Circulating miR-18a and miR-532 Levels in Extrahepatic Cholangiocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Ethics
2.2. Patient Selection
2.3. Sample Collection and miRNA Analysis
2.4. Data Analysis
2.5. Statistical Analysis
3. Results
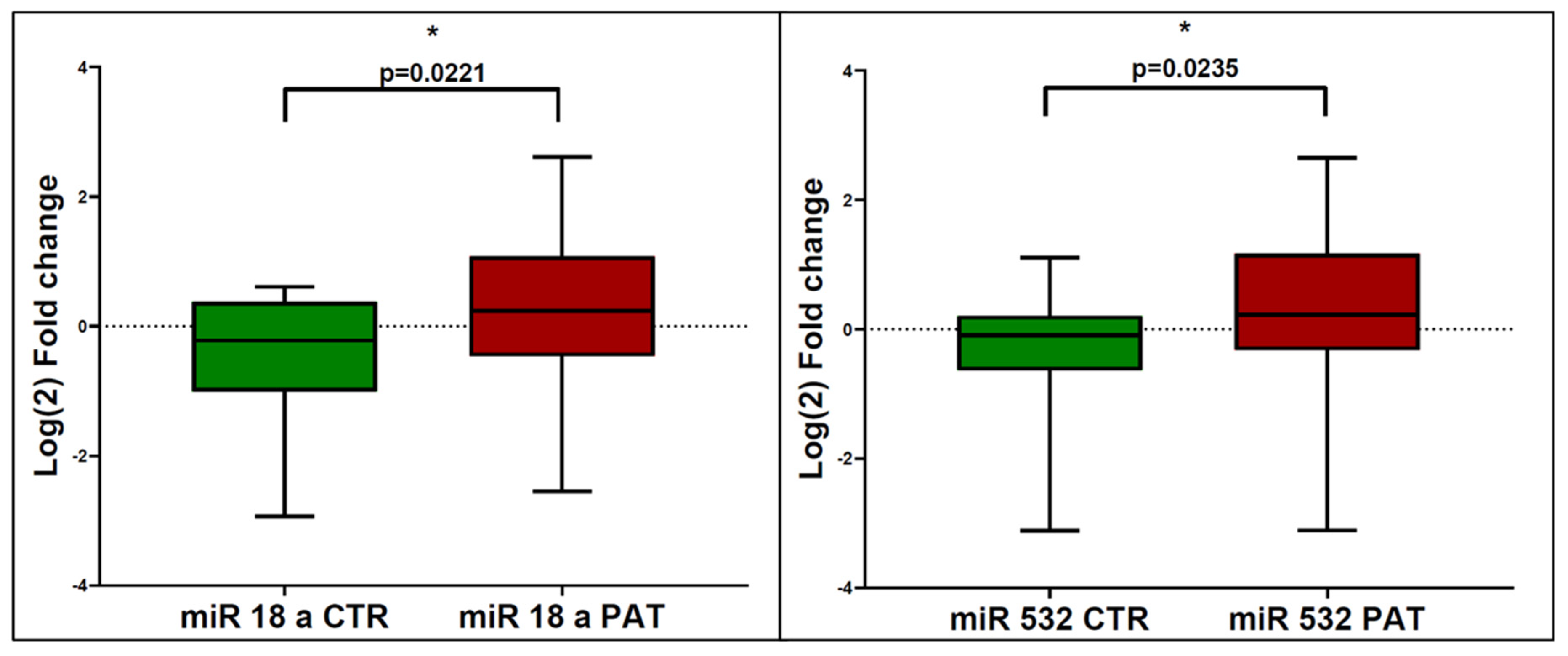
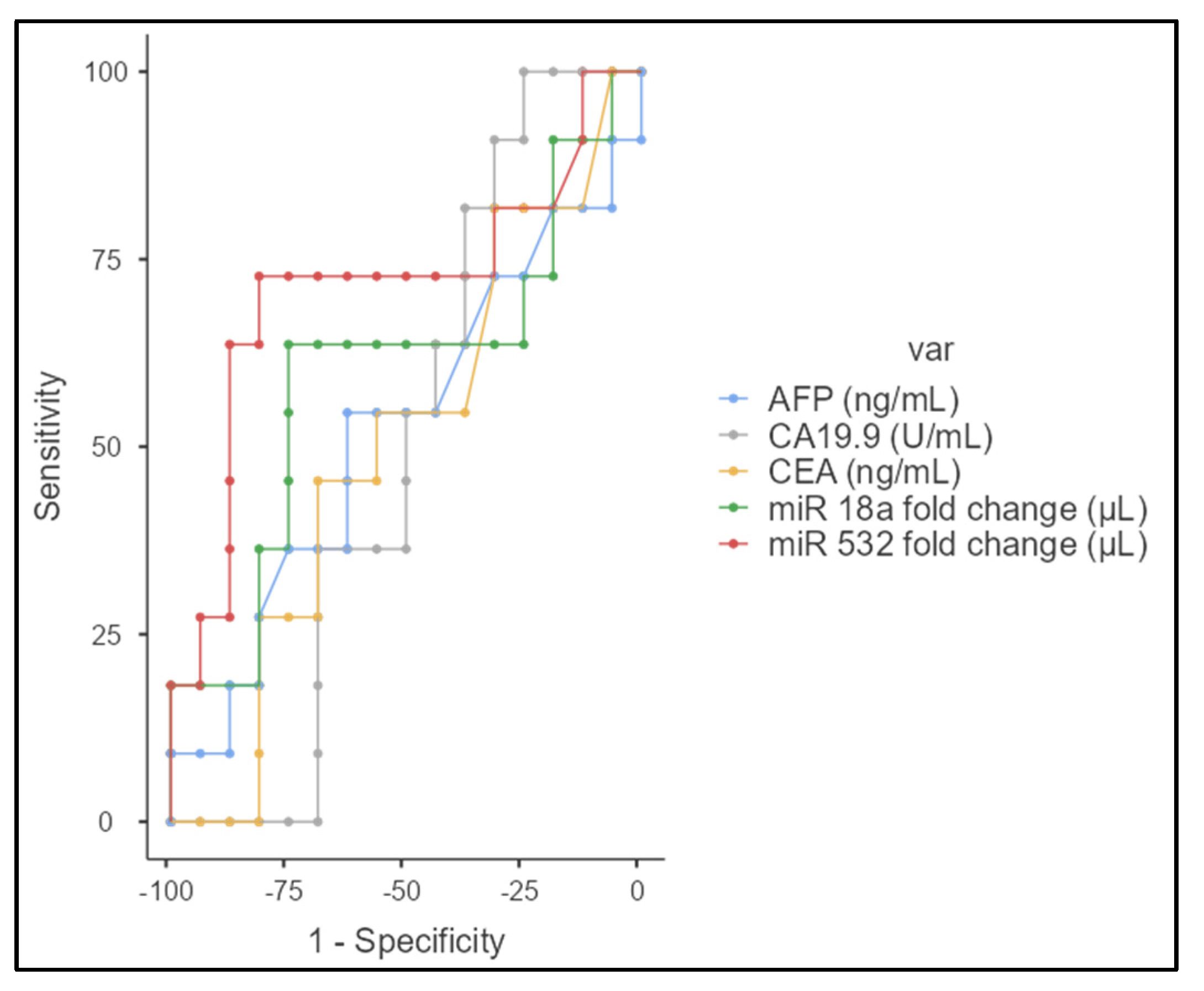
3.1. miRNA Expression in eCCA Patients and Controls
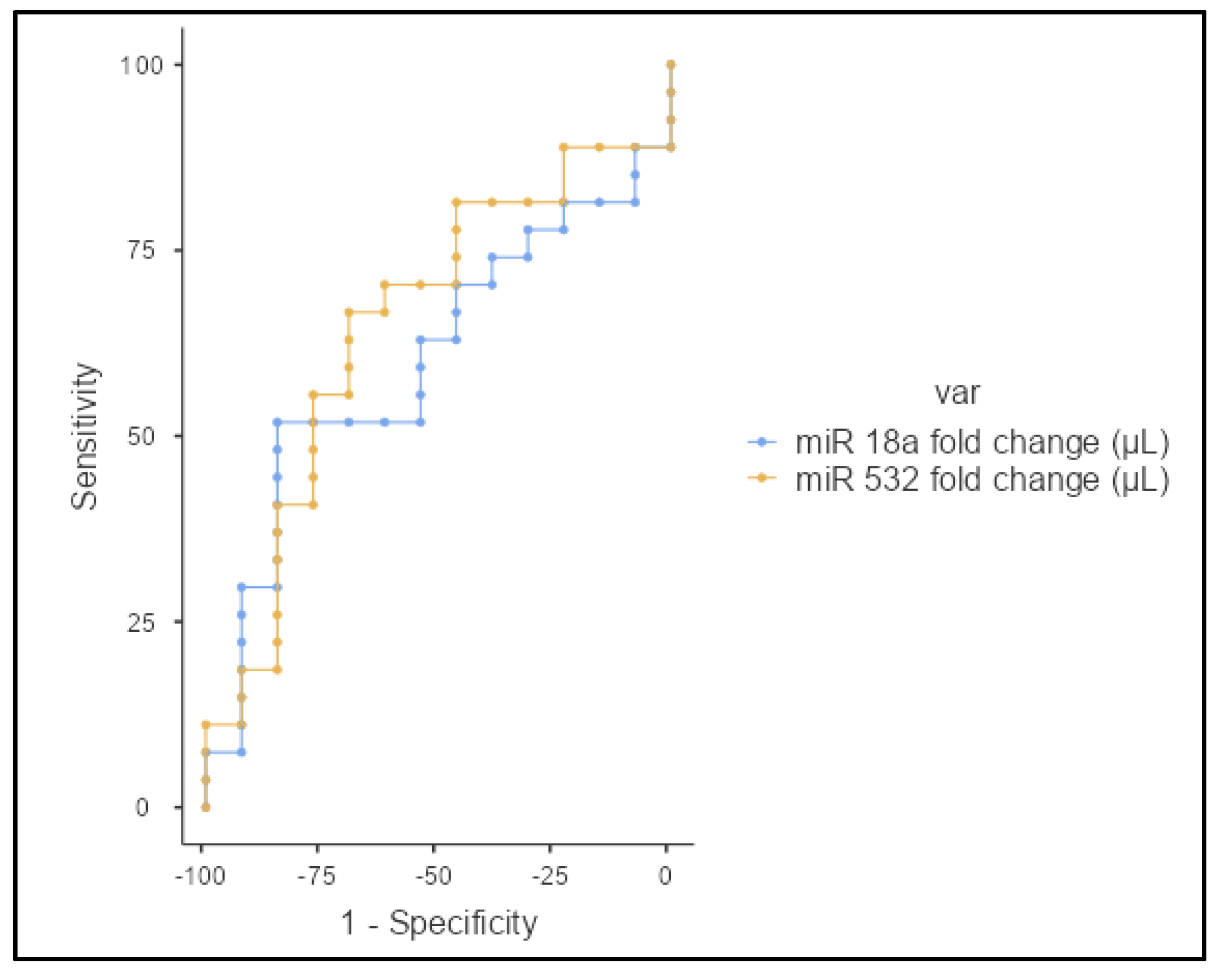
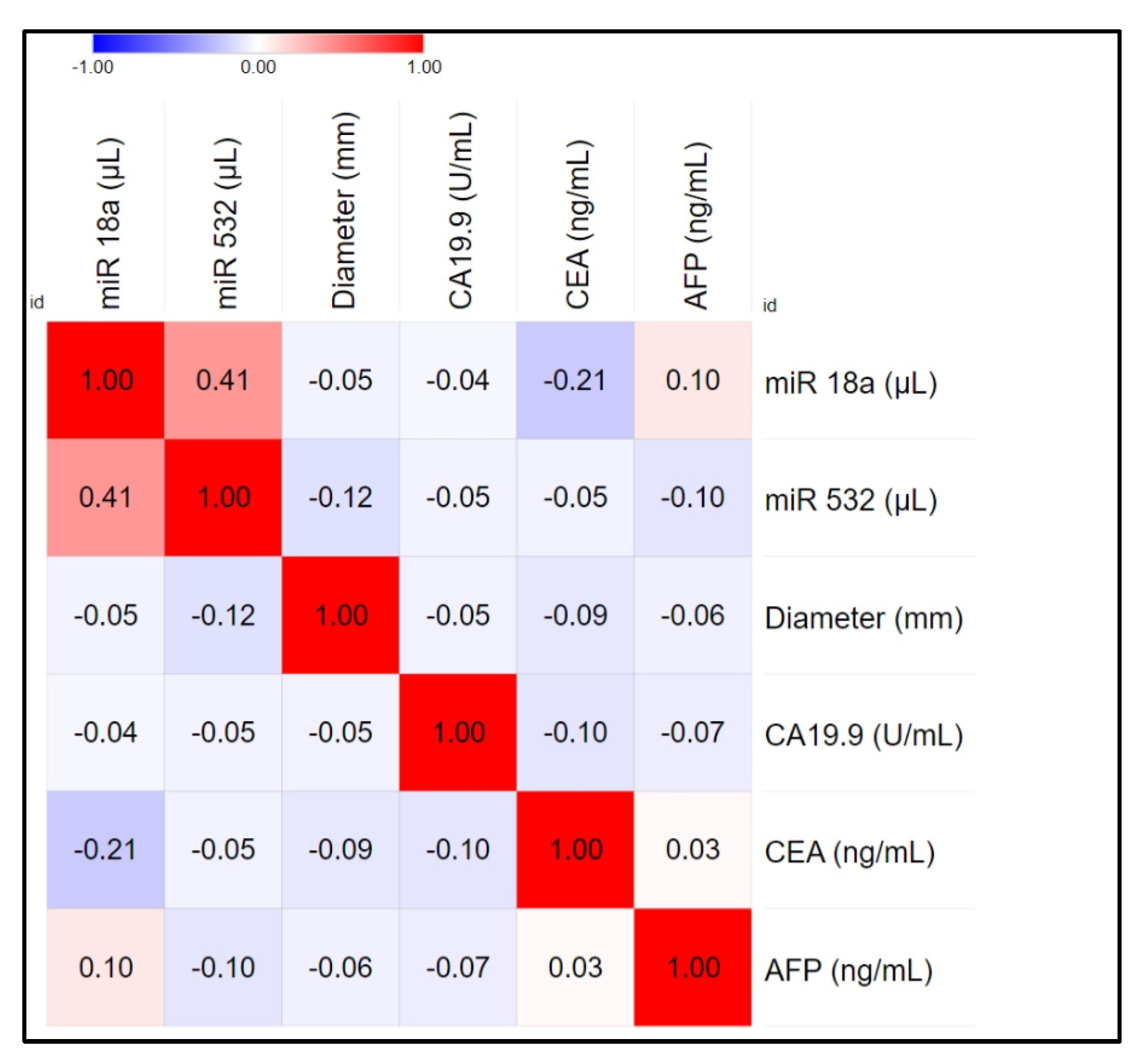
3.2. Correlation and Statistical Analysis
3.3. Vascular Invasion
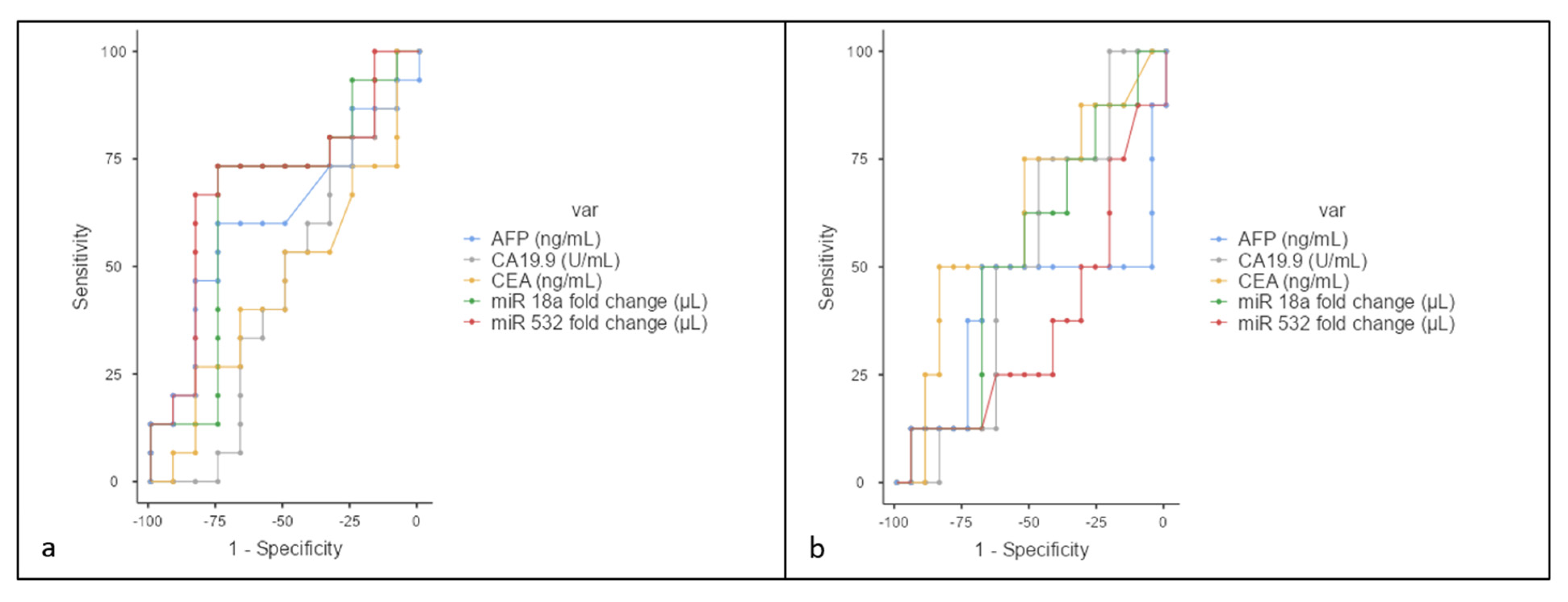
3.4. N Stage Analysis
3.5. Resectability Evaluation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Prim. 2021, 7, 65. [Google Scholar] [CrossRef]
