Asthma Inflammatory Phenotypes: How Can We Distinguish Them?
Abstract
1. Introduction
2. Materials and Methods
Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Anderson, G.P. Endotyping asthma: New insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet 2008, 372, 1107–1119. [Google Scholar] [CrossRef]
- Wenzel, S.E. Asthma phenotypes: The evolution from clinical to molecular approaches. Nat. Med. 2012, 18, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Kuruvilla, M.E.; Lee, F.E.; Lee, G.B. Understanding asthma phenotypes, endotypes, and mechanism of disease. Clin. Rev. Allergy Immunol. 2019, 56, 219–293. [Google Scholar] [CrossRef] [PubMed]
- Canonica, G.W.; Fernando, I.; Biardini, F.; Puggioni, F.; Racca, F.; Passalacqua, G.; Heffler, E. Asthma: Personalized and precision medicine. Curr. Opin. Allergy Clin. Immunol. 2018, 18, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Ozdemir, C.; Kucukser, C.U.; Akdis, M.; Akdis, C.A. The concept of asthma endotypes and phenotypes to guide current and novel treatment strategies. Expert Rev. Respir. Med. 2018, 12, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Ciprandi, G.; Tosca, M.A.; Silvestri, M.; Ricciardolo, F.L.M. Inflammatory biomarkers in asthma endotypes and consequent personalized therapy. Expert Rev. Clin. Immunol. 2017, 13, 715–721. [Google Scholar] [CrossRef]
- Kaur, R.; Chupp, G. Phenotypes and endotypes in adult asthma: Moving toward precision medicine. J. Allergy Clin. Immunol. 2019, 144, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, M.; Ray, A.; Wenzel, S.A. Evolving Concepts of Asthma. Am. J. Respir. Crit. Care Med. 2015, 192, 660–668. [Google Scholar] [CrossRef]
- Simpson, J.L.; Scott, R.; Boyle, M.J.; Gibson, P.G. Inflammatory subtypes in asthma: Assessment and identification using induced sputum. Respirology 2006, 11, 54–61. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, X.; Wang, Y.; Du, R.; Mao, H. Delineating asthma according to inflammation phenotypes with a focus on paucigranulocytic asthma. Chin. Med. J. 2023, 136, 1513–1522. [Google Scholar] [CrossRef]
- Esteban-Gorgojo, I.; Antolín-Amérigo, D.; Domínguez-Ortega, J.; Quirce, S. Non-eosinophilic asthma: Current perspectives. J. Asthma Allergy 2018, 11, 267–281. [Google Scholar] [CrossRef]
- Svenningsen, S.; Nair, P. Asthma endotypes and an overview of targeted therapy for asthma. Front. Med. 2017, 4, 158. [Google Scholar] [CrossRef] [PubMed]
- Gibson, P.G. Inflammatory phenotypes in adult asthma. Clin. Appl. Clin. Respir. J. 2009, 3, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Diamant, Z.; Vijverberg, S.; Alving, K.; Bakirtas, A.; Bjermer, L.; Custovic, A.; Dahlen, S.E.; Gaga, M.; Gerth van Wijk, R.; Del Giacco, S.; et al. Toward clinically applicable biomarkers for asthma: An EAACI position paper. Allergy 2019, 74, 1835–1851. [Google Scholar] [CrossRef] [PubMed]
- Canonica, G.W.; Spanevello, A.; de Llano, L.P.; Domingo Ribas, C.; Blakey, J.D.; Garcia, G.; Inoue, H.; Dalcolmo, M.; Yang, D.; Mokashi, S.; et al. Is asthma control more than just an absence of symptoms? An expert consensus statement. Respir. Med. 2022, 202, 106942. [Google Scholar] [CrossRef] [PubMed]
