Pancreatic Neuroendocrine Tumor: The Case Report of a Patient with Germline FANCD2 Mutation and Tumor Analysis Using Single-Cell RNA Sequencing
Abstract
1. Introduction
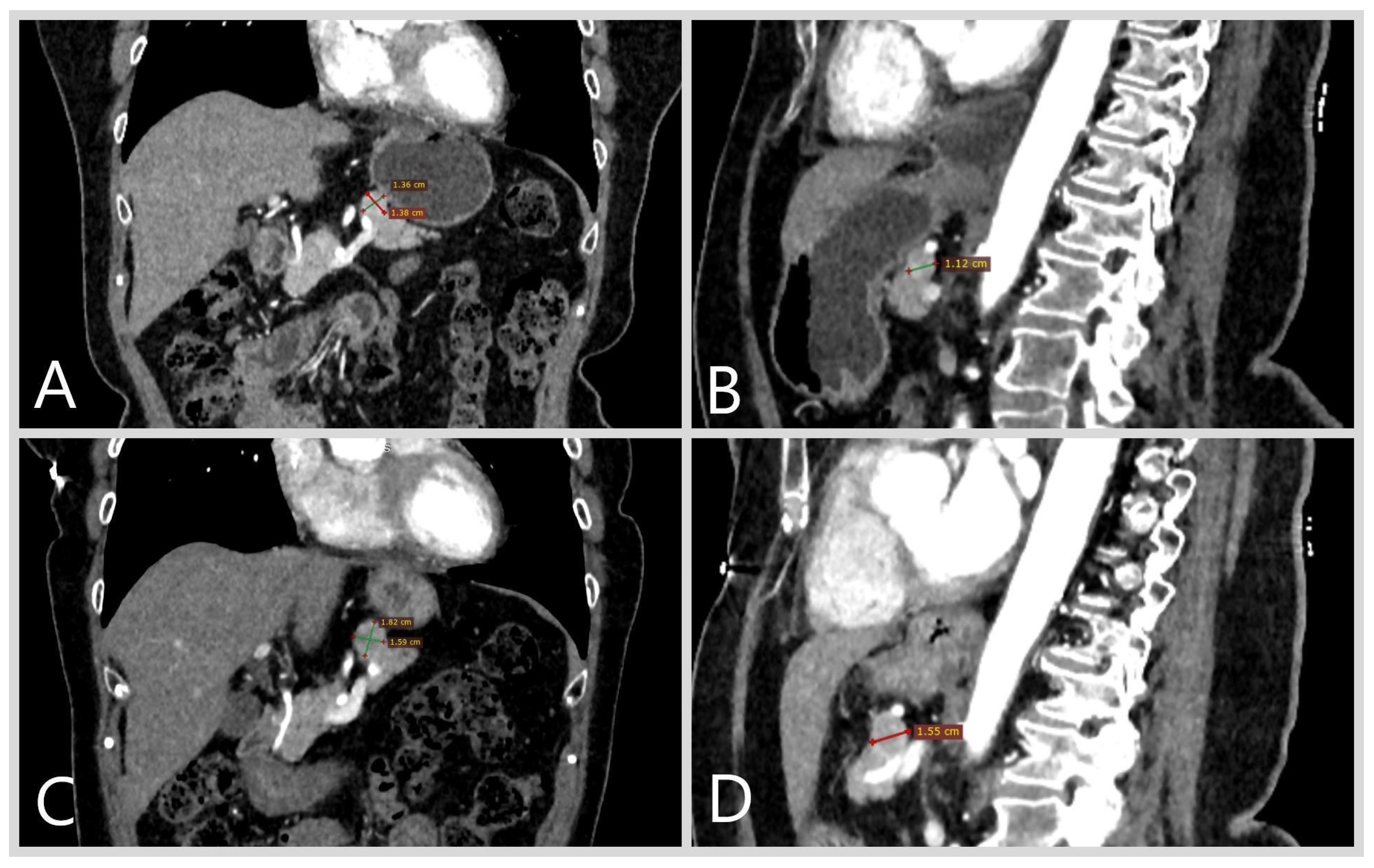
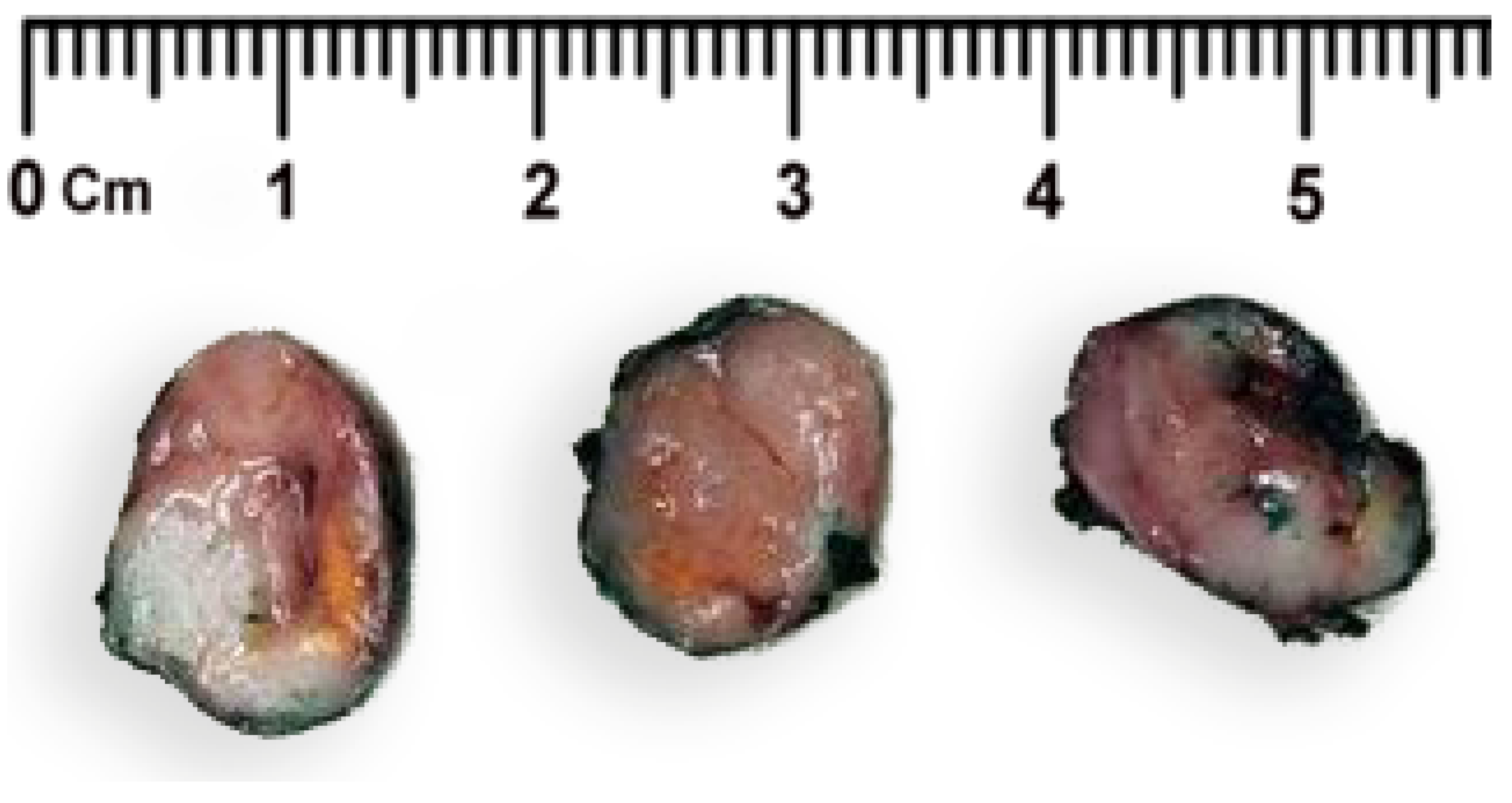
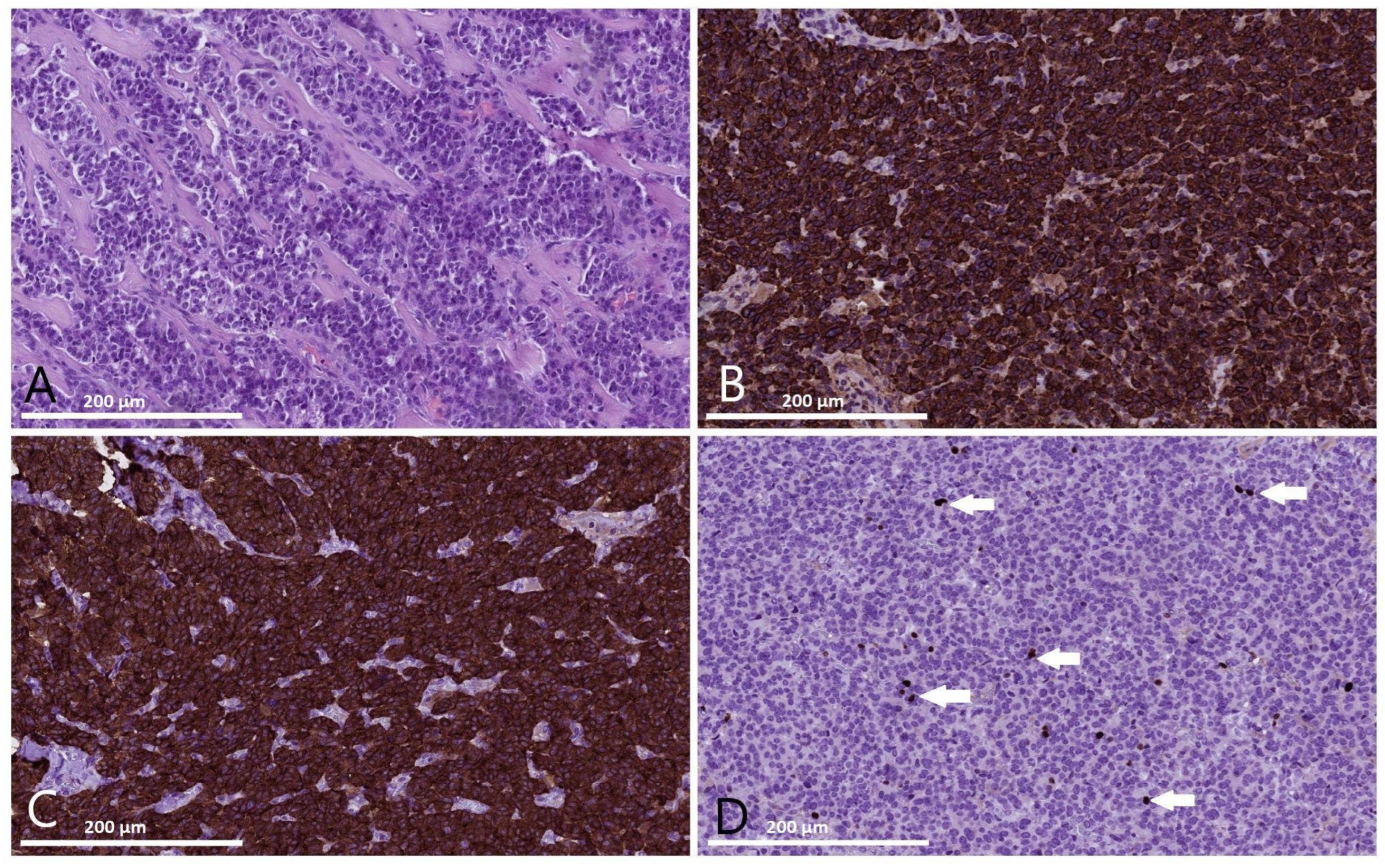
2. Case Report
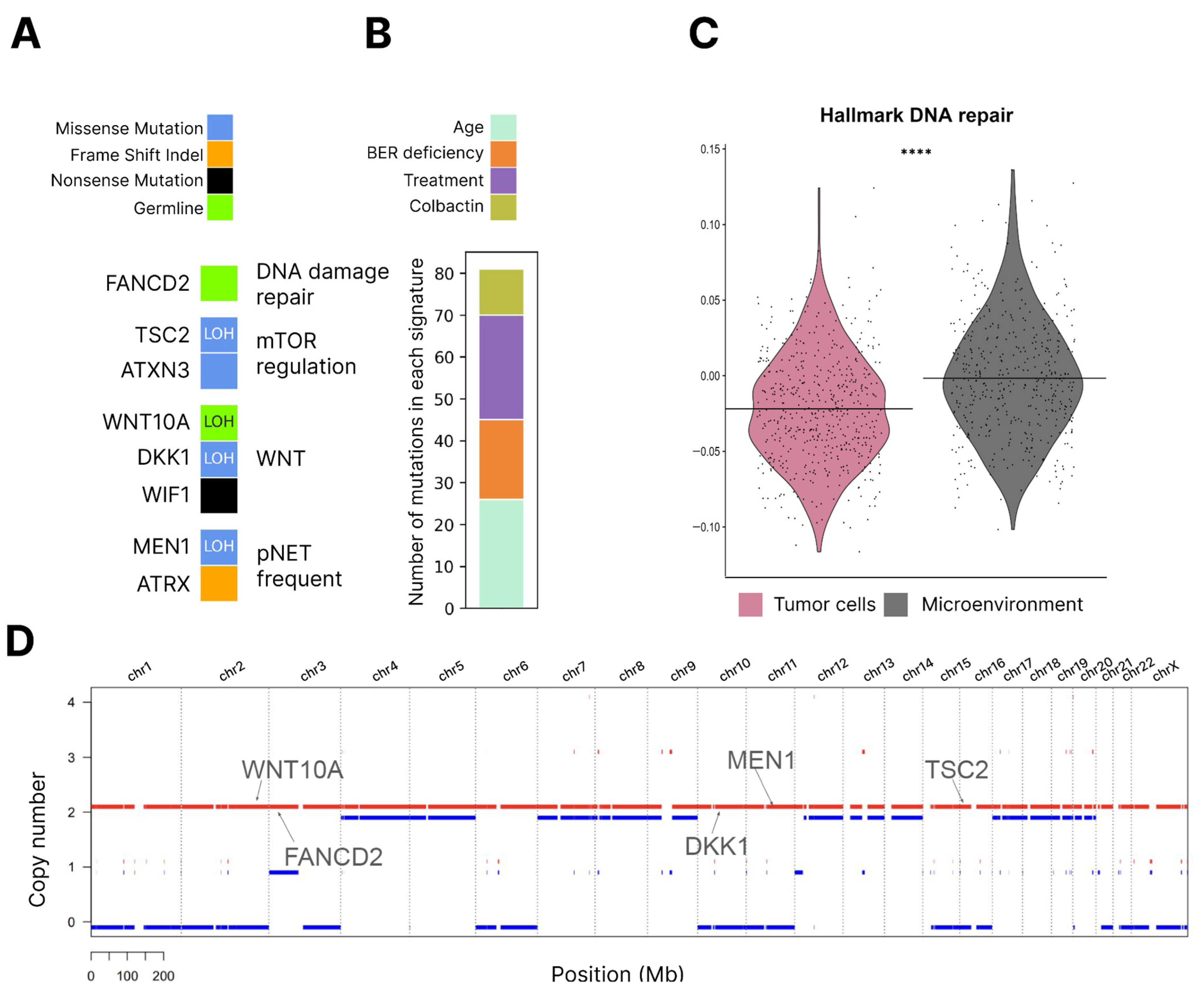
3. Discussion
Limitations
4. Materials and Methods
4.1. Sources of the Human Tissue and Blood Samples
4.2. Histopathology and Immunohistochemistry
4.3. Preparation of a Single-Cell Suspension, 10× Library Preparation and Sequencing
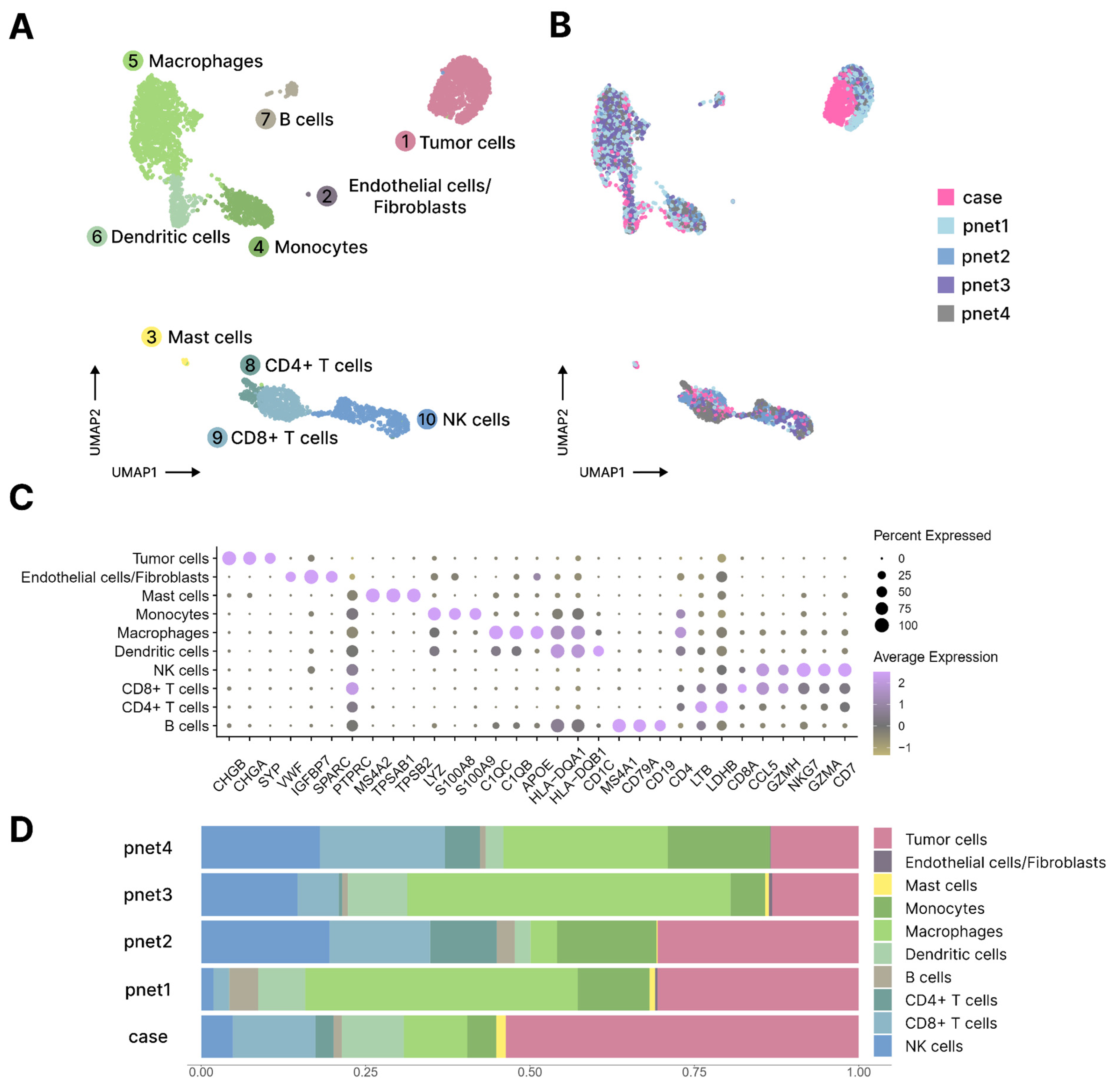
4.4. Single-Cell Transcriptome Data Preprocessing and Analysis
4.5. Hallmark Signature Scoring
4.6. Whole Exome Sequencing and Analysis
4.7. scRNA-Seq CNV Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTH | adrenocorticotropic hormone |
| AFP | alpha-fetoprotein |
| ALT | alternative lengthening of telomeres |
| ATRX | α-thalassemia/mental retardation syndrome X-linked protein |
| BER | base excision repair |
| BRCA1 | breast cancer-associated 1 protein |
| CA 19-9 | cancer antigen 19-9 |
| CCA | canonical correlation analysis |
| CEA | carcinoembryonic antigen |
| CNV | copy number variation |
| COSMIC | catalogue of somatic mutations in cancer |
| DAXX | death domain-associated protein |
| EUS-FNB | endoscopic-ultrasound fine-needle biopsy |
| GSEA | gene set enrichment analysis |
| HR | homologous recombination |
| HRD-score | homologous recombination deficiency score |
| ICL | interstrand crosslink |
| IHC | immunohistochemical |
| KEGG | Kyoto encyclopedia of genes and genomes |
| LOH | loss of heterozygosity |
| MEN1 | multiple endocrine neoplasia type 1 gene |
| MSI-score | microsatellite instability score |
| MSCT | multispiral computed tomography |
| NGS | next-generation sequencing |
| NECs | neuroendocrine carcinomas |
| NF1 | neurofibromatosis type 1 gene |
| PanNENs | pancreatic neuroendocrine neoplasms |
| PanNETs | pancreatic neuroendocrine tumors |
| SSTR2 | somatostatin receptor type 2 |
| scRNA-seq | single-cell RNA-sequencing |
| TCGA | The Cancer Genome Atlas |
| TMB | tumor mutational burden |
| TSC | tuberous sclerosis complex gene |
| UMI | unique molecular identifier |
| VAF | variant allele frequency |
| VHL | von Hippel-Lindau syndrome gene |
| WES | whole exome sequencing |
References
- Sun, J. Pancreatic neuroendocrine tumors. Intractable Rare Dis. Res. 2017, 6, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Cloyd, J.M.; Poultsides, G.A. Non-functional neuroendocrine tumors of the pancreas: Advances in diagnosis and management. World J. Gastroenterol. 2015, 21, 9512–9525. [Google Scholar] [CrossRef] [PubMed]
- Chatani, P.D.; Agarwal, S.K.; Sadowski, S.M. Molecular Signatures and Their Clinical Utility in Pancreatic Neuroendocrine Tumors. Front. Endocrinol. 2021, 11, 575620. [Google Scholar] [CrossRef]
- Basturk, O.; Tang, L.; Hruban, R.H.; Adsay, V.; Yang, Z.; Krasinskas, A.M.; Vakiani, E.; La Rosa, S.; Jang, K.T.; Frankel, W.L.; et al. Poorly differentiated neuroendocrine carcinomas of the pancreas: A clinicopathologic analysis of 44 cases. Am. J. Surg. Pathol. 2014, 38, 437–447. [Google Scholar] [CrossRef]
