Landscape of NRXN1 Gene Variants in Phenotypic Manifestations of Autism Spectrum Disorder: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
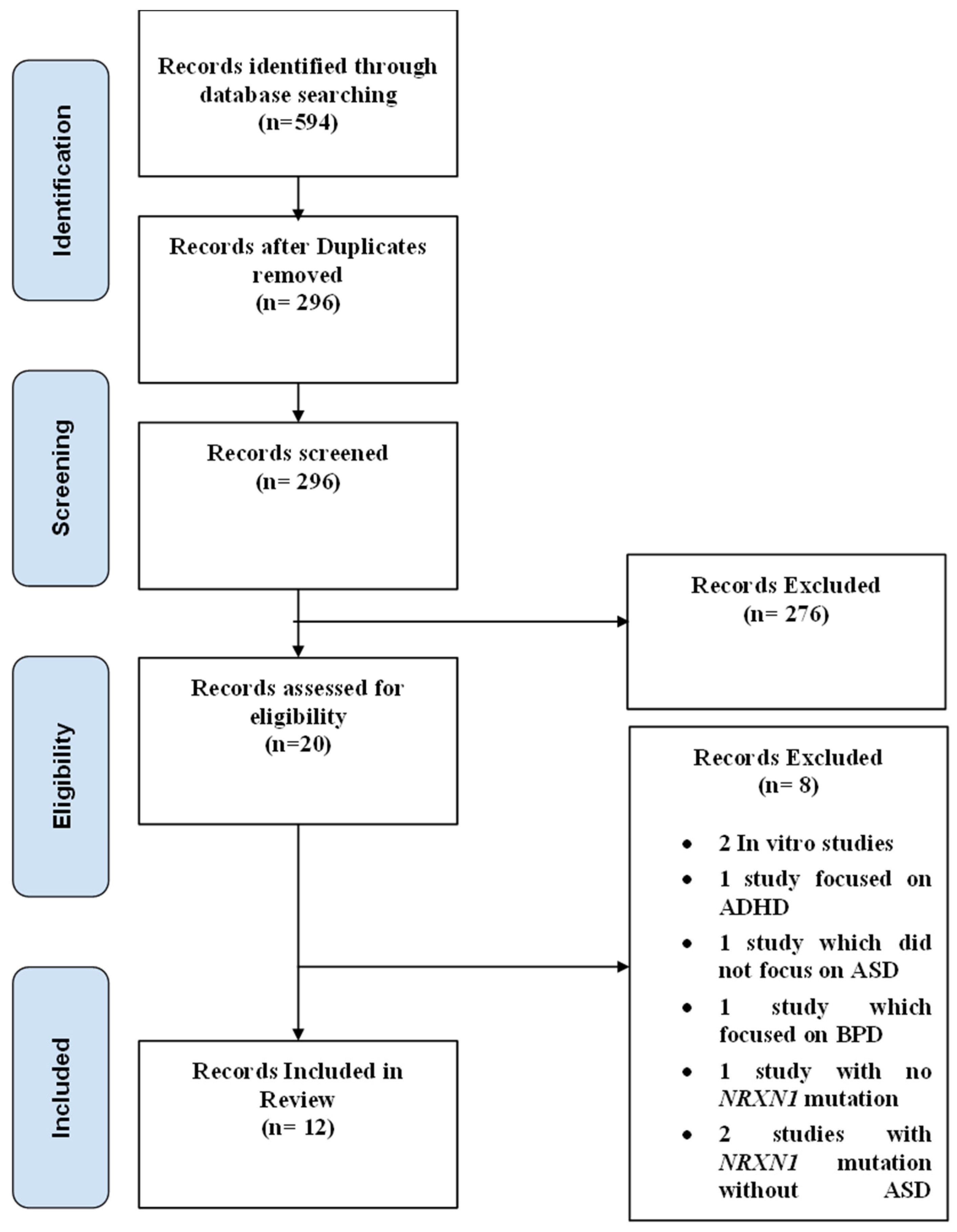
2.2. Study Selection
2.3. Data Extraction
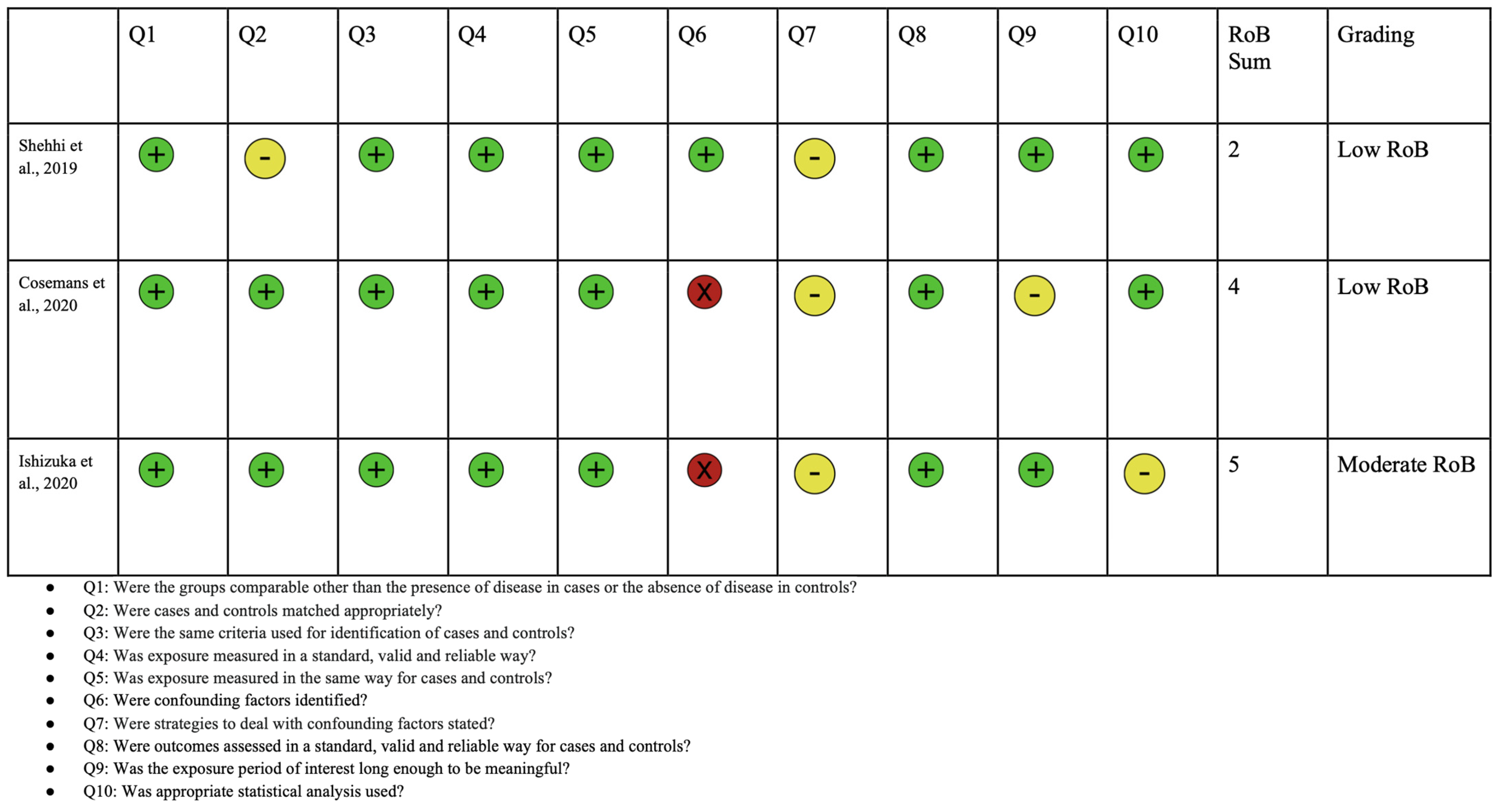
2.4. Quality Assessment
3. Results and Discussion
3.1. Patient Population and Diagnosis
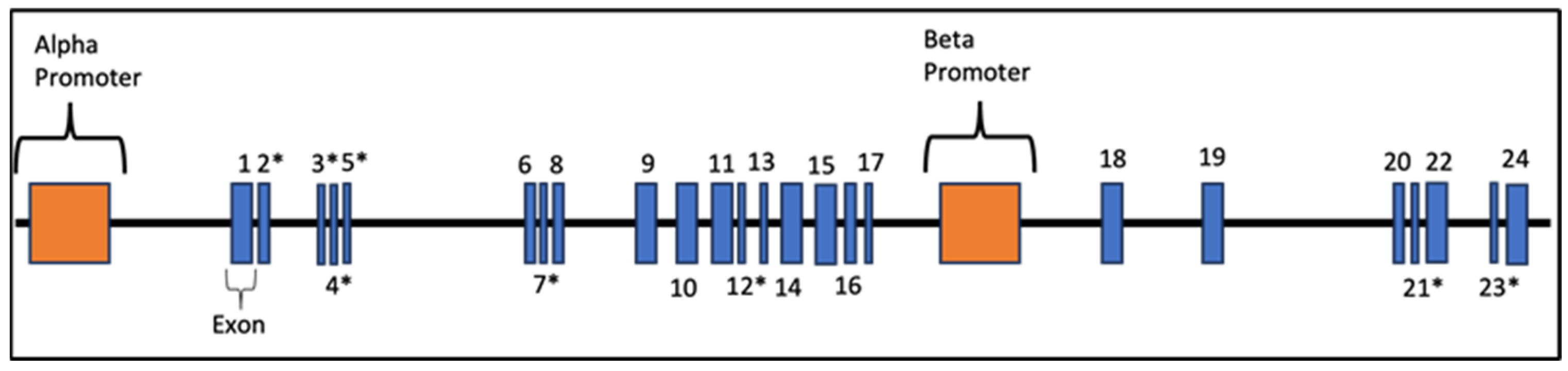
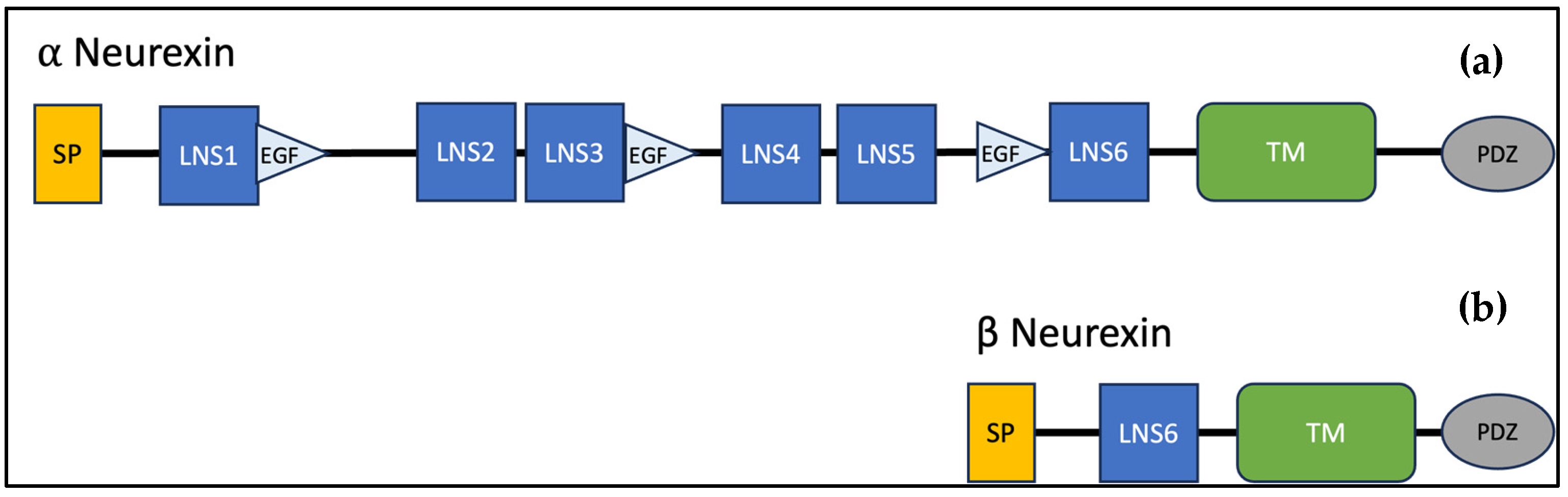
3.2. Genotypic Variants of NRXN1 in Individuals with ASD
3.3. Phenotypic Features of NRXN1 Mutations and ASD
3.3.1. Intellectual Abilities
3.3.2. Speech Abilities
3.3.3. Behavior/Neuropsychiatric Diagnosis
3.3.4. Physical Characteristics
4. Limitations
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirota, T.; King, B.H. Autism Spectrum Disorder: A Review. JAMA 2023, 329, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Hodges, H.; Fealko, C.; Soares, N. Autism spectrum disorder: Definition, epidemiology, causes, and clinical evaluation. Transl. Pediatr. 2020, 9, S55–S65. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.; Elsabbagh, M.; Baird, G.; Veenstra-Vanderweele, J. Autism spectrum disorder. Lancet 2018, 392, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Nakai, N.; Fujima, S.; Choe, K.Y.; Takumi, T. Social circuits and their dysfunction in autism spectrum disorder. Mol. Psychiatry 2023, 28, 3194–3206. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.; Moosa, M.; McKenna, K.; Mittal, J.; Memis, I.; Mittal, R.; Eshraghi, A.A. Quality of Life, Neurosensory Disorders and Co-Occurring Medical Conditions in Individuals on the Spectrum, with a Special Focus on Females Diagnosed with Autism: A Systematic Review. J. Clin. Med. 2023, 12, 927. [Google Scholar] [CrossRef]
- Anderson, G.R.; Aoto, J.; Tabuchi, K.; Földy, C.; Covy, J.; Yee, A.X.; Wu, D.; Lee, S.J.; Chen, L.; Malenka, R.C.; et al. β-Neurexins Control Neural Circuits by Regulating Synaptic Endocannabinoid Signaling. Cell 2015, 162, 593–606. [Google Scholar] [CrossRef] [PubMed]
- Eckes, T.; Buhlmann, U.; Holling, H.-D.; Möllmann, A. Comprehensive ABA-based interventions in the treatment of children with autism spectrum disorder—A meta-analysis. BMC Psychiatry 2023, 23, 133. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, A.K.; Paterson, C.; Wang, Y.; Hyde, T.M.; Kleinman, J.E.; Law, A.J. Neurexin 1 (NRXN1) splice isoform expression during human neocortical development and aging. Mol. Psychiatry 2016, 21, 701–706. [Google Scholar] [CrossRef] [PubMed]
- Béna, F.; Bruno, D.L.; Eriksson, M.; van Ravenswaaij-Arts, C.; Stark, Z.; Dijkhuizen, T.; Gerkes, E.; Gimelli, S.; Ganesamoorthy, D.; Thuresson, A.C.; et al. Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2013, 162, 388–403. [Google Scholar] [CrossRef]
- Castronovo, P.; Baccarin, M.; Ricciardello, A.; Picinelli, C.; Tomaiuolo, P.; Cucinotta, F.; Frittoli, M.; Lintas, C.; Sacco, R.; Persico, A.M. Phenotypic spectrum of NRXN1 mono- and bi-allelic deficiency: A systematic review. Clin. Genet. 2020, 97, 125–137. [Google Scholar] [CrossRef]
- Kirov, G.; Gumus, D.; Chen, W.; Norton, N.; Georgieva, L.; Sari, M.; O’Donovan, M.C.; Erdogan, F.; Owen, M.J.; Ropers, H.-H. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008, 17, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Hu, Z.; Xun, G.; Peng, Y.; Lu, L.; Xu, X.; Xiong, Z.; Xia, L.; Liu, D.; Li, W.; et al. Mutation analysis of the NRXN1 gene in a Chinese autism cohort. J. Psychiatr. Res. 2012, 46, 630–634. [Google Scholar] [CrossRef]
- Reissner, C.; Runkel, F.; Missler, M. Neurexins. Genome Biol. 2013, 14, 213. [Google Scholar] [CrossRef]
- Roberts, J.L.; Hovanes, K.; Dasouki, M.; Manzardo, A.M.; Butler, M.G. Chromosomal microarray analysis of consecutive individuals with autism spectrum disorders or learning disability presenting for genetic services. Gene 2014, 535, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Styles, M.; Alsharshani, D.; Samara, M.; Alsharshani, M.; Khattab, A.; Qoronfleh, M.W.; Al-Dewik, N.I. Risk factors, diagnosis, prognosis and treatment of autism. Front. Biosci. (Landmark Ed.) 2020, 25, 1682–1717. [Google Scholar] [CrossRef]
- Tromp, A.; Mowry, B.; Giacomotto, J. Neurexins in autism and schizophrenia—A review of patient mutations, mouse models and potential future directions. Mol. Psychiatry 2021, 26, 747–760. [Google Scholar] [CrossRef] [PubMed]
- Varghese, M.; Keshav, N.; Jacot-Descombes, S.; Warda, T.; Wicinski, B.; Dickstein, D.L.; Harony-Nicolas, H.; De Rubeis, S.; Drapeau, E.; Buxbaum, J.D.; et al. Autism spectrum disorder: Neuropathology and animal models. Acta Neuropathol. 2017, 134, 537–566. [Google Scholar] [CrossRef]
- Bourgeron, T. The possible interplay of synaptic and clock genes in autism spectrum disorders. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2007; pp. 645–654. [Google Scholar]
