Potential Impact of HLA DQB1*05 on Identical Sibling Hematopoietic Stem Cell Transplantation Outcome †
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Inclusion and Exclusion Criteria
2.3. Outcomes
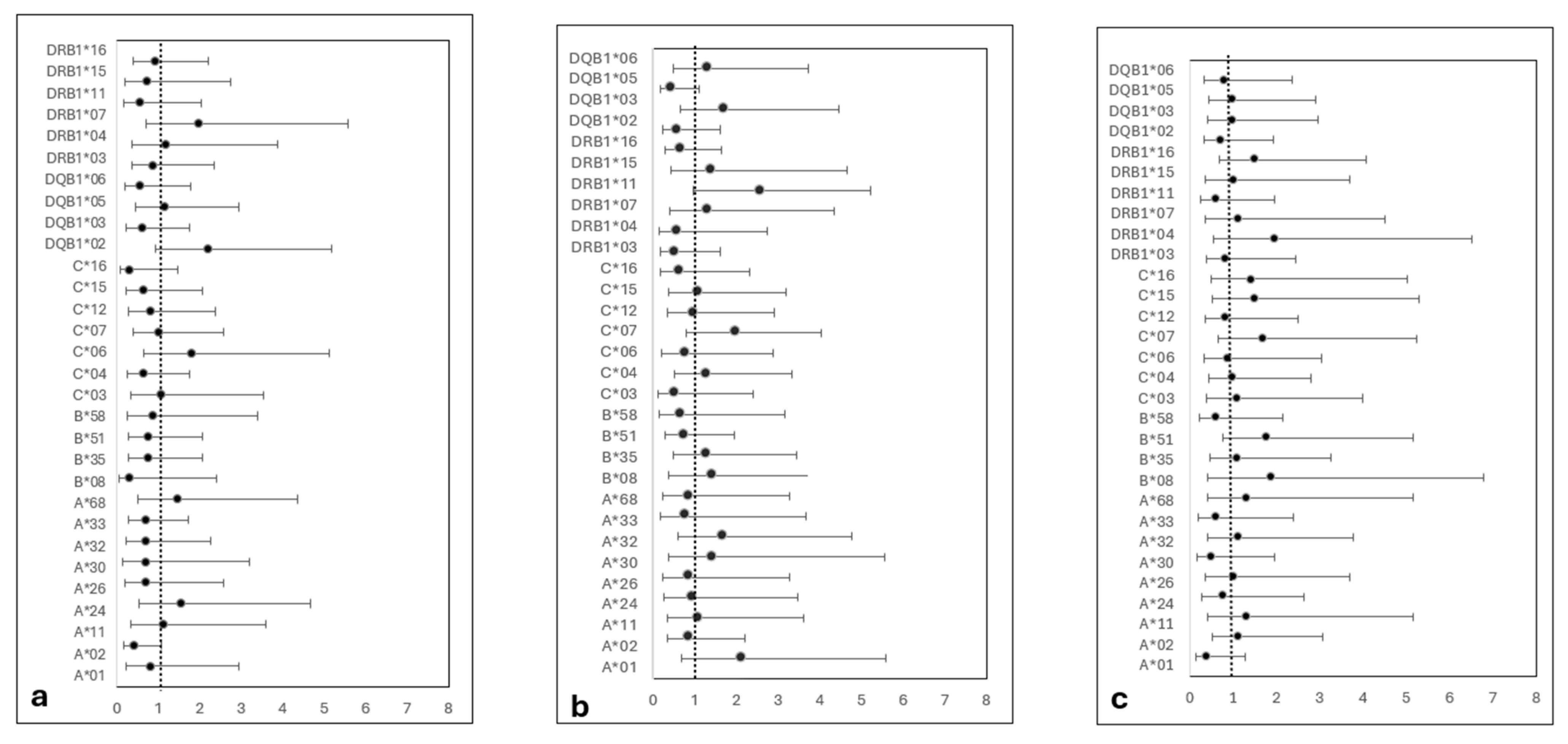
3. Results
3.1. Baseline Characteristics
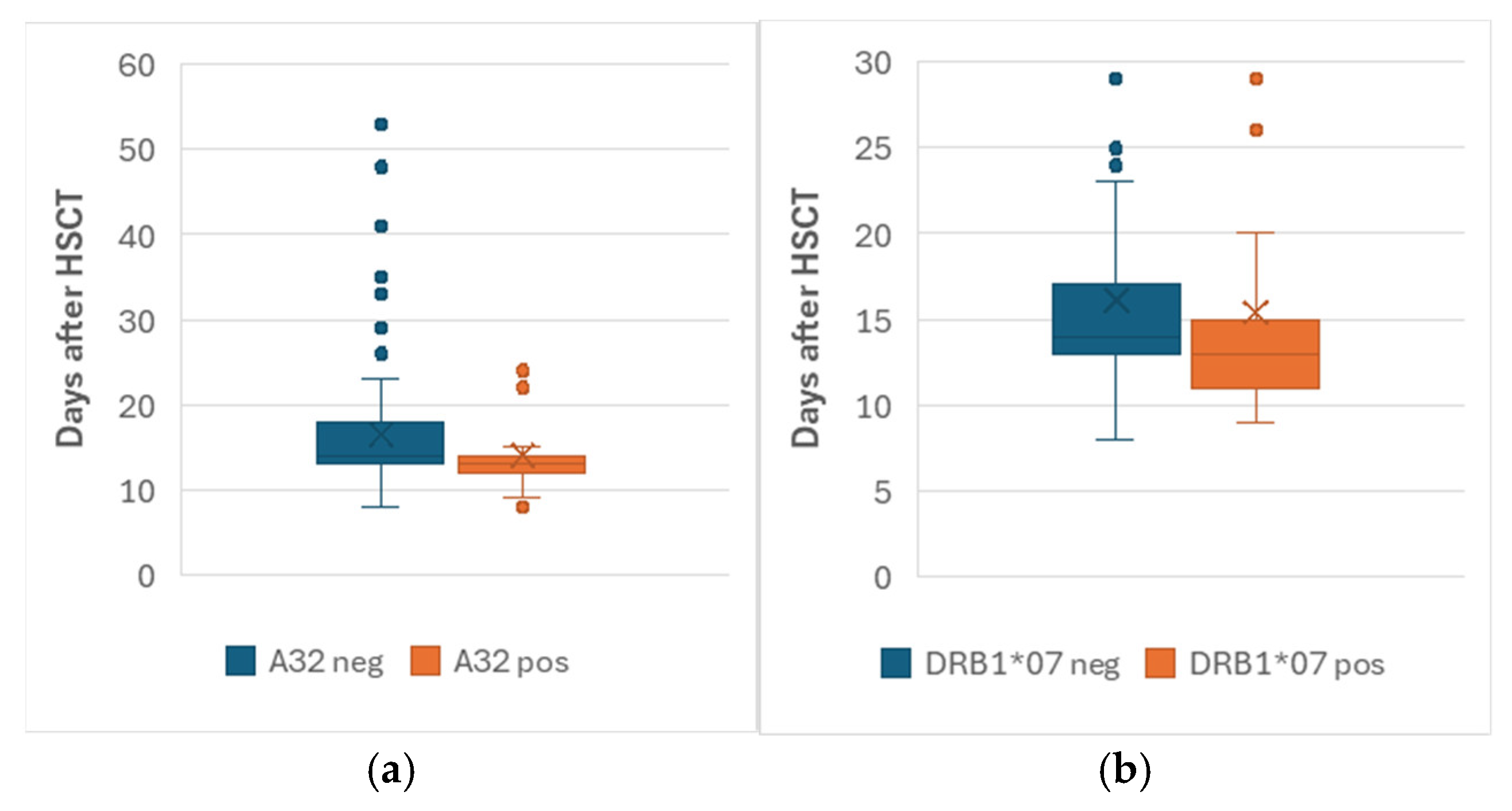
3.2. GVHD, Engraftment and Chimerism
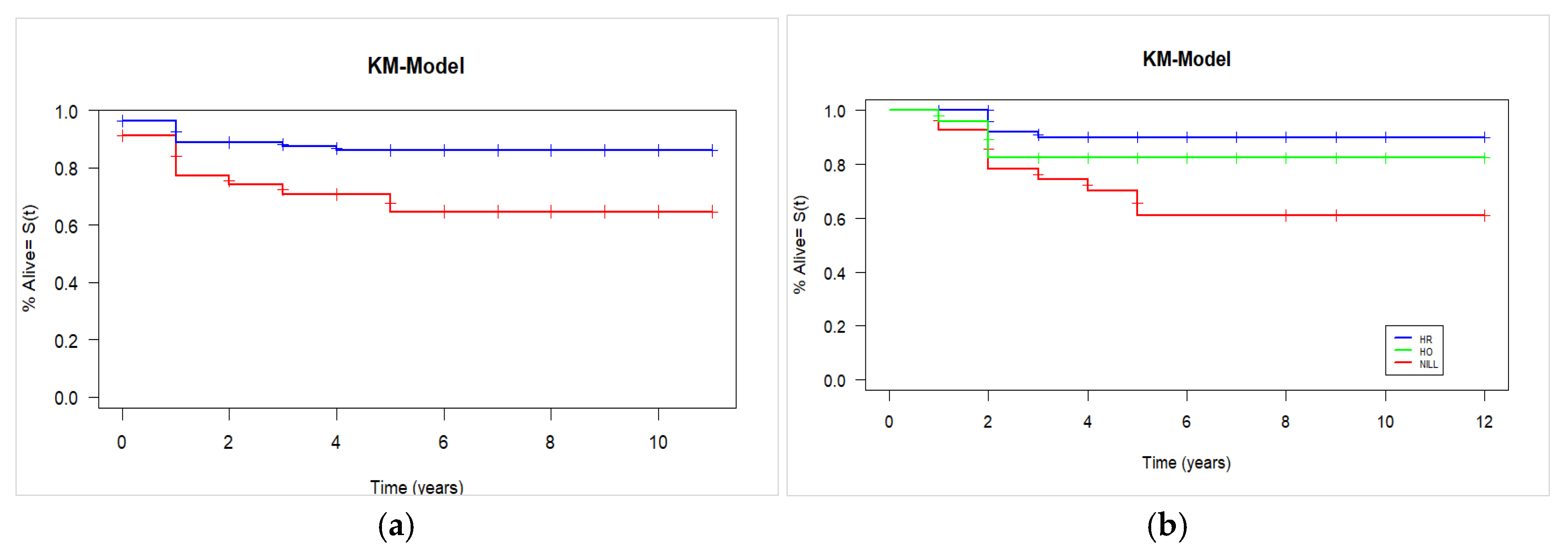
3.3. Overall Survival
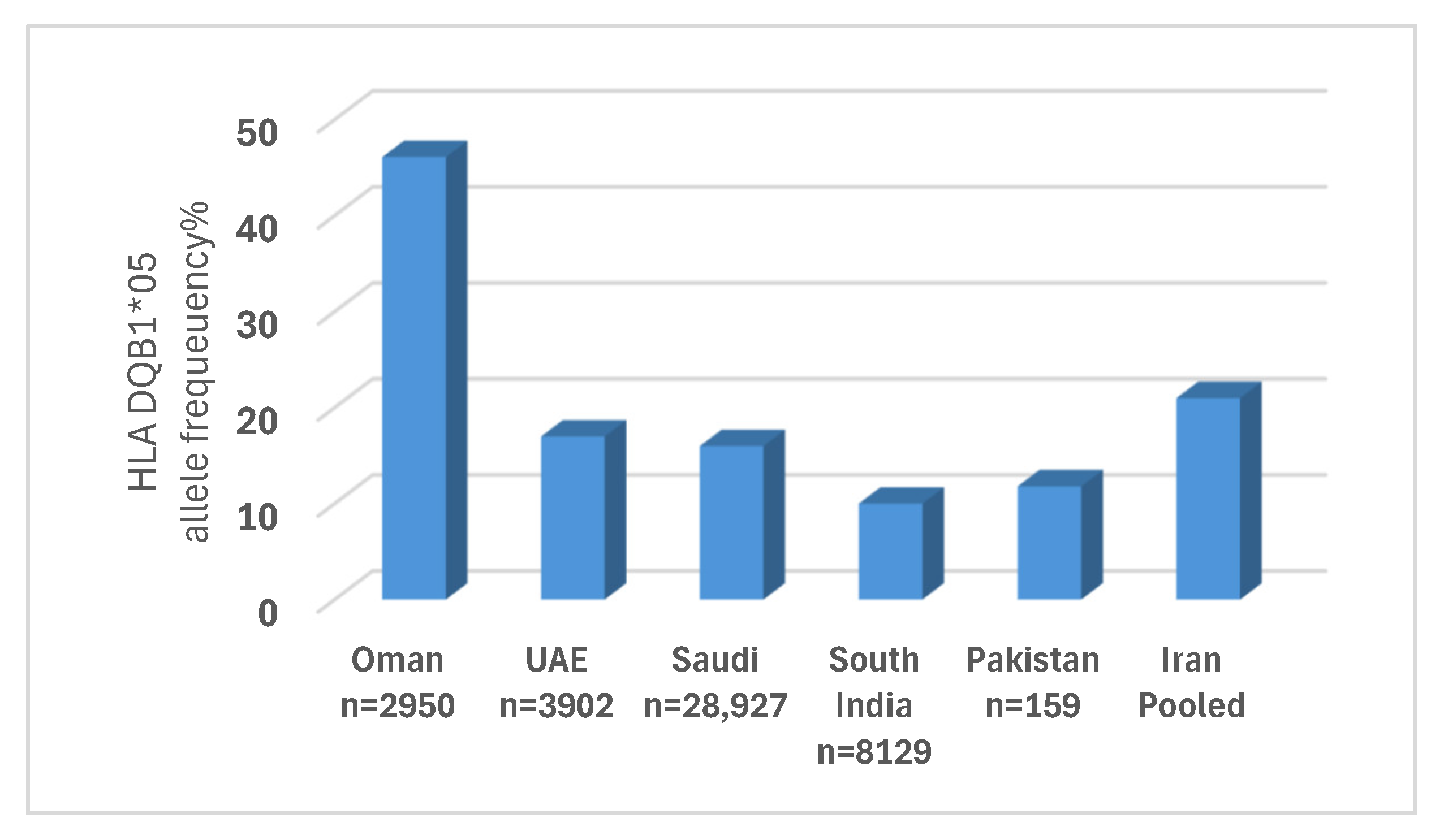
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Allo-HSCT | Allogenic Hematopoietic Stem Cell Transplantation |
| ISD | Identical Sibling Donor |
| HLA | Human Leukocyte Antigen |
| SQUH | Sultan Qaboos University Hospital |
| GVHD | Graft versus Host disease |
| cGVHD | Chronic Graft versus Host Disease |
| aGVHD | Acute Graft versus Host Disease |
| OS | Overall Survival |
| ATL | Adult T cell Leukemia/Lymphoma |
| CMV | Cytomegalo Virus |
| WHO | World Health Organisation |
| SSO | Single Strand Oligonucleotide |
| CSA | Cyclosporin A |
| OR | Odd Ratio |
References
- Bertaina, A.; Andreani, M. Major Histocompatibility Complex and Hematopoietic Stem Cell Transplantation: Beyond the Clas-sical HLA Polymorphism. Int. J. Mol. Sci. 2018, 19, 621. [Google Scholar] [CrossRef] [PubMed]
- Níchonghaile, M. Chapter 3 Donor Selection. In The European Blood and Marrow Transplantation Textbook for Nurses: Under the Auspices of EBMT; Kenyon, M., Babic, A., Eds.; Springer: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Morishima, S.; Fukuda, T.; Doki, N.; Mori, T.; Onizuka, M.; Kawakita, T.; Kato, C.; Ozawa, Y.; Tanaka, M.; Kurokawa, M.; et al. Individual HLAs influence immuno-logical events in allogeneic stem cell transplantation from HLA-identical sibling donors. Bone Marrow Transplant. 2021, 56, 646–654. [Google Scholar] [CrossRef]