- Dasari, A.; Shen, C.; Halperin, D.; Zhao, B.; Zhou, S.; Xu, Y.; Shih, T.; Yao, J.C. Trends in the Incidence, Prevalence, and Survival Outcomes in Patients With Neuroendocrine Tumors in the United States. JAMA Oncol. 2017, 3, 1335–1342. [Google Scholar] [CrossRef]
- Sonbol, M.B.; Mazza, G.L.; Mi, L.; Oliver, T.; Starr, J.; Gudmundsdottir, H.; Cleary, S.P.; Hobday, T.; Halfdanarson, T.R. Survival and incidence patterns of pancreatic neuroendocrine tumors over the last 2 decades: A SEER database analysis. Oncologist 2022, 27, 573–578. [Google Scholar] [CrossRef]
- Gosain, R.; Ball, S.; Rana, N.; Groman, A.; Gage-Bouchard, E.; Dasari, A.; Mukherjee, S. Geographic and demographic features of neuroendocrine tumors in the United States of America: A population-based study. Cancer 2020, 126, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Sherman, S.K.; Howe, J.R.; Sahai, V. Progress in the Management of Pancreatic Neuroendocrine Tumors. Annu. Rev. Med. 2022, 73, 213–229. [Google Scholar] [CrossRef]
- de Ponthaud, C.; Menegaux, F.; Gaujoux, S. Updated Principles of Surgical Management of Pancreatic Neuroendocrine Tumours (pNETs): What Every Surgeon Needs to Know. Cancers 2021, 13, 5969. [Google Scholar] [CrossRef]
- Jutric, Z.; Grendar, J.; Hoen, H.M.; Cho, S.W.; Cassera, M.A.; Newell, P.H.; Hammill, C.W.; Hansen, P.D.; Wolf, R.F. Regional Metastatic Behavior of Nonfunctional Pancreatic Neuroendocrine Tumors: Impact of Lymph Node Positivity on Survival. Pancreas 2017, 46, 898–903. [Google Scholar] [CrossRef]
- Masui, T.; Sato, A.; Nakano, K.; Uchida, Y.; Yogo, A.; Anazawa, T.; Nagai, K.; Kawaguchi, Y.; Takaori, K.; Uemoto, S. Predictive value of the Ki67 index for lymph node metastasis of small non-functioning pancreatic neuroendocrine neoplasms. Surg. Today 2019, 49, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Ricci, C.; Casadei, R.; Taffurelli, G.; Pacilio, C.A.; Campana, D.; Ambrosini, V.; Donatella, S.; Minni, F. Sporadic Small (≤20 mm) Nonfunctioning Pancreatic Neuroendocrine Neoplasm: Is the Risk of Malignancy Negligible When Adopting a More Conservative Strategy? A Systematic Review and Meta-analysis. Ann. Surg. Oncol. 2017, 24, 2603–2610. [Google Scholar] [CrossRef] [PubMed]
- Mei, W.; Cao, F.; Lu, J.; Qu, C.; Fang, Z.; Li, J.; Li, F. Characteristics of small pancreatic neuroendocrine tumors and risk factors for invasion and metastasis. Front. Endocrinol. 2023, 14, 1140873. [Google Scholar] [CrossRef]
- Mohindroo, C.; McAllister, F.; De Jesus-Acosta, A. Genetics of Pancreatic Neuroendocrine Tumors. Hematol. Oncol. Clin. North Am. 2022, 36, 1033–1051. [Google Scholar] [CrossRef]
- Ciobanu, O.A.; Martin, S.C.; Herlea, V.; Fica, S. Insights into Epigenetic Changes Related to Genetic Variants and Cells-of-Origin of Pancreatic Neuroendocrine Tumors: An Algorithm for Practical Workup. Cancers 2022, 14, 4444. [Google Scholar] [CrossRef]
- Saleh, Z.; Moccia, M.C.; Ladd, Z.; Joneja, U.; Li, Y.; Spitz, F.; Hong, Y.K.; Gao, T. Pancreatic Neuroendocrine Tumors: Signaling Pathways and Epigenetic Regulation. Int. J. Mol. Sci. 2024, 25, 1331. [Google Scholar] [CrossRef]
- Shen, X.; Wang, X.; Lu, X.; Zhao, Y.; Guan, W. Molecular biology of pancreatic neuroendocrine tumors: From mechanism to translation. Front. Oncol. 2022, 12, 967071. [Google Scholar] [CrossRef]