- Bourgeron, T. Genes, synapses and autism spectrum disorders. In Synaptic Plasticity and the Mechanism of Alzheimer’s Disease; Springer: Berlin/Heidelberg, Germany, 2008; pp. 169–179. [Google Scholar]
- Bourgeron, T. A synaptic trek to autism. Curr. Opin. Neurobiol. 2009, 19, 231–234. [Google Scholar] [CrossRef]
- Keller, R.; Basta, R.; Salerno, L.; Elia, M. Autism, epilepsy, and synaptopathies: A not rare association. Neurol. Sci. 2017, 38, 1353–1361. [Google Scholar] [CrossRef]
- Lepeta, K.; Lourenco, M.V.; Schweitzer, B.C.; Martino Adami, P.V.; Banerjee, P.; Catuara-Solarz, S.; de La Fuente Revenga, M.; Guillem, A.M.; Haidar, M.; Ijomone, O.M.; et al. Synaptopathies: Synaptic dysfunction in neurological disorders—A review from students to students. J. Neurochem. 2016, 138, 785–805. [Google Scholar] [CrossRef]
- Sauer, A.K.; Stanton, J.E.; Hans, S.; Grabrucker, A.M. Autism Spectrum Disorders: Etiology and Pathology. In Autism Spectrum Disorders; Grabrucker, A.M., Ed.; Exon Publications Copyright: Brisbane, Australia, 2021. [Google Scholar]
- Smith, R.; Sadee, W. Synaptic Signaling and Aberrant RNA Splicing in Autism Spectrum Disorders. Front. Synaptic Neurosci. 2011, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Südhof, T.C. Synaptic neurexin complexes: A molecular code for the logic of neural circuits. Cell 2017, 171, 745–769. [Google Scholar] [CrossRef] [PubMed]
- Buxbaum, J.D. Multiple rare variants in the etiology of autism spectrum disorders. Dialogues Clin. Neurosci. 2009, 11, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Cooke, J.; Molloy, C.J.; Cáceres, A.S.J.; Dinneen, T.; Bourgeron, T.; Murphy, D.; Gallagher, L.; Loth, E. The Synaptic Gene Study: Design and Methodology to Identify Neurocognitive Markers in Phelan-McDermid Syndrome and NRXN1 Deletions. Front. Neurosci. 2022, 16, 806990. [Google Scholar] [CrossRef] [PubMed]
- Curran, S.; Ahn, J.W.; Grayton, H.; Collier, D.A.; Ogilvie, C.M. NRXN1 deletions identified by array comparative genome hybridisation in a clinical case series—Further understanding of the relevance of NRXN1 to neurodevelopmental disorders. J. Mol. Psychiatry 2013, 1, 4. [Google Scholar] [CrossRef] [PubMed]
- Cuttler, K.; Hassan, M.; Carr, J.; Cloete, R.; Bardien, S. Emerging evidence implicating a role for neurexins in neurodegenerative and neuropsychiatric disorders. Open Biol. 2021, 11, 210091. [Google Scholar] [CrossRef]
- Dean, C.; Scholl, F.G.; Choih, J.; DeMaria, S.; Berger, J.; Isacoff, E.; Scheiffele, P. Neurexin mediates the assembly of presynaptic terminals. Nat. Neurosci. 2003, 6, 708–716. [Google Scholar] [CrossRef]
- Gauthier, J.; Siddiqui, T.J.; Huashan, P.; Yokomaku, D.; Hamdan, F.F.; Champagne, N.; Lapointe, M.; Spiegelman, D.; Noreau, A.; Lafrenière, R.G. Truncating mutations in NRXN2 and NRXN1 in autism spectrum disorders and schizophrenia. Hum. Genet. 2011, 130, 563–573. [Google Scholar] [CrossRef]
- Huang, A.Y.; Yu, D.; Davis, L.K.; Sul, J.H.; Tsetsos, F.; Ramensky, V.; Zelaya, I.; Ramos, E.M.; Osiecki, L.; Chen, J.A. Rare copy number variants in NRXN1 and CNTN6 increase risk for Tourette syndrome. Neuron 2017, 94, 1101–1111.e7. [Google Scholar] [CrossRef]
- Kember, R.; Ji, X.; Zhang, J.; Brown, C.; Rader, D.; Almasy, L.; Bucan, M. Spectrum of common and rare mutations contributing to autism risk in families. Eur. Neuropsychopharmacol. 2019, 29, S962–S963. [Google Scholar] [CrossRef]
- Kim, H.G.; Kishikawa, S.; Higgins, A.W.; Seong, I.S.; Donovan, D.J.; Shen, Y.; Lally, E.; Weiss, L.A.; Najm, J.; Kutsche, K.; et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 2008, 82, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Krueger, D.D.; Tuffy, L.P.; Papadopoulos, T.; Brose, N. The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr. Opin. Neurobiol. 2012, 22, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Leblond, C.S.; Cliquet, F.; Carton, C.; Huguet, G.; Mathieu, A.; Kergrohen, T.; Buratti, J.; Lemière, N.; Cuisset, L.; Bienvenu, T.; et al. Both rare and common genetic variants contribute to autism in the Faroe Islands. npj Genom. Med. 2019, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Rujescu, D.; Ingason, A.; Cichon, S.; Pietiläinen, O.P.; Barnes, M.R.; Toulopoulou, T.; Picchioni, M.; Vassos, E.; Ettinger, U.; Bramon, E. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum. Mol. Genet. 2009, 18, 988–996. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gong, J.; Li, L.; Chen, Y.; Liu, L.; Gu, H.; Luo, X.; Hou, F.; Zhang, J.; Song, R. Neurexin gene family variants as risk factors for autism spectrum disorder. Autism Res. 2018, 11, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Aoto, J.; Südhof, T.C. Alternative splicing of presynaptic neurexins differentially controls postsynaptic NMDA and AMPA receptor responses. Neuron 2019, 102, 993–1008.e5. [Google Scholar] [CrossRef] [PubMed]