- Reddel, H.K.; Taylor, D.R.; Bateman, E.D.; Boulet, L.P.; Boushey, H.A.; Busse, W.W.; Casale, T.B.; Chanez, P.; Enright, P.L.; Gibson, P.G.; et al. Asthma control and exacerbations: Standardizing endpoints for clinical asthma trials and clinical practice. Am. J. Respir. Crit. Care Med. 2009, 180, 59–99. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.L.; Bacharier, L.B.; Bateman, E.; Boulet, L.P.; Brightling, C.; Buhl, R.; Brusselle, G.; Cruz, A.A.; Drazen, J.M.; Duijts, L.; et al. Key recommendations for primary care from the 2022 Global Initiative for Asthma (GINA) update. NPJ Prim. Care Respir. Med. 2023, 33, 7. [Google Scholar] [CrossRef]
- Samitas, K.; Zervas, E.; Gaga, M. T2-low asthma: Current approach to diagnosis and therapy. Curr. Opin. Pulm. Med. 2017, 23, 48–55. [Google Scholar] [CrossRef]
- Fitzpatrick, A.M.; Chipps, B.E.; Holguin, F.; Woodruff, P.G. T2-”Low” Asthma: Overview and Management Strategies. J. Allergy Clin. Immunol. Pract. 2020, 8, 452–463. [Google Scholar] [CrossRef]
- Engelkes, M.; Janssens, H.M.; de Jongste, J.C.; Sturkenboom, M.C.; Verhamme, K.M. Medication adherence and the risk of severe asthma exacerbations: A systematic review. Eur. Respir. J. 2015, 45, 396–407. [Google Scholar] [CrossRef]
- Chung, K.F.; Wenzel, S.E.; Brozek, J.L. International ERS/ATS guidelines on definition, evaluation and treatment of severe asthma. Eur. Respir. J. 2014, 43, 343. [Google Scholar] [CrossRef] [PubMed]
- Nathan, R.A.; Sorkness, C.A.; Kosinski, M.; Schatz, M.; Li, J.T.; Marcus, P.; Murray, J.J.; Pendergraft, T.B. Development of the asthma control test: A survey for assessing asthma control. J. Allergy Clin. Immunol. 2004, 113, 59–65. [Google Scholar] [CrossRef]
- Juniper, E.F.; Bousquet, J.; Abetz, L.; Bateman, E.D.; GOAL Committee. Identifying ‘well-controlled’ and ‘not well-controlled’ asthma using the Asthma Control Questionnaire. Respir. Med. 2006, 100, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, I.L.; Li, J.T.; Bernstein, D.I.; Hamilton, R.; Spector, S.L.; Tan, R.; Sicherer, S.; Golden, D.B.; Khan, D.A.; Nicklas, R.A.; et al. Allergy diagnostic testing: An updated practice parameter. Ann. Allergy Asthma Immunol. 2008, 100 (Suppl. S3), S1–S148. [Google Scholar] [CrossRef] [PubMed]
- Exhaled, N.O. ATS/ERS recommendations for standardized procedures for the online and offline measurement of exhaled lower respiratory nitric oxide and nasal nitric oxide. Am. J. Respir. Crit. Care Med. 2005, 171, 912–930. [Google Scholar]
- King, G.G.; Bates, J.; Berger, K.I.; Calverley, P.; de Melo, P.L.; Dellaca, R.L.; Farre, R.; Hall, G.L.; Ioan, I.; Irvin, C.G.; et al. Technical standards for respiratory oscillometry. Eur. Respir. J. 2020, 55, 1900753. [Google Scholar] [CrossRef]
- Djukanovic, R.; Sterk, P.J.; Fahy, J.V.; Hargreave, F.E. Standardized methodology of sputum induction and processing. Eur. Respir. J. Suppl. 2002, 37, 1s–2s. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; McElduff, P.; Gibson, P.G. Assessment and reproducibility of non-eosinophilic asthma using induced sputum. Respiration 2010, 79, 147–151. [Google Scholar] [CrossRef]
- Thomson, N.C.; Chaudhuri, R.; Heaney, L.G.; Bucknall, C.; Niven, R.M.; Brightling, C.E.; Menzies-Gow, A.N.; Mansur, A.H.; McSharry, C. Clinical outcomes and inflammatory biomarkers in current smokers and exsmokers with severe asthma. J. Allergy Clin. Immunol. 2013, 131, 1008–1016. [Google Scholar] [CrossRef]