- Banales, J.M.; Cardinale, V.; Carpino, G.; Marzioni, M.; Andersen, J.B.; Invernizzi, P.; Lind, G.E.; Folseraas, T.; Forbes, S.J.; Fouassier, L.; et al. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 261–280. [Google Scholar] [CrossRef]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 557–588. [Google Scholar] [CrossRef]
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. Off. J. Int. Assoc. Study Liver 2019, 39 (Suppl. 1), 19–31. [Google Scholar] [CrossRef]
- Strijker, M.; Belkouz, A.; van der Geest, L.G.; van Gulik, T.M.; van Hooft, J.E.; de Meijer, V.E.; Mohammad, N.H.; de Reuver, P.R.; Verheij, J.; de Vos-Geelen, J.; et al. Treatment and survival of resected and unresected distal cholangiocarcinoma: A nationwide study. Acta Oncol. 2019, 58, 1048–1055. [Google Scholar] [CrossRef]
- Luvira, V.; Suwanrungruang, K.; Kamsa-Ard, S.; Luvira, V.; Santong, C.; Srisuk, T.; Pugkhem, A.; Bhudhisawasdi, V.; Pairojkul, C. Cholangiocarcinoma Trends, Incidence, and Relative Survival in Khon Kaen, Thailand From 1989 Through 2013: A Population-Based Cancer Registry Study. J. Epidemiol. 2019, 29, 197–204. [Google Scholar] [CrossRef]
- Cambridge, W.A.; Fairfield, C.M.; Powell, J.J.M.; Harrison, E.M.P.; Søreide, K.M.; Wigmore, S.J.M.; Guest, R.V.P. Meta-analysis