- Flaherty, E.; Zhu, S.; Barretto, N.; Cheng, E.; Deans, P.J.M.; Fernando, M.B.; Schrode, N.; Francoeur, N.; Antoine, A.; Alganem, K.; et al. Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat. Genet. 2019, 51, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Missler, M.; Südhof, T.C. Neurexins: Three genes and 1001 products. Trends Genet. 1998, 14, 20–26. [Google Scholar] [CrossRef]
- Tabuchi, K.; Südhof, T.C. Structure and evolution of neurexin genes: Insight into the mechanism of alternative splicing. Genomics 2002, 79, 849–859. [Google Scholar] [CrossRef]
- Miller, M.T.; Mileni, M.; Comoletti, D.; Stevens, R.C.; Harel, M.; Taylor, P. The crystal structure of the α-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure 2011, 19, 767–778. [Google Scholar] [CrossRef]
- Zambonino, M.; Pereira, P. The structure of Neurexin 1α (n1α) and its role as synaptic organizer. Bionatura 2019, 4, 883–886. [Google Scholar] [CrossRef]
- Camacho-Garcia, R.J.; Planelles, M.I.; Margalef, M.; Pecero, M.L.; Martínez-Leal, R.; Aguilera, F.; Vilella, E.; Martinez-Mir, A.; Scholl, F.G. Mutations affecting synaptic levels of neurexin-1β in autism and mental retardation. Neurobiol. Dis. 2012, 47, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Dachtler, J.; Ivorra, J.L.; Rowland, T.E.; Lever, C.; Rodgers, R.J.; Clapcote, S.J. Heterozygous deletion of α-neurexin I or α-neurexin II results in behaviors relevant to autism and schizophrenia. Behav. Neurosci. 2015, 129, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Esclassan, F.; Francois, J.; Phillips, K.G.; Loomis, S.; Gilmour, G. Phenotypic characterization of nonsocial behavioral impairment in neurexin 1α knockout rats. Behav. Neurosci. 2015, 129, 74. [Google Scholar] [CrossRef] [PubMed]
- Ching, M.S.; Shen, Y.; Tan, W.H.; Jeste, S.S.; Morrow, E.M.; Chen, X.; Mukaddes, N.M.; Yoo, S.Y.; Hanson, E.; Hundley, R. Deletions of NRXN1 (neurexin-1) predispose to a wide spectrum of developmental disorders. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2010, 153, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Dabell, M.P.; Rosenfeld, J.A.; Bader, P.; Escobar, L.F.; El-Khechen, D.; Vallee, S.E.; Dinulos, M.B.P.; Curry, C.; Fisher, J.; Tervo, R. Investigation of NRXN1 deletions: Clinical and molecular characterization. Am. J. Med. Genet. Part A 2013, 161, 717–731. [Google Scholar] [CrossRef] [PubMed]
- Janz, P.; Bainier, M.; Marashli, S.; Schoenenberger, P.; Valencia, M.; Redondo, R.L. Neurexin1α knockout rats display oscillatory abnormalities and sensory processing deficits back-translating key endophenotypes of psychiatric disorders. Transl. Psychiatry 2022, 12, 455. [Google Scholar] [CrossRef] [PubMed]
- Pak, C.; Danko, T.; Zhang, Y.; Aoto, J.; Anderson, G.; Maxeiner, S.; Yi, F.; Wernig, M.; Südhof, T.C. Human Neuropsychiatric Disease Modeling using Conditional Deletion Reveals Synaptic Transmission Defects Caused by Heterozygous Mutations in NRXN1. Cell Stem Cell 2015, 17, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Torres, V.I.; Vallejo, D.; Inestrosa, N.C. Emerging Synaptic Molecules as Candidates in the Etiology of Neurological Disorders. Neural Plast. 2017, 2017, 8081758. [Google Scholar] [CrossRef]
- Zweier, C.; de Jong, E.K.; Zweier, M.; Orrico, A.; Ousager, L.B.; Collins, A.L.; Bijlsma, E.K.; Oortveld, M.A.; Ekici, A.B.; Reis, A.; et al. CNTNAP2 and NRXN1 are mutated in autosomal-recessive Pitt-Hopkins-like mental retardation and determine the level of a common synaptic protein in Drosophila. Am. J. Hum. Genet. 2009, 85, 655–666. [Google Scholar] [CrossRef]
- Graf, E.R.; Kang, Y.; Hauner, A.M.; Craig, A.M. Structure Function and Splice Site Analysis of the Synaptogenic Activity of the Neurexin-1β LNS Domain. J. Neurosci. 2006, 26, 4256–4265. [Google Scholar] [CrossRef] [PubMed]
- Graf, E.R.; Zhang, X.; Jin, S.X.; Linhoff, M.W.; Craig, A.M. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 2004, 119, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Missler, M.; Zhang, W.; Rohlmann, A.; Kattenstroth, G.; Hammer, R.E.; Gottmann, K.; Südhof, T.C. Alpha-neurexins couple Ca2+ channels to synaptic vesicle exocytosis. Nature 2003, 423, 939–948. [Google Scholar] [CrossRef]
- Nam, C.I.; Chen, L. Postsynaptic assembly induced by neurexin-neuroligin interaction and neurotransmitter. Proc. Natl. Acad. Sci. USA 2005, 102, 6137–6142. [Google Scholar] [CrossRef] [PubMed]
- Sons, M.S.; Busche, N.; Strenzke, N.; Moser, T.; Ernsberger, U.; Mooren, F.C.; Zhang, W.; Ahmad, M.; Steffens, H.; Schomburg, E.D.; et al. alpha-Neurexins are required for efficient transmitter release and synaptic homeostasis at the mouse neuromuscular junction. Neuroscience 2006, 138, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Uchigashima, M.; Cheung, A.; Suh, J.; Watanabe, M.; Futai, K. Differential expression of neurexin genes in the mouse brain. J. Comp. Neurol. 2019, 527, 1940–1965. [Google Scholar] [CrossRef]