- Jensen, R.T.; Berna, M.J.; Bingham, D.B.; Norton, J.A. Inherited pancreatic endocrine tumor syndromes: Advances in molecular pathogenesis, diagnosis, management, and controversies. Cancer 2008, 113 (Suppl. Sl), 1807–1843. [Google Scholar] [CrossRef] [PubMed]
- Lowney, J.K.; Frisella, M.M.; Lairmore, T.C.; Doherty, G.M. Pancreatic islet cell tumor metastasis in multiple endocrine neoplasia type 1: Correlation with primary tumor size. Surgery 1998, 124, 1043–1049. [Google Scholar] [CrossRef]
- Kouvaraki, M.A.; Shapiro, S.E.; Cote, G.J.; Lee, J.E.; Yao, J.C.; Waguespack, S.G.; Gagel, R.F.; Evans, D.B.; Perrier, N.D. Management of pancreatic endocrine tumors in multiple endocrine neoplasia type 1. World J. Surg. 2006, 30, 643–653. [Google Scholar] [CrossRef]
- Triponez, F.; Dosseh, D.; Goudet, P.; Cougard, P.; Bauters, C.; Murat, A.; Cadiot, G.; Niccoli-Sire, P.; Chayvialle, J.A.; Calender, A.; et al. Epidemiology data on 108 MEN 1 patients from the GTE with isolated nonfunctioning tumors of the pancreas. Ann. Surg. 2006, 243, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Libutti, S.K.; Choyke, P.L.; Alexander, H.R.; Glenn, G.; Bartlett, D.L.; Zbar, B.; Lubensky, I.; McKee, S.A.; Maher, E.R.; Linehan, W.M.; et al. Clinical and genetic analysis of patients with pancreatic neuroendocrine tumors associated with von Hippel-Lindau disease. Surgery 2000, 128, 1022–1028. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, I.; Nishimori, I.; Ashida, S.; Kohsaki, T.; Onishi, S.; Shuin, T. Clinical characteristics of pancreatic neuroendocrine tumors in Japanese patients with von Hippel-Lindau disease. Pancreas 2006, 33, 382–385. [Google Scholar] [CrossRef]
- Blansfield, J.A.; Choyke, L.; Morita, S.Y.; Choyke, P.L.; Pingpank, J.F.; Alexander, H.R.; Seidel, G.; Shutack, Y.; Yuldasheva, N.; Eugeni, M.; et al. Clinical, genetic and radiographic analysis of 108 patients with von Hippel-Lindau disease (VHL) manifested by pancreatic neuroendocrine neoplasms (PNETs). Surgery 2007, 142, 814–818.e8182. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, T.; Osuga, T.; Maeda, M.; Sakamoto, N.; Maeda, T.; Sakaguchi, K.; Onishi, Y.; Toyoda, M.; Maeda, H.; Miyamoto, K.; et al. Malignant endocrine tumor of the pancreas associated with von Recklinghausen’s disease. J. Gastroenterol. 2002, 37, 59–67. [Google Scholar] [CrossRef]
- Perren, A.; Wiesli, P.; Schmid, S.; Montani, M.; Schmitt, A.; Schmid, C.; Moch, H.; Komminoth, P. Pancreatic endocrine tumors are a rare manifestation of the neurofibromatosis type 1 phenotype: Molecular analysis of a malignant insulinoma in a NF-1 patient. Am. J. Surg. Pathol. 2006, 30, 1047–1051. [Google Scholar] [CrossRef]
- Nishi, T.; Kawabata, Y.; Hari, Y.; Imaoka, H.; Ishikawa, N.; Yano, S.; Maruyama, R.; Tajima, Y. A case of pancreatic neuroendocrine tumor in a patient with neurofibromatosis-1. World J. Surg. Oncol. 2012, 10, 153. [Google Scholar] [CrossRef]
- Eledrisi, M.S.; Stuart, C.A.; Alshanti, M. Insulinoma in a patient with tuberous sclerosis: Is there an association? Endocr. Pract. 2002, 8, 109–112. [Google Scholar] [CrossRef]