- Ntontsi, P.; Loukides, S.; Bakakos, P.; Kostikas, K.; Papatheodorou, G.; Papathanassiou, E.; Hillas, G.; Koulouris, N.; Papiris, S.; Papaioannou, A.I. Clinical, functional and inflammatory characteristics in patients with paucigranulocytic stable asthma: Comparison with different sputum phenotypes. Allergy 2017, 72, 1761–1767. [Google Scholar] [CrossRef]
- Crespo-Lessmann, A.; Curto, E.; Mateus Medina, E.F.; Palones, E.; Belda Soler, A.; Sánchez Maza, S.; Soto-Retes, L.; Plaza, V. Characteristics of Induced-Sputum Inflammatory Phenotypes in Adults with Asthma: Predictors of Bronchial Eosinophilia. J. Asthma Allergy 2023, 19, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Han, G.J.; Zhu, Y.J.; Mao, D.; Hu, H. Clinical characteristics and biomarkers analysis of asthma inflammatory phenotypes. Biomark. Med. 2020, 14, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Westerhof, G.A.; Korevaar, D.A.; Amelink, M.; de Nijs, S.B.; de Groot, J.C.; Wang, J.; Weersink, E.J.; ten Brinke, A.; Bossuyt, P.M.; Bel, E.H. Biomarkers to identify sputum eosinophilia in different adult asthma phenotypes. Eur. Respir. J. 2015, 46, 688–696. [Google Scholar] [CrossRef] [PubMed]
- Wagener, A.H.; De Nijs, S.B.; Lutter, R.; Sousa, A.R.; Weersink, E.J.; Bel, E.H.; Sterk, P.J. External validation of blood eosinophils, FE(NO), and serum periostin as surrogates for sputum eosinophils in asthma. Thorax 2015, 70, 115–120. [Google Scholar] [CrossRef]
- Schleich, F.N.; Manise, M.; Sele, J.; Henket, M.; Seidel, L.; Louis, R. Distribution of sputum cellular phenotype in a large asthma cohort: Predicting factors for eosinophilic vs neutrophilic inflammation. BMC Pulm. Med. 2013, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Yap, E.; Chua, W.M.; Layaram, L.; Zeng, I.; Vandal, A.C.; et Garrett, J. Can we predict sputum eosinophilia from clinical assessment in patients referred to an adult asthma clinic? Intern. Med. J. 2013, 43, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Korevaar, D.A.; Westerhof, G.A.; Wang, J.; Cohen, J.F.; Spijker, R.; Sterk, P.J.; Bel, E.H.; Bossuyt, P.M. Diagnostic accuracy of minimally invasive markers for detection of airway eosinophilia in asthma: A systematic review and meta-analysis. Lancet Respir. Med. 2015, 3, 290–300. [Google Scholar] [CrossRef]
- Hastie, A.T.; Moore, W.C.; Li, H.; Rector, B.M.; Ortega, V.E.; Pascual, R.M.; Peters, S.P.; Meyers, D.A.; Bleecker, E.R.; Heart, N. Biomarker surrogates do not accurately predict sputum eosinophil and neutrophil percentages in asthmatic subjects. J. Allergy Clin. Immunol. 2013, 132, 72–80. [Google Scholar] [CrossRef]
- Comou, H.; Bel, E.H. Improving the diagnosis of eosinophilic asthma. Expert Rev. Respir. Med. 2016, 10, 1093–1103. [Google Scholar] [CrossRef]
- Guida, G.; Bagnasco, D.; Carriero, V.; Bertolini, F.; Ricciardolo, F.L.M.; Nicola, S.; Brussino, L.; Nappi, E.; Paoletti, G.; Canonica, G.W.; et al. Critical evolution of asthma biomarkers in clinical practice. Front. Med. 2022, 10, 969243. [Google Scholar] [CrossRef]
- Petsky, H.L.; Cates, C.J.; Kew, K.M.; Chang, A.B. Tailoring asthma treatment on eosinophilic markers (exhaled nitric oxide or sputum eosinophils): A systematic review and meta-analysis. Thorax 2018, 73, 1110–1119. [Google Scholar] [CrossRef]