and Meta-regression of Survival After Liver Transplantation for Unresectable Perihilar Cholangiocarcinoma. Ann. Surg. 2021, 273, 240–250. [Google Scholar] [CrossRef]
- Chakrabortty, A.; Patton, D.J.; Smith, B.F.; Agarwal, P. miRNAs: Potential as Biomarkers and Therapeutic Targets for Cancer. Genes 2023, 14, 1375. [Google Scholar] [CrossRef]
- Shi, T.; Morishita, A.; Kobara, H.; Masaki, T. The Role of microRNAs in Cholangiocarcinoma. Int. J. Mol. Sci. 2021, 22, 7627. [Google Scholar] [CrossRef]
- Kim, Y.-K. Extracellular microRNAs as Biomarkers in Human Disease. Chonnam Med. J. 2015, 51, 51–57. [Google Scholar] [CrossRef]
- Barbato, A.; Piscopo, F.; Salati, M.; Reggiani-Bonetti, L.; Franco, B.; Carotenuto, P. Micro-RNA in Cholangiocarcinoma: Implications for Diagnosis, Prognosis, and Therapy. J. Mol. Pathol. 2022, 3, 88–103. [Google Scholar] [CrossRef]
- Montal, R.; Sia, D.; Montironi, C.; Leow, W.Q.; Esteban-Fabró, R.; Pinyol, R.; Torres-Martin, M.; Bassaganyas, L.; Moeini, A.; Peix, J.; et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J. Hepatol. 2020, 73, 315–327. [Google Scholar] [CrossRef]
- Fox, J.; Marquez, M.M.; Bouchet-Valat, M. Rcmdr: R Commander. R Package Version 2.9-2. 2024. Available online: https://cran.r-project.org/bin/windows/base/old/ (accessed on 24 July 2024).