- Cives, M.; Partelli, S.; Palmirotta, R.; Lovero, D.; Mandriani, B.; Quaresmini, D.; Pelle’, E.; Andreasi, V.; Castelli, P.; Strosberg, J.; et al. DAXX mutations as potential genomic markers of malignant evolution in small nonfunctioning pancreatic neuroendocrine tumors. Sci. Rep. 2019, 9, 18614. [Google Scholar] [CrossRef]
- Singhi, A.D.; Liu, T.C.; Roncaioli, J.L.; Cao, D.; Zeh, H.J.; Zureikat, A.H.; Tsung, A.; Marsh, J.W.; Lee, K.K.; Hogg, M.E.; et al. Alternative Lengthening of Telomeres and Loss of DAXX/ATRX Expression Predicts Metastatic Disease and Poor Survival in Patients with Pancreatic Neuroendocrine Tumors. Clin. Cancer Res. 2017, 23, 600–609. [Google Scholar] [CrossRef]
- Marinoni, I.; Kurrer, A.S.; Vassella, E.; Dettmer, M.; Rudolph, T.; Banz, V.; Hunger, F.; Pasquinelli, S.; Speel, E.J.; Perren, A. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology 2014, 146, 453–460.e5. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, C.M.; Singhi, A.D. The diagnostic and prognostic utility of incorporating DAXX, ATRX, and alternative lengthening of telomeres to the evaluation of pancreatic neuroendocrine tumors. Hum. Pathol. 2022, 129, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Mastrosimini, M.G.; Manfrin, E.; Remo, A.; De Bellis, M.; Parisi, A.; Pedron, S.; Luchini, C.; Brunelli, M.; Ammendola, S.; Bernardoni, L.; et al. Endoscopic ultrasound fine-needle biopsy to assess DAXX/ATRX expression and alternative lengthening of telomeres status in non-functional pancreatic neuroendocrine tumors. Pancreatology 2023, 23, 429–436. [Google Scholar] [CrossRef]
- Park, J.K.; Paik, W.H.; Lee, K.; Ryu, J.K.; Lee, S.H.; Kim, Y.T. DAXX/ATRX and MEN1 genes are strong prognostic markers in pancreatic neuroendocrine tumors. Oncotarget 2017, 8, 49796–49806. [Google Scholar] [CrossRef]
- Hackeng, W.M.; Brosens, L.A.A.; Kim, J.Y.; O’Sullivan, R.; Sung, Y.N.; Liu, T.C.; Cao, D.; Heayn, M.; Brosnan-Cashman, J.; An, S.; et al. Non-functional pancreatic neuroendocrine tumours: ATRX/DAXX and alternative lengthening of telomeres (ALT) are prognostically independent from ARX/PDX1 expression and tumour size. Gut 2022, 71, 961–973. [Google Scholar] [CrossRef]
- Knies, K.; Schuster, B.; Ameziane, N.; Rooimans, M.; Bettecken, T.; de Winter, J.; Schindler, D. Genotyping of fanconi anemia patients by whole exome sequencing: Advantages and challenges. PLoS ONE 2012, 7, e52648. [Google Scholar] [CrossRef]
- Zhang, J.H.; Tasaki, T.; Tsukamoto, M.; Wang, K.Y.; Azuma, K. Deficiency of Wnt10a causes female infertility via the β-catenin/Cyp19a1 pathway in mice. Int. J. Med. Sci. 2022, 19, 701–710. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Liu, W.; Xu, Q.; Wang, C. Clinical significance and biological role of Wnt10a in ovarian cancer. Oncol. Lett. 2017, 14, 6611–6617. [Google Scholar] [CrossRef]
- Zhang, J.H.; Tasaki, T.; Tsukamoto, M.; Wang, K.Y.; Kubo, K.Y.; Azuma, K. Deletion of Wnt10a Is Implicated in Hippocampal Neurodegeneration in Mice. Biomedicines 2022, 10, 1500. [Google Scholar] [CrossRef]
- Volante, M.; Brizzi, M.P.; Faggiano, A.; La Rosa, S.; Rapa, I.; Ferrero, A.; Mansueto, G.; Righi, L.; Garancini, S.; Capella, C.; et al. Somatostatin receptor type 2A immunohistochemistry in neuroendocrine tumors: A proposal of scoring system correlated with somatostatin receptor scintigraphy. Mod. Pathol. 2007, 20, 1172–1182. [Google Scholar] [CrossRef]