- Dweik, R.A.; Boggs, P.B.; Erzurum, S.C.; Irvin, C.G.; Leigh, M.W.; Lundberg, J.O.; Olin, A.C.; Plummer, A.L.; Taylor, D.R.; American Thoracic Society Committee on Interpretation of Exhaled Nitric Oxide Levels (FENO) for Clinical Applications. An Official ATS Clinical Practice Guideline: Interpretation of Exhaled Nitric Oxide Levels (FENO) for Clinical Applications. Am. J. Respir. Crit. Care Med. 2011, 184, 602–615. [Google Scholar] [CrossRef] [PubMed]
- Cottini, M.; Bondi, B.; Bagnasco, D.; Braido, F.; Passalacqua, G.; Licini, A.; Lombardi, C.; Berti, A.; Comberiati, P.; Landi, M.; et al. Impulse oscillometry defined small airway dysfunction in asthmatic patients with normal spirometry: Prevalence, clinical associations, and impact on asthma control. Respir. Med. 2023, 218, 107391. [Google Scholar] [CrossRef]
- Marseglia, G.L.; Cirillo, I.; Vizzaccaro, A.; Klersy, C.; Tosca, M.A.; La Rosa, M.; Marseglia, A.; Licari, A.; Leone, M.; Ciprandi, G. Role of forced expiratory flow at 25–75% as an early marker of small airway impairment in subjects with allergic rhinitis. Allergy Asthma Proc. 2007, 28, 74–78. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, I.; Klersy, C.; Marseglia, G.L.; Vizzaccaro, A.; Pallestrini, E.; Tosca, M.; Ciprandi, G. Role of FEF 25–75% as a predictor of bronchial hyperreactivity in allergic patients. Ann. Allergy Asthma Immunol. 2006, 96, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Postma, D.S.; Brightling, C.; Baldi, S.; Van den Berge, M.; Fabbri, L.M.; Gagnatelli, A.; Papi, A.; Van der Molen, T.; Rabe, K.F.; Siddiqui, S.; et al. Exploring the relevance and extent of small airways dysfunction in asthma (ATLANTIS): Baseline data from a prospective cohort study. Lancet Respir. Med. 2019, 7, 402–416. [Google Scholar] [CrossRef] [PubMed]
- Cottini, M.; Lombardi, C.; Passalacqua, G.; Bagnasco, D.; Berti, A.; Comberiati, P.; Imeri, G.; Landi, M.; Heffler, E. Small Airways: The “Silent Zone” of 2021 GINA Report? Front. Med. 2022, 23, 884679. [Google Scholar] [CrossRef]
- Abdo, M.; Pedersen, F.; Kirsten, A.M.; Veith, V.; Biller, H.; Trinkmann, F.; von Mutius, E.; Kopp, M.; Hansen, G.; Rabe, K.F.; et al. Longitudinal Impact of Sputum Inflammatory Phenotypes on Small Airway Dysfunction and Disease Outcomes in Asthma. J. Allergy Clin. Immunol. Pract. 2022, 10, 1545–1553. [Google Scholar] [CrossRef]
- Lu, L.; Peng, J.; Zhao, N.; Wu, F.; Tian, H.; Yang, H.; Deng, Z.; Wang, Z.; Xiao, S.; Wen, X.; et al. Discordant Spirometry and Impulse Oscillometry Assessments in the Diagnosis of Small Airway Dysfunction. Front. Physiol. 2022, 13, 892448. [Google Scholar] [CrossRef]
- Liwsrisakun, C.; Chaiwong, W.; Pothirat, C. Comparative assessment of small airway dysfunction by impulse oscillometry and spirometry in chronic obstructive pulmonary disease and asthma with and without fixed airflow obstruction. Front. Med. 2023, 10, 1181188. [Google Scholar] [CrossRef]
- Ciółkowski, J.; Emeryk, A.; Hydzik, P.; Emeryk-Maksymiuk, J.; Kosmala, E.; Stasiowska, B. Eosinophilic airway inflammation is a main feature of unstable asthma in adolescents. Respir. Med. 2019, 147, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Erickson, R.W.; Choy, D.F.; Mosesova, S.; Wu, L.C.; Solberg, O.D.; Shikotra, A.; Carter, R.; Audusseau, S.; Hamid, Q.; et al. Periostin is a systemic biomarker of eosinophilic airway inflammation in asthmatic patients. J. Allergy Clin. Immunol. 2012, 130, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.L.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Jenkins, C.; Peters, M.J.; Jia, G.; Holweg, C.T.; et al. Periostin levels and eosinophilic inflammation in poorly controlled asthma. BMC Pulm. Med. 2016, 16, 67. [Google Scholar] [CrossRef]