- Zhao, W.; Gupta, A.; Krawczyk, J.; Gupta, S. The miR-17-92 cluster: Yin and Yang in human cancers. Cancer Treat. Res. Commun. 2022, 33, 100647. [Google Scholar] [CrossRef]
- Shen, K.; Cao, Z.; Zhu, R.; You, L.; Zhang, T. The dual functional role of MicroRNA-18a (miR-18a) in cancer development. Clin. Transl. Med. 2019, 8, 32. [Google Scholar] [CrossRef]
- Egeland, N.G.; Jonsdottir, K.; Aure, M.R.; Sahlberg, K.; Kristensen, V.N.; Cronin-Fenton, D.; Skaland, I.; Gudlaugsson, E.; Baak, J.P.A.; Janssen, E.A.M. MiR-18a and miR-18b are expressed in the stroma of oestrogen receptor alpha negative breast cancers. BMC Cancer 2020, 20, 377. [Google Scholar] [CrossRef]
- Yau, T.O.; Wu, C.W.; Dong, Y.; Tang, C.-M.; Ng, S.S.M.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br. J. Cancer 2014, 111, 1765–1771. [Google Scholar] [CrossRef]
- Xu, X.; Bhandari, K.; Xu, C.; Morris, K.; Ding, W.-Q. miR-18a and miR-106a Signatures in Plasma Small EVs Are Promising Biomarkers for Early Detection of Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2023, 24, 7215. [Google Scholar] [CrossRef]
- Yun, J.H.; Baek, M.-J.; Jung, H.I. Expression of miR-221 and miR-18a in patients with hepatocellular carcinoma and its clinical significance. Korean J. Clin. Oncol. 2022, 18, 17–26. [Google Scholar] [CrossRef]
- Wu, W.; Takanashi, M.; Borjigin, N.; Ohno, S.-I.; Fujita, K.; Hoshino, S.; Osaka, Y.; Tsuchida, A.; Kuroda, M. MicroRNA-18a modulates STAT3 activity through negative regulation of PIAS3 during gastric adenocarcinogenesis. Br. J. Cancer 2013, 108, 653–661. [Google Scholar] [CrossRef]
- Hsu, T.-I.; Hsu, C.-H.; Lee, K.-H.; Lin, J.-T.; Chen, C.-S.; Chang, K.-C.; Su, C.-Y.; Hsiao, M.; Lu, P.-J. MicroRNA-18a is elevated in prostate cancer and promotes tumorigenesis through suppressing STK4 in vitro and in vivo. Oncogenesis 2014, 3, e99. [Google Scholar] [CrossRef]
- Zhao, W.; Zhao, J.; Guo, X.; Feng, Y.; Zhang, B.; Tian, L. LncRNA MT1JP plays a protective role in intrahepatic cholangiocarcinoma by regulating miR-18a-5p/FBP1 axis. BMC Cancer 2021, 21, 142. [Google Scholar] [CrossRef]
- Peng, L.; Liu, Y.; Nie, S.; Gao, M. LncRNA CASC2 inhibits cell proliferation, metastasis and EMT through miR-18a/SOCS5 axis in cholangiocarcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8367–8376. [Google Scholar] [CrossRef]
- Karimkhanloo, H.; Mohammadi-Yeganeh, S.; Ahsani, Z.; Paryan, M. Bioinformatics prediction and experimental validation of microRNA-20a targeting Cyclin D1 in hepatocellular carcinoma. Tumor Biol. 2017, 39, 1010428317698361. [Google Scholar] [CrossRef]
- Gao, S.; Bu, X.; Gao, Y.; Bao, Z.; Shi, W.; Luan, L.; Chen, H.; Zhang, B.; Tian, Q.; Guan, W.; et al. The miR-532-E2F1 feedback loop contributes to gastric cancer progression. Cell Death Dis. 2022, 13, 376. [Google Scholar] [CrossRef]