- Hoffman, S.E.; Dowrey, T.W.; Villacorta Martin, C.; Bi, K.; Titchen, B.; Johri, S.; DelloStritto, L.; Patel, M.; Mackichan, C.; Inga, S.; et al. Intertumoral lineage diversity and immunosuppressive transcriptional programs in well-differentiated gastroenteropancreatic neuroendocrine tumors. Sci. Adv. 2023, 9, eadd9668. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, A.; Chang, D.K.; Nones, K.; Corbo, V.; Patch, A.M.; Bailey, P.; Lawlor, R.T.; Johns, A.L.; Miller, D.K.; Mafficini, A.; et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 2017, 543, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, B.; Blenkiron, C.; Parker, K.; Tsai, P.; Fitzgerald, S.; Shields, P.; Robb, T.; Yeong, M.L.; Kramer, N.; James, S.; et al. Recurrent loss of heterozygosity correlates with clinical outcome in pancreatic neuroendocrine cancer. NPJ Genom. Med. 2018, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Drost, J.; van Boxtel, R.; Blokzijl, F.; Mizutani, T.; Sasaki, N.; Sasselli, V.; de Ligt, J.; Behjati, S.; Grolleman, J.E.; van Wezel, T.; et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 2017, 358, 234–238. [Google Scholar] [CrossRef]
- Nepal, M.; Che, R.; Ma, C.; Zhang, J.; Fei, P. FANCD2 and DNA Damage. Int. J. Mol. Sci. 2017, 18, 1804. [Google Scholar] [CrossRef]
- Zhao, S.; Huang, C.; Yang, Y.; Xu, W.; Yu, Y.; Wen, C.; Cao, L.; Gao, F.; Qin, Y.; Chen, Z.J.; et al. DNA repair protein FANCD2 has both ubiquitination-dependent and ubiquitination-independent functions during germ cell development. J. Biol. Chem. 2023, 299, 102905. [Google Scholar] [CrossRef]
- Olazabal-Herrero, A.; He, B.; Kwon, Y.; Gupta, A.K.; Dutta, A.; Huang, Y.; Boddu, P.; Liang, Z.; Liang, F.; Teng, Y.; et al. The FANCI/FANCD2 complex links DNA damage response to R-loop regulation through SRSF1-mediated mRNA export. Cell Rep. 2024, 43, 113610. [Google Scholar] [CrossRef]
- Pilati, C.; Shinde, J.; Alexandrov, L.B.; Assié, G.; André, T.; Hélias-Rodzewicz, Z.; Ducoudray, R.; Le Corre, D.; Zucman-Rossi, J.; Emile, J.F.; et al. Mutational signature analysis identifies MUTYH deficiency in colorectal cancers and adrenocortical carcinomas. J. Pathol. 2017, 242, 10–15. [Google Scholar] [CrossRef]
- Liu, W.; Polaczek, P.; Roubal, I.; Meng, Y.; Choe, W.C.; Caron, M.C.; Sedgeman, C.A.; Xi, Y.; Liu, C.; Wu, Q.; et al. FANCD2 and RAD51 recombinase directly inhibit DNA2 nuclease at stalled replication forks and FANCD2 acts as a novel RAD51 mediator in strand exchange to promote genome stability. Nucleic. Acids. Res. 2023, 51, 9144–9165. [Google Scholar] [CrossRef]
- Ahn, V.E.; Chu, M.L.; Choi, H.J.; Tran, D.; Abo, A.; Weis, W.I. Structural basis of Wnt signaling inhibition by Dickkopf binding to LRP5/6. Dev. Cell 2011, 21, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Poggi, L.; Casarosa, S.; Carl, M. An Eye on the Wnt Inhibitory Factor Wif1. Front. Cell Dev. Biol. 2018, 6, 167. [Google Scholar] [CrossRef] [PubMed]