- Al Shehhi, M.; Forman, E.B.; Fitzgerald, J.E.; McInerney, V.; Krawczyk, J.; Shen, S.; Betts, D.R.; Ardle, L.M.; Gorman, K.M.; King, M.D.; et al. NRXN1 deletion syndrome; phenotypic and penetrance data from 34 families. Eur. J. Med. Genet. 2019, 62, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Girirajan, S.; Dennis, M.Y.; Baker, C.; Malig, M.; Coe, B.P.; Campbell, C.D.; Mark, K.; Vu, T.H.; Alkan, C.; Cheng, Z.; et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 2013, 92, 221–237. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Xiao, X.; Zhang, Z.; Li, M. Genetic insights and neurobiological implications from NRXN1 in neuropsychiatric disorders. Mol. Psychiatry 2019, 24, 1400–1414. [Google Scholar] [CrossRef]
- Kasem, E.; Kurihara, T.; Tabuchi, K. Neurexins and neuropsychiatric disorders. Neurosci. Res. 2018, 127, 53–60. [Google Scholar] [CrossRef]
- Levy, D.; Ronemus, M.; Yamrom, B.; Lee, Y.H.; Leotta, A.; Kendall, J.; Marks, S.; Lakshmi, B.; Pai, D.; Ye, K.; et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron 2011, 70, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.J.; Ercan-Sencicek, A.G.; Hus, V.; Luo, R.; Murtha, M.T.; Moreno-De-Luca, D.; Chu, S.H.; Moreau, M.P.; Gupta, A.R.; Thomson, S.A.; et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 2011, 70, 863–885. [Google Scholar] [CrossRef] [PubMed]
- Viñas-Jornet, M.; Esteba-Castillo, S.; Gabau, E.; Ribas-Vidal, N.; Baena, N.; San, J.; Ruiz, A.; Coll, M.D.; Novell, R.; Guitart, M. A common cognitive, psychiatric, and dysmorphic phenotype in carriers of NRXN1 deletion. Mol. Genet. Genom. Med. 2014, 2, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Craig, A.M.; Kang, Y. Neurexin–neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 2007, 17, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Ichtchenko, K.; Nguyen, T.; Südhof, T.C. Structures, alternative splicing, and neurexin binding of multiple neuroligins. J. Biol. Chem. 1996, 271, 2676–2682. [Google Scholar] [CrossRef] [PubMed]
- Guang, S.; Pang, N.; Deng, X.; Yang, L.; He, F.; Wu, L.; Chen, C.; Yin, F.; Peng, J. Synaptopathology Involved in Autism Spectrum Disorder. Front. Cell. Neurosci. 2018, 12, 470. [Google Scholar] [CrossRef] [PubMed]
- Culotta, L.; Penzes, P. Exploring the mechanisms underlying excitation/inhibition imbalance in human iPSC-derived models of ASD. Mol. Autism 2020, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Alkanli, S.S.; Alkanli, N.; Ay, A.; Albeniz, I. CRISPR/Cas9 Mediated Therapeutic Approach in Huntington’s Disease. Mol. Neurobiol. 2023, 60, 1486–1498. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.; Peng, H.; Crapper, L.; Kolobova, I.; Maussion, G.; Vasuta, C.; Yerko, V.; Wong, T.P.; Ernst, C. A Rapid Pipeline to Model Rare Neurodevelopmental Disorders with Simultaneous CRISPR/Cas9 Gene Editing. Stem Cells Transl. Med. 2017, 6, 886–896. [Google Scholar] [CrossRef]
- Hatada, I.; Morita, S.; Horii, T. CRISPR/Cas9. Methods Mol. Biol. 2023, 2637, 41–47. [Google Scholar] [CrossRef]
- Ricci, R.; Colasante, G. CRISPR/dCas9 as a Therapeutic Approach for Neurodevelopmental Disorders: Innovations and Limitations Compared to Traditional Strategies. Dev. Neurosci. 2021, 43, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Tyumentseva, M.; Tyumentsev, A.; Akimkin, V. CRISPR/Cas9 Landscape: Current State and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 16077. [Google Scholar] [CrossRef] [PubMed]
- Nele, C.; Laura, V.; Annick, V.; Koenraad, D.; Hilde Van, E.; Griet Van, B.; Hilde, O.; de Thomy, R.; Els, O.; Eric, L.; et al. The clinical relevance of intragenic NRXN1 deletions. J. Med. Genet. 2020, 57, 347. [Google Scholar] [CrossRef]
- Ishizuka, K.; Yoshida, T.; Kawabata, T.; Imai, A.; Mori, H.; Kimura, H.; Inada, T.; Okahisa, Y.; Egawa, J.; Usami, M.; et al. Functional characterization of rare NRXN1 variants identified in autism spectrum disorders and schizophrenia. J. Neurodev. Disord. 2020, 12, 25. [Google Scholar] [CrossRef]
- Alfieri, P.; Scibelli, F.; Sinibaldi, L.; Valeri, G.; Caciolo, C.; Novello, R.L.; Novelli, A.; Digilio, M.C.; Tartaglia, M.; Vicari, S. Further insight into the neurobehavioral pattern of children carrying the 2p16.3 heterozygous deletion involving NRXN1: Report of five new cases. Genes Brain Behav. 2020, 19, e12687. [Google Scholar] [CrossRef]
- Calderoni, S.; Ricca, I.; Balboni, G.; Cagiano, R.; Cassandrini, D.; Doccini, S.; Cosenza, A.; Tolomeo, D.; Tancredi, R.; Santorelli, F.M.; et al. Evaluation of Chromosome Microarray Analysis in a Large Cohort of Females with Autism Spectrum Disorders: A Single Center Italian Study. J. Pers. Med. 2020, 10, 160. [Google Scholar] [CrossRef] [PubMed]