- Gu, C.; Cai, J.; Xu, Z.; Zhou, S.; Ye, L.; Yan, Q.; Zhang, Y.; Fang, Y.; Liu, Y.; Tu, C.; et al. MiR-532-3p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated Wnt/β-catenin signaling. Cell Death Dis. 2019, 10, 739. [Google Scholar] [CrossRef]
- Wang, Q. CircANTXR1 Contributes to the Malignant Progression of Hepatocellular Carcinoma by Promoting Proliferation and Metastasis. J. Hepatocell. Carcinoma 2021, 2021, 1339–1353. [Google Scholar] [CrossRef]
- El-Daly, S.M.; Gouhar, S.A.; Elmageed, Z.Y.A. Circulating microRNAs as Reliable Tumor Biomarkers: Opportunities and Challenges Facing Clinical Application. J. Pharmacol. Exp. Ther. 2023, 384, 35–51. [Google Scholar] [CrossRef]
- Li, L.M.; Hu, Z.B.; Zhou, Z.X.; Chen, X.; Liu, F.Y.; Zhang, J.F.; Shen, H.B.; Zhang, C.Y.; Zen, K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010, 70, 9798–9807. [Google Scholar] [CrossRef]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef]
- McDermott, A.M.; Kerin, M.J.; Miller, N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS ONE 2013, 8, e83718. [Google Scholar] [CrossRef]
| MIR | Mature miRNA Sequence |
|---|---|
| HSA-miR-18a | ACUGCCCUAAGUGCUCCUUCU |
| HSA-miR-532 | CCUCCCACACCCAAGGCUUGCA |
| RNU6B | CGCAAGGATGACACGCAAATTCGTGAAGCGTTCCATATTTTT |
| Characteristic | eCCA Patients (n = 27) |
|---|---|
| Age (years), mean ± SD | 62.14 (±10.97) |
| Sex (Male/Female) | 14:13 |
| Final diagnosis | |
| dCCA | 10 (37.0%) |
| pCCA | 17 (62.9%) |
| Histological grading | |
| G1 (low grade) | 9 (33.3%) |
| G2 (intermediate grade) | 13 (48.1%) |
| G3 (high grade) | 5 (18.5%) |
| Vascular invasion | |
| None | 15 (55.5%) |
| Present | 12 (44.4%) |
| Visceral invasion | 10 (37.0%) |
| Metastases | 4 (14.8%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Orzan, R.I.; Țigu, A.B.; Nechita, V.-I.; Nistor, M.; Agoston, R.; Gonciar, D.; Pojoga, C.; Seicean, A. Circulating miR-18a and miR-532 Levels in Extrahepatic Cholangiocarcinoma. J. Clin. Med. 2024, 13, 6177. https://doi.org/10.3390/jcm13206177
Orzan RI, Țigu AB, Nechita V-I, Nistor M, Agoston R, Gonciar D, Pojoga C, Seicean A. Circulating miR-18a and miR-532 Levels in Extrahepatic Cholangiocarcinoma. Journal of Clinical Medicine. 2024; 13(20):6177. https://doi.org/10.3390/jcm13206177
Chicago/Turabian StyleOrzan, Rares Ilie, Adrian Bogdan Țigu, Vlad-Ionuț Nechita, Madalina Nistor, Renata Agoston, Diana Gonciar, Cristina Pojoga, and Andrada Seicean. 2024. "Circulating miR-18a and miR-532 Levels in Extrahepatic Cholangiocarcinoma" Journal of Clinical Medicine 13, no. 20: 6177. https://doi.org/10.3390/jcm13206177
APA StyleOrzan, R. I., Țigu, A. B., Nechita, V.-I., Nistor, M., Agoston, R., Gonciar, D., Pojoga, C., & Seicean, A. (2024). Circulating miR-18a and miR-532 Levels in Extrahepatic Cholangiocarcinoma. Journal of Clinical Medicine, 13(20), 6177. https://doi.org/10.3390/jcm13206177