- Missiaglia, E.; Dalai, I.; Barbi, S.; Beghelli, S.; Falconi, M.; della Peruta, M.; Piemonti, L.; Capurso, G.; Di Florio, A.; delle Fave, G.; et al. Pancreatic endocrine tumors: Expression profiling evidences a role for AKT-mTOR pathway. J. Clin. Oncol. 2010, 28, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Smetsers, S.; Muter, J.; Bristow, C.; Patel, L.; Chandler, K.; Bonney, D.; Wynn, R.F.; Whetton, A.D.; Will, A.M.; Rockx, D.; et al. Heterozygote FANCD2 mutations associated with childhood T Cell ALL and testicular seminoma. Fam. Cancer 2012, 11, 661–665. [Google Scholar] [CrossRef]
- Akbari, M.R.; Malekzadeh, R.; Lepage, P.; Roquis, D.; Sadjadi, A.R.; Aghcheli, K.; Yazdanbod, A.; Shakeri, R.; Bashiri, J.; Sotoudeh, M.; et al. Mutations in Fanconi anemia genes and the risk of esophageal cancer. Hum. Genet. 2011, 129, 573–582. [Google Scholar] [CrossRef]
- Pejovic, T.; Yates, J.E.; Liu, H.Y.; Hays, L.E.; Akkari, Y.; Torimaru, Y.; Keeble, W.; Rathbun, R.K.; Rodgers, W.H.; Bale, A.E.; et al. Cytogenetic instability in ovarian epithelial cells from women at risk of ovarian cancer. Cancer Res. 2006, 66, 9017–9025. [Google Scholar] [CrossRef]
- Alanazi, N.; Siyal, A.; Basit, S.; Shammas, M.; Al-Mukhaylid, S.; Aleem, A.; Mahmood, A.; Iqbal, Z. Clinical Validation of the Somatic FANCD2 Mutation (c.2022-5C>T) as a Novel Molecular Biomarker for Early Disease Progression in Chronic Myeloid Leukemia: A Case-Control Study. Hematol. Rep. 2024, 16, 465–478. [Google Scholar] [CrossRef]
- Duan, X.; Zhao, M.; Yin, X.; Mi, L.; Shi, J.; Li, N.; Han, X.; Han, G.; Wang, J.; Hou, J.; et al. Molecular typing and mutational characterization of rectal neuroendocrine neoplasms. Cancer Med. 2023, 12, 16207–16220. [Google Scholar] [CrossRef]
- Banck, M.S.; Kanwar, R.; Kulkarni, A.A.; Boora, G.K.; Metge, F.; Kipp, B.R.; Zhang, L.; Thorland, E.C.; Minn, K.T.; Tentu, R.; et al. The genomic landscape of small intestine neuroendocrine tumors. J. Clin. Investig. 2013, 123, 2502–2508. [Google Scholar] [CrossRef]
- Ji, S.; Yang, W.; Liu, J.; Zhao, J.; Chen, L.; Ni, Q.; Long, J.; Yu, X. High throughput gene sequencing reveals altered landscape in DNA damage responses and chromatin remodeling in sporadic pancreatic neuroendocrine tumors. Pancreatology 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Zhao, Z.; Wang, R.; Wang, R.; Song, J.; Ma, F.; Pan, H.; Gao, C.; Wang, D.; Chen, X.; Fan, X. Pancancer analysis of the prognostic and immunological role of FANCD2: A potential target for carcinogenesis and survival. BMC Med. Genom. 2024, 17, 69. [Google Scholar] [CrossRef] [PubMed]
- Fagerholm, R.; Sprott, K.; Heikkinen, T.; Bartkova, J.; Heikkilä, P.; Aittomäki, K.; Bartek, J.; Weaver, D.; Blomqvist, C.; Nevanlinna, H. Overabundant FANCD2, alone and combined with NQO1, is a sensitive marker of adverse prognosis in breast cancer. Ann. Oncol. 2013, 24, 2780–2785. [Google Scholar] [CrossRef] [PubMed]
- van der Groep, P.; Hoelzel, M.; Buerger, H.; Joenje, H.; de Winter, J.P.; van Diest, P.J. Loss of expression of FANCD2 protein in sporadic and hereditary breast cancer. Breast Cancer Res. Treat. 2008, 107, 41–47. [Google Scholar] [CrossRef]
- Zheng, C.; Ren, Z.; Chen, H.; Yuan, X.; Suye, S.; Yin, H.; Zhou, Z.; Fu, C. FANCD2 promotes the malignant behavior of endometrial cancer cells and its prognostic value. Exp. Cell Res. 2022, 421, 113388. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, H.; Iwatsuki, M.; Mimori, K.; Sato, T.; Johansson, F.; Toh, H.; Watanabe, M.; Mori, M. FANCD2 mRNA overexpression is a bona fide indicator of lymph node metastasis in human colorectal cancer. Ann. Surg. Oncol. 2010, 17, 2341–2348. [Google Scholar] [CrossRef]