- Davidson, J.; Tate, D.; Poulton, K.; Lucas, G.; Yin, J.; Liakopoulou, E.; Wynn, R. HLA-DR15, reduced relapse rate and improved survival after HLA identical sibling hemopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2007, 13, 493–494. [Google Scholar] [CrossRef]
- Stern, M.; Passweg, J.; Tiercy, J.-M.; Genitsch, A.; Meyer-Monard, S.; Heim, D.; Tichelli, A.; Gratwohl, A.; Nissen-Druey, C. Human leukocyte antigen DR15 is associated with reduced relapse rate and improved survival after human leukocyte antigen-identical sibling hematopoietic stem cell trans-plantation. Biol. Blood Marrow Transplant. 2006, 12, 1169–1175. [Google Scholar] [CrossRef]
- Battiwalla, M.; Wang, T.; Carreras, J.; Deeg, H.J.; Ayas, M.; Bajwa, R.P.; George, B.; Gupta, V.; Pasquini, R.; Schrezenmeier, H.; et al. HLA-matched sibling transplantation for severe aplastic anemia: Impact of HLA DR15 antigen status on engraftment, graft-versus-host disease, and overall survival. Biol. Blood Marrow Transplant. 2012, 18, 1401–1406. [Google Scholar] [CrossRef]
- Hamidpour, M.; Roshandel, E.; Nazari, H.G.; Sankanian, G.; Bonakchi, H.; Salimi, M.; Salari, S. Association Between Human Leu-kocyte Antigens and Graft-Versus-Host Disease Occurrence in Allogeneic Hematopoietic Stem Cell Transplantation—A 10-Year Experience on Iranian Patients. Arch. Iran. Med. 2022, 25, 798–806. [Google Scholar] [CrossRef]
