoui, M.; Ciritsis, A.; Rossi, C.; Gut, A.; Hejduk, P.; Boss, A. Deep learning for the standardized classification of Ki-67 in vulva carcinoma: A feasibility study. Heliyon 2021, 7, e07577. [Google Scholar] [CrossRef] [PubMed]
- Erul, E.; Aktekin, Y.; Danışman, F.B.; Gümüştaş, Ş.A.; Aktekin, B.S.; Yekedüz, E.; Ürün, Y. Perceptions, Attitudes, and Concerns on Artificial Intelligence. Appl. Patients Cancer Cancer Control 2025, 32, 10732748251343245. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).