- Lei, L.C.; Yu, V.Z.; Ko, J.M.Y.; Ning, L.; Lung, M.L. FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression. Cancers 2020, 12, 2545. [Google Scholar] [CrossRef]
- Yang, Z.; Song, Y.; Li, Y.; Mao, Y.; Du, G.; Tan, B.; Zhang, H. Integrative analyses of prognosis, tumor immunity, and ceRNA network of the ferroptosis-associated gene FANCD2 in hepatocellular carcinoma. Front. Genet. 2022, 13, 955225. [Google Scholar] [CrossRef]
- Ye, C.; Lu, Y.; Yuan, Z.; Mi, M.; Qi, L.; Yuan, Y.; Weng, S. Ferroptosis regulator FANCD2 is associated with immune infiltration and predicts worse prognosis in lung adenocarcinoma. Front. Genet. 2022, 13, 922914. [Google Scholar] [CrossRef]
- Xie, X.; Zhao, Y.; Du, F.; Cai, B.; Fang, Z.; Liu, Y.; Sang, Y.; Ma, C.; Liu, Z.; Yu, X.; et al. Pan-cancer analysis of the tumorigenic role of Fanconi anemia complementation group D2 (FANCD2) in human tumors. Genomics 2024, 116, 110762. [Google Scholar] [CrossRef]
- Rudland, P.S.; Platt-Higgins, A.M.; Davies, L.M.; de Silva Rudland, S.; Wilson, J.B.; Aladwani, A.; Winstanley, J.H.; Barraclough, D.L.; Barraclough, R.; West, C.R.; et al. Significance of the Fanconi anemia FANCD2 protein in sporadic and metastatic human breast cancer. Am. J. Pathol. 2010, 176, 2935–2947. [Google Scholar] [CrossRef]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Incorvaia, L.; Perez, A.; Marchetti, C.; Brando, C.; Gristina, V.; Cancelliere, D.; Pivetti, A.; Contino, S.; Di Giovanni, E.; Barraco, N.; et al. Theranostic biomarkers and PARP-inhibitors effectiveness in patients with non-BRCA associated homologous recombination deficient tumors: Still looking through a dirty glass window? Cancer Treat. Rev. 2023, 121, 102650. [Google Scholar] [CrossRef] [PubMed]
- Mani, C.; Tripathi, K.; Chaudhary, S.; Somasagara, R.R.; Rocconi, R.P.; Crasto, C.; Reedy, M.; Athar, M.; Palle, K. Hedgehog/GLI1 Transcriptionally Regulates FANCD2 in Ovarian Tumor Cells: Its Inhibition Induces HR-Deficiency and Synergistic Lethality with PARP Inhibition. Neoplasia 2021, 23, 1002–1015. [Google Scholar] [CrossRef] [PubMed]
- Fallah, J.; Xu, J.; Weinstock, C.; Gao, X.; Heiss, B.L.; Maguire, W.F.; Chang, E.; Agrawal, S.; Tang, S.; Amiri-Kordestani, L.; et al. Efficacy of Poly(ADP-ribose) Polymerase Inhibitors by Individual Genes in Homologous Recombination Repair Gene-Mutated Metastatic Castration-Resistant Prostate Cancer: A US Food and Drug Administration Pooled Analysis. J. Clin. Oncol. 2024, 42, 1687–1698. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Carreira, S.; Sandhu, S.; Miranda, S.; Mossop, H.; Perez-Lopez, R.; Nava Rodrigues, D.; Robinson, D.; Omlin, A.; Tunariu, N.; et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N. Engl. J. Med. 2015, 373, 1697–1708. [Google Scholar] [CrossRef] [PubMed]
- Barkas, N.; Petukhov, V.; Kharchenko, P.; Steiger, S.; Rydbirk, R.; Biederstedt, E. pagoda2: Single Cell Analysis and Differential Expression; R Package Version 1.0.12; 2021. Available online: https://cran.r-project.org/web/packages/pagoda2/pagoda2.pdf (accessed on 27 October 2024).