- Aksu Uzunhan, T.; Ayaz, A. Homozygous exonic and intragenic NRXN1 deletion presenting as either West syndrome or autism spectrum disorder in two siblings. Clin. Neurol. Neurosurg. 2022, 214, 107141. [Google Scholar] [CrossRef]
- Annunziata, S.; Bulgheroni, S.; D’Arrigo, S.; Esposito, S.; Taddei, M.; Saletti, V.; Alfei, E.; Sciacca, F.L.; Rizzo, A.; Pantaleoni, C.; et al. CGH Findings in Children with Complex and Essential Autistic Spectrum Disorder. J. Autism Dev. Disord. 2023, 53, 615–623. [Google Scholar] [CrossRef]
- Cameli, C.; Viggiano, M.; Rochat, M.J.; Maresca, A.; Caporali, L.; Fiorini, C.; Palombo, F.; Magini, P.; Duardo, R.C.; Ceroni, F.; et al. An increased burden of rare exonic variants in NRXN1 microdeletion carriers is likely to enhance the penetrance for autism spectrum disorder. J. Cell. Mol. Med. 2021, 25, 2459–2470. [Google Scholar] [CrossRef]
- Williams, S.M.; An, J.Y.; Edson, J.; Watts, M.; Murigneux, V.; Whitehouse, A.J.O.; Jackson, C.J.; Bellgrove, M.A.; Cristino, A.S.; Claudianos, C. An integrative analysis of non-coding regulatory DNA variations associated with autism spectrum disorder. Mol. Psychiatry 2019, 24, 1707–1719. [Google Scholar] [CrossRef]
- Zarrei, M.; Burton, C.L.; Engchuan, W.; Young, E.J.; Higginbotham, E.J.; MacDonald, J.R.; Trost, B.; Chan, A.J.S.; Walker, S.; Lamoureux, S.; et al. A large data resource of genomic copy number variation across neurodevelopmental disorders. npj Genom. Med. 2019, 4, 26. [Google Scholar] [CrossRef] [PubMed]
- Lowther, C.; Speevak, M.; Armour, C.M.; Goh, E.S.; Graham, G.E.; Li, C.; Zeesman, S.; Nowaczyk, M.J.; Schultz, L.A.; Morra, A.; et al. Molecular characterization of NRXN1 deletions from 19,263 clinical microarray cases identifies exons important for neurodevelopmental disease expression. Genet. Med. 2017, 19, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Skiba, A.; Talarowska, M.; Szemraj, J.; Gałecki, P. Is NRXN1 Gene Expression an Important Marker of Treatment of Depressive Disorders? A Pilot Study. J. Pers. Med. 2021, 11, 637. [Google Scholar] [CrossRef] [PubMed]
- Todarello, G.; Feng, N.; Kolachana, B.S.; Li, C.; Vakkalanka, R.; Bertolino, A.; Weinberger, D.R.; Straub, R.E. Incomplete penetrance of NRXN1 deletions in families with schizophrenia. Schizophr. Res. 2014, 155, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, B.; Campbell, D.B. Contribution of long noncoding RNAs to autism spectrum disorder risk. Int. Rev. Neurobiol. 2013, 113, 35–59. [Google Scholar] [CrossRef] [PubMed]
- Duong, L.T.; Hoeffding, L.K.; Petersen, K.B.; Knudsen, C.D.; Thygesen, J.H.; Klitten, L.L.; Tommerup, N.; Ingason, A.; Werge, T. Two rare deletions upstream of the NRXN1 gene (2p16.3) affecting the non-coding mRNA AK127244 segregate with diverse psychopathological phenotypes in a family. Eur. J. Med. Genet. 2015, 58, 650–653. [Google Scholar] [CrossRef]
- Rizzo, A.; Alfei, E.; Zibordi, F.; Saletti, V.; Zorzi, G.; Freri, E.; Estienne, M.; Girgenti, V.; D’Arrigo, S.; Esposito, S.; et al. The noncoding RNA AK127244 in 2p16.3 locus: A new susceptibility region for neuropsychiatric disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2018, 177, 557–562. [Google Scholar] [CrossRef]
- López-Cruz, C.; Cano-López, I.; Aliño, M.; Puig-Pérez, S. West Syndrome and associated Autism Spectrum Disorder: Proposal for a neuropsychological assessment and intervention protocol. Papeles Psicól. 2022, 43, 125–132. [Google Scholar] [CrossRef]
- Ouss, L.; Saint-Georges, C.; Robel, L.; Bodeau, N.; Laznik, M.-C.; Crespin, G.C.; Chetouani, M.; Bursztejn, C.; Golse, B.; Nabbout, R.; et al. Infant’s engagement and emotion as predictors of autism or intellectual disability in West syndrome. Eur. Child Adolesc. Psychiatry 2014, 23, 143–149. [Google Scholar] [CrossRef]
- Strasser, L.; Downes, M.; Kung, J.; Cross, J.H.; De Haan, M. Prevalence and risk factors for autism spectrum disorder in epilepsy: A systematic review and meta-analysis. Dev. Med. Child Neurol. 2018, 60, 19–29. [Google Scholar] [CrossRef]
- Schuck, R.K.; Flores, R.E.; Fung, L.K. Brief Report: Sex/Gender Differences in Symptomology and Camouflaging in Adults with Autism Spectrum Disorder. J. Autism Dev. Disord. 2019, 49, 2597–2604. [Google Scholar] [CrossRef] [PubMed]
- Taşkıran, E.Z.; Karaosmanoğlu, B.; Koşukcu, C.; Ürel-Demir, G.; Akgün-Doğan, Ö.; Şimşek-Kiper, P.; Alikaşifoğlu, M.; Boduroğlu, K.; Utine, G.E. Diagnostic yield of whole-exome sequencing in non-syndromic intellectual disability. J. Intellect. Disabil. Res. 2021, 65, 577–588. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhu, J.; You, L.; Wang, J.; Zhang, S.; Liu, Z.; Xu, Q.; Yuan, X.; Yang, L.; Wang, W.; et al. NRXN1 depletion in the medial prefrontal cortex induces anxiety-like behaviors and abnormal social phenotypes along with impaired neurite outgrowth in rat. J. Neurodev. Disord. 2023, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Hu, Z.; Zhang, L.; Liu, H.; Cheng, Y.; Xia, K.; Zhang, X. Not all neuroligin 3 and 4X missense variants lead to significant functional inactivation. Brain Behav. 2017, 7, e00793. [Google Scholar] [CrossRef] [PubMed]