- Shawkatová, I.; Bojtárová, E.; Kováčová, M.; Klučková, K.; Kušíková, M.; Mistrík, M.; Homolová, M. Individual HLA alleles and risk of graft-versus-host disease after haematopoietic stem cell transplantation from HLA-identical siblings. Biologia 2020, 75, 2045–2052. [Google Scholar] [CrossRef]
- Morishima, S.; Yoshimitsu, M.; Shindo, T.; Utsunomiya, A.; Ishida, T.; Ito, A.; Nakano, N.; Kawakita, T.; Eto, T.; Suehiro, Y.; et al. Individual HLAs affect survival after allogeneic stem cell transplantation in adult T-cell leukaemia/lymphoma. HLA 2024, 103, e15555. [Google Scholar] [CrossRef] [PubMed]
- Al-Khabori, M.; Al-Huneini, M. Hematopoietic stem cell transplantation in the Sultanate of Oman. Hematol. Stem Cell Ther. 2017, 10, 305–307. [Google Scholar] [CrossRef] [PubMed]
- Toubai, T.; Sun, Y.; Reddy, P. GVHD pathophysiology: Is acute different from chronic? Best Pract. Res. Clin. Haematol. 2008, 21, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, A.; Mancini, G. Current Approaches for the Prevention and Treatment of Acute and Chronic GVHD. Cells 2024, 13, 1524. [Google Scholar] [CrossRef]
- Baron, F.; Sandmaier, B.M. Chimerism and outcomes after allogeneic hematopoietic cell transplantation following nonmyeloablative conditioning. Leukemia 2006, 20, 1690–1700. [Google Scholar] [CrossRef]
- Wolff, S. Second hematopoietic stem cell transplantation for the treatment of graft failure, graft rejection or relapse after al-logeneic transplantation. Bone Marrow Transplant. 2002, 29, 545–552. [Google Scholar] [CrossRef]
- Teltschik, H.; Heinzelmann, F.; Gruhn, B.; Feuchtinger, T.; Schlegel, P.; Schumm, M.; Kremens, B.; Müller, I.; Ebinger, M.; Schwarze, C.P.; et al. Treatment of graft failure with TNI-based reconditioning and haploidentical stem cells in paediatric patients. Br. J. Haematol. 2016, 175, 115–122. [Google Scholar] [CrossRef]