- Wolock, S.L.; Lopez, R.; Klein, A.M. Scrublet Computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019, 8, 281–291.e9. [Google Scholar] [CrossRef]
- Yang, S.; Corbett, S.E.; Koga, Y.; Wang, Z.; Johnson, W.E.; Yajima, M.; Campbell, J.D. Decontamination of ambient RNA in single-cell RNA-seq with DecontX. Genome Biol. 2020, 21, 57. [Google Scholar] [CrossRef]
- Franzén, O.; Gan, L.-M.; Björkegren, J.L.M. PanglaoDB: A web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Díaz-Gay, M.; Vangara, R.; Barnes, M.; Wang, X.; Islam, S.M.A.; Vermes, I.; Narasimman, N.B.; Yang, T.; Jiang, Z.; Moody, S.; et al. Assigning mutational signatures to individual samples and individual somatic mutations with SigProfilerAssignment. Bioinformatics 2023, 39, btad756. [Google Scholar] [CrossRef]
- Deng, W.; Murugan, S.; Lindberg, J.; Chellappa, V.; Shen, X.; Pawitan, Y.; Vu, T.N. Fusion Gene Detection Using Whole-Exome Sequencing Data in Cancer Patients. Front. Genet. 2022, 13, 820493. [Google Scholar] [CrossRef] [PubMed]
- Favero, F.; Joshi, T.; Marquard, A.M.; Birkbak, N.J.; Krzystanek, M.; Li, Q.; Szallasi, Z.; Eklund, A.C. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2015, 26, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Jia, P.; Yang, X.; Guo, L.; Liu, B.; Lin, J.; Liang, H.; Sun, J.; Zhang, C.; Ye, K. MSIsensor-pro: Fast, Accurate, and Matched-normal-sample-free Detection of Microsatellite Instability. Genom. Proteom. Bioinform. 2020, 18, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Sztupinszki, Z.; Diossy, M.; Krzystanek, M.; Reiniger, L.; Csabai, I.; Favero, F.; Birkbak, N.J.; Eklund, A.C.; Syed, A.; Szallasi, Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer 2018, 4, 16. [Google Scholar] [CrossRef]
- Gao, T.; Soldatov, R.; Sarkar, H.; Kurkiewicz, A.; Biederstedt, E.; Loh, P.R.; Kharchenko, P.V. Haplotype-aware analysis of somatic copy number variations from single-cell transcriptomes. Nat. Biotechnol. 2023, 41, 417–426. [Google Scholar] [CrossRef]


| Test | Result | Normal Range |
|---|---|---|
| CA 19-9 (cancer antigen 19-9) | 9.89 U/mL | <37 U/mL |
| CEA (carcinoembryonic antigen) | 3.4 ng/mL | <5.0 ng/mL |
| AFP (alpha-fetoprotein) | 2.26 ng/mL | 0–7 ng/mL |
| gastrin | 59.0 ng/L | 13–115 ng/L |
| chromogranin A | 41.0 ng/mL | 0–100 ng/mL |
| calcitonin | 0.71 ng/L | 0.0–6.40 ng/L |
| serotonin | 7.93 µmol/L | 1.85–8.16 µmol/L |
| ACTH (adrenocorticotropic hormone) | 5 pg/mL | 0–46 pg/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avsievich, E.; Salimgereeva, D.; Maluchenko, A.; Antysheva, Z.; Voloshin, M.; Feidorov, I.; Glazova, O.; Abramov, I.; Maksimov, D.; Kaziakhmedova, S.; et al. Pancreatic Neuroendocrine Tumor: The Case Report of a Patient with Germline FANCD2 Mutation and Tumor Analysis Using Single-Cell RNA Sequencing. J. Clin. Med. 2024, 13, 7621. https://doi.org/10.3390/jcm13247621
Avsievich E, Salimgereeva D, Maluchenko A, Antysheva Z, Voloshin M, Feidorov I, Glazova O, Abramov I, Maksimov D, Kaziakhmedova S, et al. Pancreatic Neuroendocrine Tumor: The Case Report of a Patient with Germline FANCD2 Mutation and Tumor Analysis Using Single-Cell RNA Sequencing. Journal of Clinical Medicine. 2024; 13(24):7621. https://doi.org/10.3390/jcm13247621
Chicago/Turabian StyleAvsievich, Ekaterina, Diana Salimgereeva, Alesia Maluchenko, Zoia Antysheva, Mark Voloshin, Ilia Feidorov, Olga Glazova, Ivan Abramov, Denis Maksimov, Samira Kaziakhmedova, and et al. 2024. "Pancreatic Neuroendocrine Tumor: The Case Report of a Patient with Germline FANCD2 Mutation and Tumor Analysis Using Single-Cell RNA Sequencing" Journal of Clinical Medicine 13, no. 24: 7621. https://doi.org/10.3390/jcm13247621
APA StyleAvsievich, E., Salimgereeva, D., Maluchenko, A., Antysheva, Z., Voloshin, M., Feidorov, I., Glazova, O., Abramov, I., Maksimov, D., Kaziakhmedova, S., Bodunova, N., Karnaukhov, N., Volchkov, P., & Krupinova, J. (2024). Pancreatic Neuroendocrine Tumor: The Case Report of a Patient with Germline FANCD2 Mutation and Tumor Analysis Using Single-Cell RNA Sequencing. Journal of Clinical Medicine, 13(24), 7621. https://doi.org/10.3390/jcm13247621