- Gregor, A.; Albrecht, B.; Bader, I.; Bijlsma, E.K.; Ekici, A.B.; Engels, H.; Hackmann, K.; Horn, D.; Hoyer, J.; Klapecki, J. Expanding the clinical spectrum associated with defects in CNTNAP2 and NRXN1. BMC Med. Genet. 2011, 12, 106. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, C.P.; Boone, P.M.; Sampath, S.; Williams, C.; Bader, P.I.; Mueller, J.M.; Shchelochkov, O.A.; Brown, C.W.; Crawford, H.P.; Phalen, J.A. Phenotypic spectrum and genotype–phenotype correlations of NRXN1 exon deletions. Eur. J. Hum. Genet. 2012, 20, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
- Pena, S.A.; Iyengar, R.; Eshraghi, R.S.; Bencie, N.; Mittal, J.; Aljohani, A.; Mittal, R.; Eshraghi, A.A. Gene therapy for neurological disorders: Challenges and recent advancements. J. Drug Target. 2020, 28, 111–128. [Google Scholar] [CrossRef]
- Preta, G. Development of New Genome Editing Tools for the Treatment of Hyperlipidemia. Cells 2023, 12, 2466. [Google Scholar] [CrossRef]
- Sandhu, A.; Kumar, A.; Rawat, K.; Gautam, V.; Sharma, A.; Saha, L. Modernising autism spectrum disorder model engineering and treatment via CRISPR-Cas9: A gene reprogramming approach. World J. Clin. Cases 2023, 11, 3114–3127. [Google Scholar] [CrossRef]
- Gutiérrez-Rodríguez, A.; Cruz-Fuentes, C.S.; Genis-Mendoza, A.D.; Nicolini, H. CRISPR/Cas9 Genome Editing Approaches for Psychiatric Research. Braz. J. Psychiatry 2023, 45, 137–145. [Google Scholar] [CrossRef]
- Karimian, A.; Azizian, K.; Parsian, H.; Rafieian, S.; Shafiei-Irannejad, V.; Kheyrollah, M.; Yousefi, M.; Majidinia, M.; Yousefi, B. CRISPR/Cas9 technology as a potent molecular tool for gene therapy. J. Cell. Physiol. 2019, 234, 12267–12277. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Jindal, D.; Singh, M. Gene Therapy, A Potential Therapeutic Tool for Neurological and Neuropsychiatric Disorders: Applications, Challenges and Future Perspective. Curr. Gene Ther. 2023, 23, 20–40. [Google Scholar] [CrossRef] [PubMed]
- Thapar, N.; Eid, M.A.F.; Raj, N.; Kantas, T.; Billing, H.S.; Sadhu, D. Application of CRISPR/Cas9 in the management of Alzheimer’s disease and Parkinson’s disease: A review. Ann. Med. Surg. 2024, 86, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Weuring, W.; Geerligs, J.; Koeleman, B.P.C. Gene Therapies for Monogenic Autism Spectrum Disorders. Genes 2021, 12, 1667. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Howard, L.; Gallagher, L.; Shen, S. Regulation and postsynaptic binding of neurexins—Drug targets for neurodevelopmental and neuropsychiatric disorders. Front. Biol. 2015, 10, 239–251. [Google Scholar] [CrossRef]
- Chen, X.; Mangala, L.S.; Rodriguez-Aguayo, C.; Kong, X.; Lopez-Berestein, G.; Sood, A.K. RNA interference-based therapy and its delivery systems. Cancer Metastasis Rev. 2018, 37, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Arya, V.; Reynolds, K.S.; Rogers, H. Clinical Pharmacology of RNA Interference–Based Therapeutics: A Summary Based on Food and Drug Administration–Approved Small Interfering RNAs. Drug Metab. Dispos. 2023, 51, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Liguori, L.; Monticelli, M.; Allocca, M.; Hay Mele, B.; Lukas, J.; Cubellis, M.V.; Andreotti, G. Pharmacological Chaperones: A Therapeutic Approach for Diseases Caused by Destabilizing Missense Mutations. Int. J. Mol. Sci. 2020, 21, 489. [Google Scholar] [CrossRef] [PubMed]
- Vavilis, T.; Stamoula, E.; Ainatzoglou, A.; Sachinidis, A.; Lamprinou, M.; Dardalas, I.; Vizirianakis, I. mRNA in the Context of Protein Replacement Therapy. Pharmaceutics 2023, 15, 166. [Google Scholar] [CrossRef]
- Weidemann, F.; Jovanovic, A.; Herrmann, K.; Vardarli, I. Chaperone therapy in Fabry disease. Int. J. Mol. Sci. 2022, 23, 1887. [Google Scholar] [CrossRef]
- Yadav, D.; Malviya, R. Vector-Mediated Delivery of Transgenes and RNA Interference-Based Gene Silencing Sequences to Astrocytes for Disease Management: Advances and Prospectives. Curr. Gene Ther. 2024, 24, 110–121. [Google Scholar] [CrossRef] [PubMed]
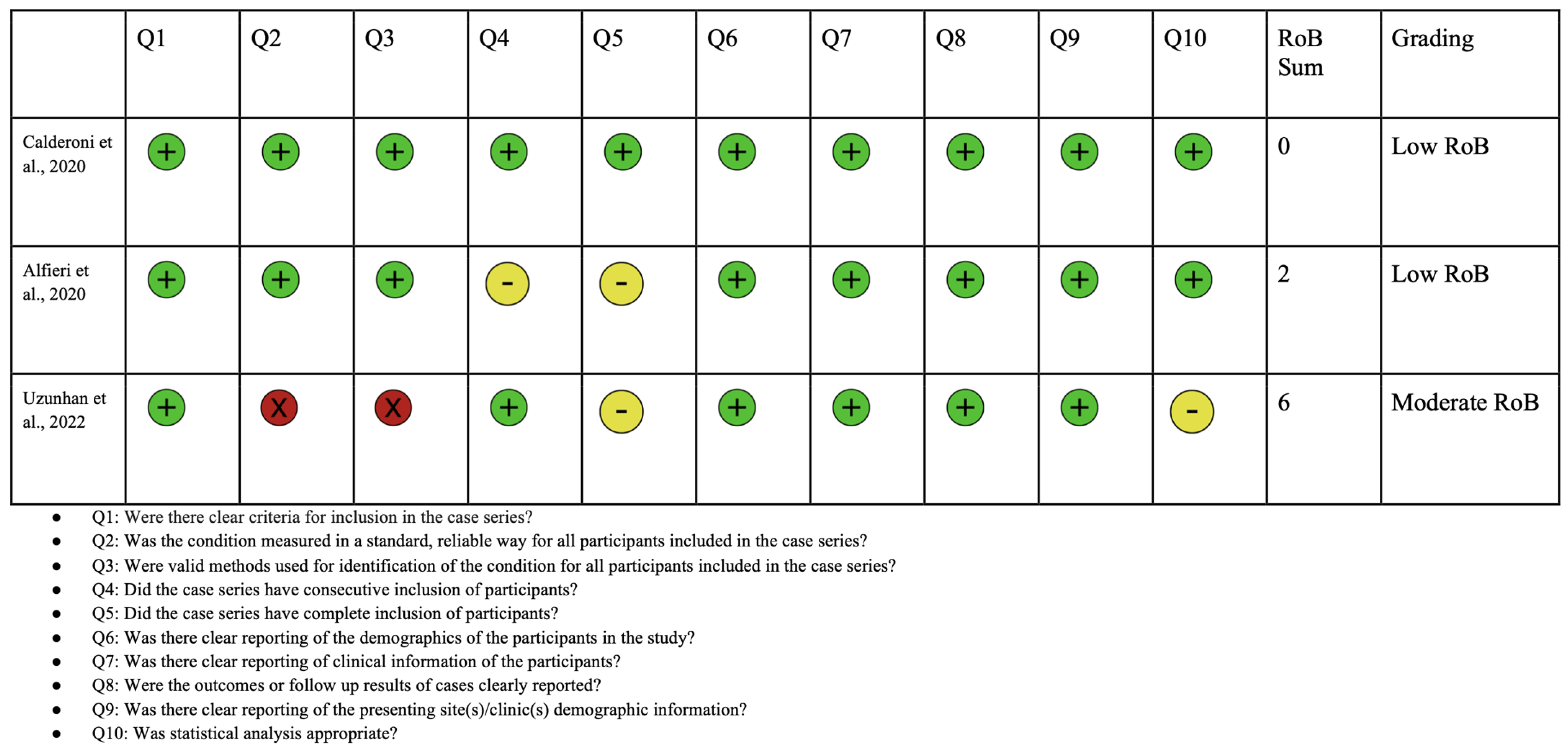
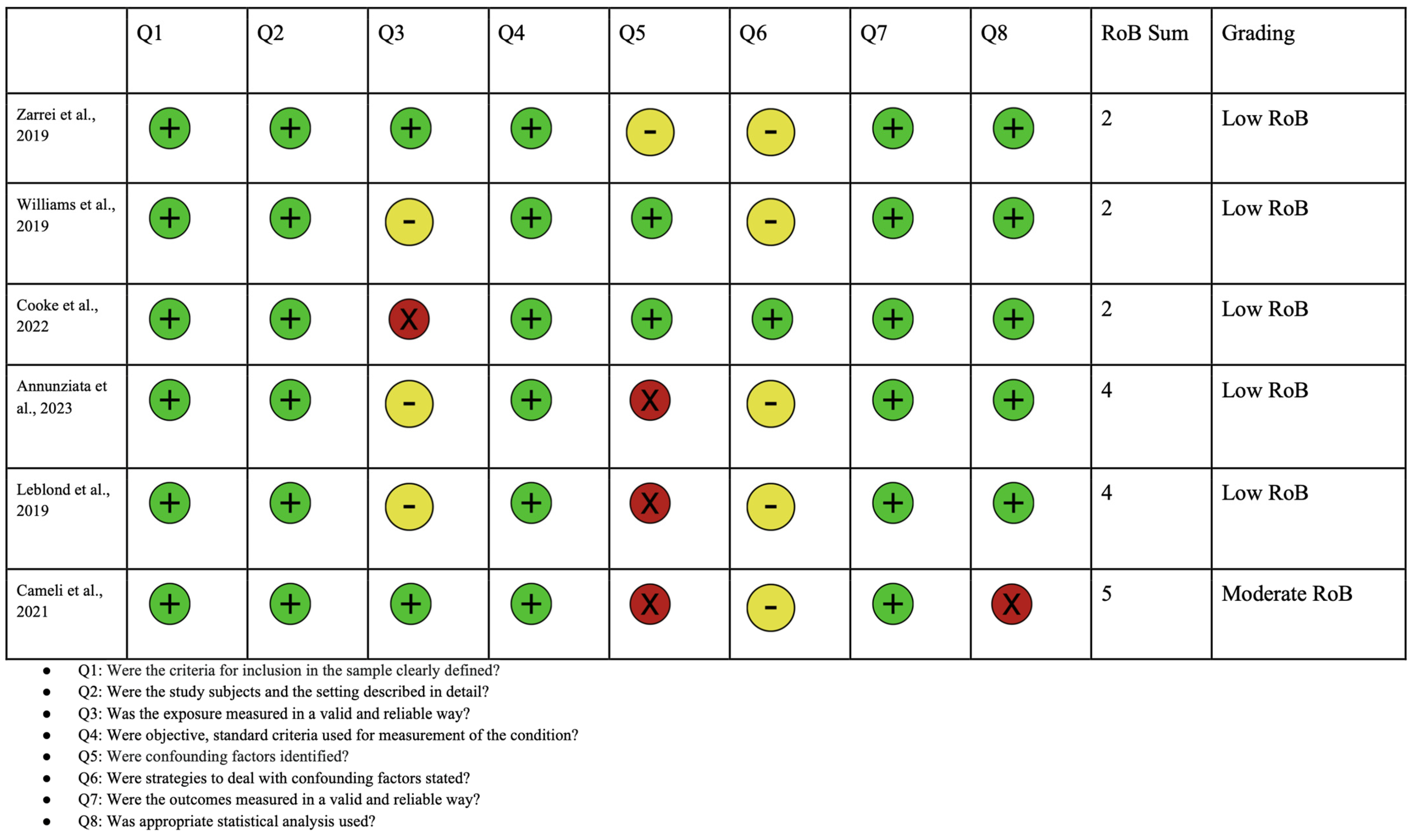
| Study | Diagnostic Test for ASD |
|---|---|
| Alfieri et al. [78], 2020 | ADOS2 |
| Annunziata et al. [81], 2023 | DSM5 |
| Calderoni et al. [79], 2020 | DSM 5 |
| Cameli et al. [82], 2021 | ADOS |
| Cooke et al. [27], 2022 | DSM 5 or ICD |
| Cosemans et al. [76], 2020 | DSM 5 |
| Ishizuka et al. [77], 2020 | DSM 5 |
| Leblond et al. [36], 2019 | ICD-10 criteria for childhood autism/autistic disorder Gillberg criteria for Asperger syndrome ICD-10 criteria for atypical autism with the added requirement that a case thus diagnosed could not meet full criteria for childhood autism or Asperger syndrome ICD-10 criteria for disintegrative disorder |