- Bodmer, J.G.; Marsh, S.G.; Albert, E.D.; Bodmer, W.F.; Bontrop, R.E.; Dupont, B.; Erlich, H.A.; Hansen, J.A.; Mach, B.; Mayr, W.R.; et al. Nomenclature for factors of the HLA system, 1998. Tissue Antigens 1999, 53 Pt 2, 407–446. [Google Scholar] [CrossRef] [PubMed]
- Rieger, M.J.; Stolz, S.M.; Müller, A.M.; Schwotzer, R.; Nair, G.; Schneidawind, D.; Manz, M.G.; Schanz, U. Haploidentical transplant with posttransplant cyclophosphamide vs. matched related and unrelated donor transplant in acute myeloid leukemia and myelodysplastic neoplasm. Bone Marrow Transplant. 2023, 58, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Hutt, D. Engraftment, Graft Failure, and Rejection. In The European Blood and Marrow Transplantation Textbook for Nurses: Under the Auspices of EBMT [Internet]; Michell, K., Alexandra, B., Eds.; Springer: New York, NY, USA, 2018; Chapter 13. [Google Scholar]
- Raj, K.; Eikema, D.-J.; Sheth, V.; Koster, L.; de Wreede, L.C.; Blaise, D.; Di Grazia, C.; Koc, Y.; Potter, V.; Chevallier, P.; et al. Comparison of outcomes for HLA-matched sibling and haplo-identical donors in Myelodysplastic syndromes: Report from the chronic malignancies work-ing party of EBMT. Blood Cancer J. 2022, 12, 140. [Google Scholar] [CrossRef]
- Spierings, E. Minor histocompatibility antigens: Past, present, and future. Tissue Antigens 2014, 84, 374-360. [Google Scholar] [CrossRef] [PubMed]
- Hamzeh, A.R.; Nair, P.; Al-Khaja, N.; Al Ali, M.T. Association of HLA-DQA1 and -DQB1 alleles with type I diabetes in Arabs: A meta-analyses. Tissue Antigens 2015, 86, 21–27. [Google Scholar] [CrossRef]