| Shehhi et. Al. [60], 2019 | Gold standard test—unspecified |
| Uzunhan et al. [80], 2022 | Gold standard test—unspecified |
| Williams et al. [83], 2019 | DSM 4 |
| Zarrei et al. [84], 2019 | ICD11 or DSMV |
| Reference | Total ASD Cases | Total ASD and NRXN1 Nutation Cases | Mutation in Alpha Isoform | Mutation in Beta Isoform | Unspecified Mutated Isoform | Number of Exonic Deletion | Number of Intronic Deletions | Frequency of Exonic Deletion | Frequency of Intronic Deletion | Homozygous Mutation | Heterozygous Mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alfieri et al. [78], 2020 | 5 | 3 | NR | NR | 3 | 0 | 0 | NR | NR | 0 | 3 |
| Annunziata et al. [81], 2023 | 209 | 3 | NR | NR | 3 | 0 | 0 | NR | NR | NR | NR |
| Calderoniet al. [79], 2020 | 93 | 2 | NR | NR | 2 | 0 | 0 | NR | NR | NR | NR |
| Cameli et al. [82], 2021 | 104 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | NR | NR |
| Cooke et al. [27], 2022 | 69 | 12 | NR | NR | 12 | 0 | 0 | NR | NR | NR | NR |
| Cosemans et al. [76], 2020 | 43 | 17 | NR | NR | 17 | 0 | 0 | NR | NR | NR | NR |
| Ishizuka et al. [77], 2020 | 192 | 5 | 3 | 0 | 2 | 3 | 0 | 0.6 | 0 | 0 | 5 |
| Leblond et al. [36], 2019 | 36 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | NR | NR |
| Shehhi et. Al. [60], 2019 | 20 | 20 | 13 | 2 | 5 | 13 | 7 | 0.65 | 0.35 | NR | NR |
| Uzunhan et al. [80], 2022 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 |
| Williams et al. [83], 2019 | 48 | 1 | 1 | 0 | 0 | 0 | 0 | NR | NR | 0 | 1 |
| Zarrei et al. [84], 2019 | 1838 | 6 | NR | NR | 6 | 6 | 0 | 1 | 0 | NR | NR |
| Reference | Mutation Location |
|---|---|
| Alfieri et al. [78], 2020 | arr[GRCh37] 2p16.3(50432664_50536137)x1 mat, arr[GRCh37] 2p16.3(51086847_51411126) x1 mat, arr [GRCh37] 2p16.3(51037104_52339655)x1 pat. |
| Annunziata et al. [81], 2023 | arr[GRCh37/hg19] 2p16.3(51066578_51100412)x1, arr[GRCh37/hg19] 2p16.3(51175725_51328842)x1 mat, arr[GRCh37/hg19] 2p16.3(50039172_50735499)x1 mat, |
| Calderoni et al. [79], 2020 | arr[GRCh37/hg19] 2p16.3 (50909765_51083469) 1x pat |
| Cameli et al. [82], 2021 | 2p16.3 (NC_000002.11:g.50170766_50982172del) |
| Cooke et al. [27], 2022 | unspecified from SynaG cohort |
| Cosemans et al. [76], 2020 | arr[GRCh37/hg19] 2p16.3 (50453695_50662935), arr[GRCh37/hg19] 2p16.3 (50879191_50953066) mat, arr[GRCh37/hg19] 2p16.3 (50620243_50970739), arr[GRCh37/hg19] 2p16.3 (51017528_51302432) mat, arr[GRCh37/hg19] 2p16.3 (50898653_51104632) pat, arr[GRCh37/hg19] 2p16.3 (50923553_51034676) pat, arr[GRCh37/hg19] 2p16.3 (51033135_51074619), arr[GRCh37/hg19] 2p16.3 (51033989_51062766), arr[GRCh37/hg19] 2p16.3 (50992089_51026709), arr[GRCh37/hg19] 2p16.3 (51027631_51390231), arr[GRCh37/hg19] 2p16.3 (51039779_51297569) mat, arr[GRCh37/hg19] 2p16.3 (50497204_50514746) mat, arr[GRCh37/hg19] 2p16.3 (50497204_50514746) mat, arr[GRCh37/hg19] 2p16.3 (51053925_51319222) mat, arr[GRCh37/hg19] 2p16.3 (50975806_51005275), arr[GRCh37/hg19] 2p16.3 (51063155_51278187), arr[GRCh37/hg19] 2p16.3 (51160878_51356269) pat |
| Ishizuka et al. [77], 2020 | rs201336161, rs201881725, rs1457374261, rs199970666 |
| Leblond et al. [36], 2019 | del(2p16:51125625-51255427) |
| Shehhi et. Al. [60], 2019 | Del(2p16.3:50,138,031–50,214,776), Del(2p16.3: Del(2p16.3:50,138,031–50,214,776), Del(2p16.3: Del(2p16.3: 50,483,652–50,495,891), Del(2p16.3: Del(2p16.3: 50,483,652–50,495,891), Del(2p16.3: Del(2p16.3: 50,690,984–50,870,064), Del(2p16.3: Del(2p16.3: 50,881,995–50,947,729), Del(2p16.3: Del(2p16.3: 50,947,670–50,964,907), Del(2p16.3: Del(2p16.3: 50,957,455–51,251,557), Del(2p16.3: Del(2p16.3: 50,964,848–51,251,557), Del(2p16.3: Del(2p16.3: 50,968,453–51,260,612), Del(2p16.3: Del(2p16.3: 50,982,113–51,446,873), Del(2p16.3: Del(2p16.3: 51,057,824–51,142,908), Del(2p16.3: Del(2p16.3: 51,083,410–51,172,182), Del(2p16.3: Del(2p16.3: 51,122,091–51,314,430), Del(2p16.3: Del(2p16.3: 51,122,091–51,382,872), Del(2p16.3: Del(2p16.3: 51,122,091–51,606,257), Del(2p16.3: Del(2p16.3: 51,137,071–51,314,430), Del(2p16.3: Del(2p16.3: 51,148,508–51,251,557), Del(2p16.3: Del(2p16.3: 51,153,052–51,260,612), Del(2p16.3: Del(2p16.3: 51,237,000–51,260,612) |
| Uzunhan et al. [80], 2022 | Del(2p16.3:chr2:51149007–51255411) |
| Williams et al. [83], 2019 | chr2:50847195; rs78540316 |
| Zarrei et al. [84], 2019 | Del(2p16.3:50,138,031–50,996,179)pat, Del(2p16.3: 50,986,743–51,644,735), Del(2p16.3: 51,125,058–51,263,149), Del(2p16.3: 51,141,571–51,363,855)pat, Del(2p16.3: 51,163,235–51,285,498)pat, Del(2p16.3: 51,163,990–51,285,498)pat |
| Reference | NRXN1 Isoform Effected | Other Molecular Findings | Parental Consanguinity | Family History | Developmental Delay | Intellectual Disability | Seizures | EEG | Motor Abnormalities (Movement, Speech) | Sensory abnormalities (Hearing, Vision) | Behavioral Abnormalities | Other |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calderoni et al. [79], 2020. Case 1 | NR | NR | NR | NR | NR | Normal IQ (>70) | NR | NR | Verbal/non-verbal | none | none | n/a |
| Calderoni et al. [79], 2020. Case 2 | NR | duplication at Xp22.33 | NR | NR | NR | Low IQ (<70) | NR | NR | none | none | none | n/a |
| Alfieri et al. [78], 2020. Case 1 | NR | None | NR | NR | NR | Below average on TDQ (30) | - | - | Hypotonia, no speech | none | Tantrums, aggression, self-injurious behavior | Trichotillomania, teeth grinding |
| Alfieri et al. [78], 2020. Case 2 | NR | None | NR | NR | NR | Below average TDQ (44) | - | - | Chewing difficulties only babbles | none | none | Smoking and medication exposure in utero |
| Alfieri et al. [78], 2020. Case 3 | NR | None | NR | NR | NR | Below average NVIQ (74) | + | - | Motor dysregulation | none | Attention problems | Multiple ear infections |
| Alfieri et al. [78], 2020. Case 4 | NR | None | NR | NR | NR | Below average FSIQ (50) | - | - | none | none | Paranoid ideation, aggressive behavior | allergies, sIgA deficiency, recurrent respiratory infections |
| Alfieri et al. [78], 2020. Case 5 | NR | None | NR | NR | NR | Below average NVIQ (72) | n/a | n/a | none | soliloquy | Shy, withdrawing, avoidant behavior | IUGR, sleep problems |
| Zarrei et al. [84], 2019 | NR | AK12724 | NR | NR | NR | NR | NR | NR | NR | NR | NR | NR |
| Cooke et al. [27], 2022 | Alpha, beta, theta | n/a | n/a | ASD, ADHD, anxiety, depression | yes | Below average to average | no | yes | Eye movements/gaze patterns | no | Repetitive and restrictive behaviors | n/a |
| Cosemans et al. [76], 2020 | beta | n/a | n/a | ASD, psychiatric problems, intellectual disability, IQ | yes | yes | no | no | Repetitive movements | no | Anxiety behaviors | n/a |
| Shehhi et. Al. [60], 2019 | Alpha and beta | n/a | n/a | Congenital heart disease, global development delay, epilepsy, intellectual disability, speech delay | yes | yes | Hallucinations, yes in some cases | yes | Gross motor delay | Sensorineural hearing loss | Speech and language delay, learning disability—32/34 had speech delay | n/a |
| Annunziata et al. [81], 2023 | n/a | Maternal inheritance in ⅘ subjects; incomplete penetrance | n/a | n/a | Developmental delay | Intellectual disability | n/a | Epileptiform discharge while sleeping or falling asleep | n/a | n/a | n/a | n/a |
| Williams et al. [83], 2019 | Alpha | Paternal inheritance of miR-873-5p variant; maternal inheritance of NRXN1 loss of function | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| Leblond et al. [36], 2019 | alpha | none | yes | NR | NR | Intellectual disability | no | NR | NR | NR | NR | Congenital torticollis and dental carries |
| Uzunhan et al. [80], 2022 | NR | alpha | yes | no | NR | NR | No | NR | Yes | NR | Yes | Macrocephaly, frontal bossing, bitemporal narrowing, wide forehead, long face, thin upper lip |
| Cameli et al. [82], 2021 | NR | Other rare variants found unspecified | NR | Maternal history of mutation—no family history of ASD | Yes | NR | NR | Predominance of a slow background activity in the R temporal region | Yes, delayed with motor stereotypies (hand flapping); limited speech (four words) | Yes, manipulating materials for visual, acoustic, tactile stimulation | Yes—hyperactivity, short attention span | |
| Ishizuka et al. [77], 2020 | Alpha | NR | NR | Maternally inherited | NR | Yes, No | NR | NR | NR | NR | NR | [ODD (oppositional defiant disorder)], [Depression, ADHD] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cooper, J.N.; Mittal, J.; Sangadi, A.; Klassen, D.L.; King, A.M.; Zalta, M.; Mittal, R.; Eshraghi, A.A. Landscape of NRXN1 Gene Variants in Phenotypic Manifestations of Autism Spectrum Disorder: A Systematic Review. J. Clin. Med. 2024, 13, 2067. https://doi.org/10.3390/jcm13072067
Cooper JN, Mittal J, Sangadi A, Klassen DL, King AM, Zalta M, Mittal R, Eshraghi AA. Landscape of NRXN1 Gene Variants in Phenotypic Manifestations of Autism Spectrum Disorder: A Systematic Review. Journal of Clinical Medicine. 2024; 13(7):2067. https://doi.org/10.3390/jcm13072067
Chicago/Turabian StyleCooper, Jaimee N., Jeenu Mittal, Akhila Sangadi, Delany L. Klassen, Ava M. King, Max Zalta, Rahul Mittal, and Adrien A. Eshraghi. 2024. "Landscape of NRXN1 Gene Variants in Phenotypic Manifestations of Autism Spectrum Disorder: A Systematic Review" Journal of Clinical Medicine 13, no. 7: 2067. https://doi.org/10.3390/jcm13072067
APA StyleCooper, J. N., Mittal, J., Sangadi, A., Klassen, D. L., King, A. M., Zalta, M., Mittal, R., & Eshraghi, A. A. (2024). Landscape of NRXN1 Gene Variants in Phenotypic Manifestations of Autism Spectrum Disorder: A Systematic Review. Journal of Clinical Medicine, 13(7), 2067. https://doi.org/10.3390/jcm13072067