- Holtan, S.G.; Pasquini, M.; Weisdorf, D.J. Acute graft-versus-host disease: A bench-to-bedside update. Blood 2014, 124, 363–373. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Q.-F.; Xu, L.-P.; Liu, K.-Y.; Zhang, X.-H.; Ma, X.; Fan, Z.-P.; Wu, D.-P.; Huang, X.-J. Haploidentical vs. identical-sibling transplant for AML in remission: A multicenter, prospective study. Blood 2015, 125, 3956–3962. [Google Scholar] [CrossRef]
- Bai, L.Y.; Chiou, T.J.; Liu, J.H.; Yen, C.C.; Wang, W.S.; Yan, M.H.; Hsiao, L.T.; Chao, T.C.; Chen, P.M. Hematopoietic stem cell transplantation for severe aplastic ane-mia–experience of an institute in Taiwan. Ann. Hematol. 2004, 83, 38–43. [Google Scholar] [CrossRef]
- Hammad, M.; Hafez, H.; Sidhom, I.; Yassin, D.; Salem, S.; Alsheshtawi, K.; Hamdy, N.; Elsharkawy, N.; Elhaddad, A. Hematopoietic stem cell transplanta-tion from HLA-matched sibling donors in children with acute lymphoblastic leukemia: A report from the Children’s Cancer Hospital Egypt. Front. Oncol. 2022, 12, 983220. [Google Scholar] [CrossRef]
- Gluckman, E.; Cappelli, B.; Bernaudin, F.; Labopin, M.; Volt, F.; Carreras, J.; Simões, B.P.; Ferster, A.; Dupont, S.; de la Fuente, J.; et al. Sickle cell disease: An international survey of results of HLA-identical sibling hematopoietic stem cell transplantation. Blood 2017, 129, 1548–1556. [Google Scholar] [CrossRef]
- Al Hudhili, I.; Al Zadjali, F.; Al Khabori, M. HLA Alleles and Haplotypes Frequencies in Omani Population and Validation of Maximum Likelihood Estimation Predictive Algorithm. Master’s Thesis, Sultan Qaboos University, Muscat, Oman, 2020. [Google Scholar]
- Kennedy, A.E.; Ozbek, U.; Dorak, M.T. What has GWAS done for HLA and disease associations? Int. J. Immunogenet. 2017, 44, 195–211. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz-Villena, A.; Al Yafei, Z.; Juarez, I.; Palacio-Gruber, J.; Al Mahri, A.; Alvares, M.; Lopez-Nares, A.; Nieto, J.; Al Seiari, M.; Martin-Villa, J.M.; et al. HLA genetic study from United Arab Emirates (UAE), Abu Dhabi. Hum. Immunol. 2019, 80, 421–422. [Google Scholar] [CrossRef] [PubMed]
- Jawdat, D.; Uyar, F.A.; Alaskar, A.; Müller, C.R.; Hajeer, A. HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1 Allele and Haplotype Frequencies of 28,927 Saudi Stem Cell Donors Typed by Next-Generation Sequencing. Front. Immunol. 2020, 11, 544768. [Google Scholar] [CrossRef]
- Seshasubramanian, V.; SathishKannan, A.D.; Naganathan, C.; Narayan, S.; Periathiruvadi, S. Molecular analysis of HLA Class I and Class II genes in five different South Indian linguistic groups. HLA 2021, 97, 399–419. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Firasat, S.; Khaliq, S.; Abid, A.; Shah, S.S.; Mehdi, S.Q.; Mohyuddin, A. HLA class I and II polymorphisms in the Gujjar population from Pakistan. Immunol. Investig. 2013, 42, 691–700. [Google Scholar] [CrossRef]
- Abedini, F.; Rahmanian, N.; Heidari, Z.; Feizi, A.; Sherkat, R.; Rezaei, M. Diversity of HLA class I and class II alleles in Iran populations: Systematic review and Meta-Analaysis. Transpl. Immunol. 2021, 69, 101472. [Google Scholar] [CrossRef] [PubMed]
- Al-Balushi, M.; Al-Badi, S.; Al-Yaarubi, S.; Al-Riyami, H.; Al-Shidhani, A.; Al-Hinai, S.; Alshirawi, A.; Hasson, S.; Said, E.; Al-Jabri, A.; et al. The Association of Human Leukocyte Antigens Complex with Type 1 Diabetes in the Omani Population. Sultan Qaboos Univ. Med. J. 2023, 23, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Johansson, T.; Partanen, J.; Saavalainen, P. HLA allele-specific expression: Methods, disease associations, and relevance in hema-topoietic stem cell transplantation. Front. Immunol. 2022, 13, 1007425. [Google Scholar] [CrossRef] [PubMed]
- The 50th Annual Meeting of the European Society for Blood and Marrow Transplantation: Lectures. Bone Marrow Transpl. 2024, 59 (Suppl. S1), 4–5. [CrossRef]
| Patient Variable | n | % | Patient Variable | n | % |
|---|---|---|---|---|---|
| Patient gender | Patient/Donor sex match | ||||
| Male | 93 | 61.0 | Yes | 69 | 45.0 |
| Female | 60 | 39.0 | No | 76 | 49.7 |
| Unknown | 8 | 5.2 | |||
| Age group | GVHD prophylaxis | ||||
| <18 | 58 | 37.9 | Cyclosporine-based | 99 | 64.7 |
| 18–60 | 95 | 62.1 | Tacrolimus-based | 38 | 24.8 |
| CSA and Tacro | 15 | 9.8 | |||
| Patient/Donor ABO compatibility | Not found | 1 | 1.3 | ||
| Compatible | 119 | 77.8 | Clinical Diagnosis | ||
| Incompatible (major) | 19 | 12.4 | AML | 26 | 17.0 |
| Incompatible (minor) | 6 | 3.9 | ALL | 17 | 11.1 |
| Incompatible (bidirectional) | 2 | 1.3 | MDS | 8 | 8.4 |
| Not recorded | 7 | 4.6 | Aplastic anemia | 13 | 5.2 |
| Patient CMV Status | Thalassemia | 8 | 5.2 | ||
| Pos | 129 | 84.3 | Sickle cell disease | 39 | 25.5 |
| Neg | 5 | 3.3 | CGD | 7 | 4.6 |
| Not reported | 19 | 12.4 | Others (malignant/nonmalignant) | 35 (12/23) | 22.8 |
| Stem Cell Source | |||||
| Bone marrow | 43 | 28.1 | Status Jun 2023 | ||
| Peripheral blood | 105 | 68.6 | Alive/Censored | 126 | 81.8 |
| Both | 5 | 3.2 | Dead | 27 | 18.2 |
| No | Parameter | Study Cohort | Published Data | References |
|---|---|---|---|---|
| 1 | Median neutrophil engraftment time | 14 days | 12–16 days | [17,18,19] |
| 2 | Median platelet engraftment time | 15 days | 14–17 days | [17,18,19] |
| 3 | Complete chimerism rate | 84.1% (malignant) 68.4% (nonmalignant) | 80–90% (malignant) 50–60% (nonmalignant) | [20,21] |
| 4 | Acute GVHD rate | 16% | 20–35% | [22] |
| 5 | Chronic GVHD rate | 15% | 20–50% | [23] |
| 6 | 5-year overall survival | 73% malignant 88.9% nonmalignant | 56–75% (malignant) 80–90% (nonmalignant) | [24,25,26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al Lawati, F.; Al Khabori, M.; Al Harrasi, S.; Al Ansari, A. Potential Impact of HLA DQB1*05 on Identical Sibling Hematopoietic Stem Cell Transplantation Outcome. J. Clin. Med. 2025, 14, 6798. https://doi.org/10.3390/jcm14196798
Al Lawati F, Al Khabori M, Al Harrasi S, Al Ansari A. Potential Impact of HLA DQB1*05 on Identical Sibling Hematopoietic Stem Cell Transplantation Outcome. Journal of Clinical Medicine. 2025; 14(19):6798. https://doi.org/10.3390/jcm14196798
Chicago/Turabian StyleAl Lawati, Fatma, Murtadha Al Khabori, Salma Al Harrasi, and Aliya Al Ansari. 2025. "Potential Impact of HLA DQB1*05 on Identical Sibling Hematopoietic Stem Cell Transplantation Outcome" Journal of Clinical Medicine 14, no. 19: 6798. https://doi.org/10.3390/jcm14196798
APA StyleAl Lawati, F., Al Khabori, M., Al Harrasi, S., & Al Ansari, A. (2025). Potential Impact of HLA DQB1*05 on Identical Sibling Hematopoietic Stem Cell Transplantation Outcome. Journal of Clinical Medicine, 14(19), 6798. https://doi.org/10.3390/jcm14196798