- Shimoda, T.; Obase, Y.; Kishikawa, R.; Iwanaga, T. Serum high-sensitivity C-reactive protein can be an airway inflammation predictor in bronchial asthma. Allergy Asthma Proc. 2015, 36, e23–e28. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Douwes, J.; Burmanje, J.; Gokhale, P.; Crane, J.; Pattemore, P.; Stanley, T.; Keenan, J.; Brooks, C. Sputum inflammatory, neural, and remodeling mediators in eosinophilic and noneosinophilic asthma. Ann. Allergy Asthma Immunol. 2023, 130, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Wood, L.G.; Baines, K.J.; Fu, J.; Scott, H.A.; Gibson, P.G. The neutrophilic inflammatory phenotype is associated with systemic inflammation in asthma. Chest 2012, 142, 86–89. [Google Scholar] [CrossRef]
- Lambrecht, B.N.; Hammad, H. The basic immunology of asthma. Cell 2021, 184, 1469–1485. [Google Scholar]
- Bullone, M.; Carriero, V.; Bertolini, F.; Folino, A.; Mannelli, A.; Di Stefano, A.; Gnemmi, I.; Torchio, R.; Ricciardolo, F.L. Elevated serum IgE, oral corticosteroid dependence, and IL-17/22 expression in highly neutrophilic asthma. Eur. Respir. J. 2019, 54, 1900068. [Google Scholar] [CrossRef]
- Gibson, P.G.; Foster, P.S. Neutrophilic asthma: Welcome back! Eur. Respir. J. 2019, 54, 1901846. [Google Scholar] [CrossRef]
- Demarche, S.; Schleich, F.; Henket, M.; Paulus, V.; Van Hees, T.; Louis, R. Detailed analysis of sputum and systemic inflammation in asthma phenotypes: Are paucigranulocytic asthmatics really non-inflammatory? BMC Pulm. Med. 2016, 16, 46. [Google Scholar] [CrossRef]
- Dimitrova, D.; Youroukova, V.; Ivanova-Todorova, E.; Tumangelova-Yuzeir, K.; Velikova, T. Serum levels of IL-5, IL-6, IL-8, IL-13 and IL-17A in pre-defined groups of adult patients with moderate and severe bronchial asthma. Respir. Med. 2019, 154, 144–154. [Google Scholar] [CrossRef]
- Luo, W.; Hu, J.; Xu, W.; Dong, J. Distinct spatial and temporal roles for Th1, Th2, and Th17 cells in asthma. Front. Immunol. 2022, 13, 974066. [Google Scholar] [CrossRef] [PubMed]
- Solidoro, P.; Nicola, S.; Ridolfi, I.; Canonica, G.W.; Blasi, F.; Paggiaro, P.; Heffler, E.; Bagnasco, D.; Patrucco, F.; Ribolla, F.; et al. Biologics in Severe Eosinophilic Asthma: Three-Year Follow-Up in a SANI Single Center. Biomedicines 2022, 10, 200. [Google Scholar] [CrossRef]
- Šokić, M.K.; Rijavec, M.; Korošec, P.; Bidovec-Stojkovič, U.; Kern, I.; Vantur, R.; Škrgat, S. Heterogeneous Response of Airway Eosinophilia to Anti-IL-5 Biologics in Severe Asthma Patients. J. Pers. Med. 2022, 12, 70. [Google Scholar] [CrossRef] [PubMed]
- Cosmi, L.; Maggi, L.; Santarlasci, V.; Capone, M.; Cardilicchia, E.; Frosali, F.; Querci, V.; Angeli, R.; Matucci, A.; Fambrini, M.; et al. Identification of a novel subset of human circulating memory CD4(+) T cells that produce both IL-17A and IL-4. J. Allergy Clin. Immunol. 2010, 125, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Irvin, C.; Zafar, I.; Good, J.; Rollins, D.; Christianson, C.; Gorska, M.M.; Martin, R.J.; Alam, R. Increased frequency of dual-positiveTH2/TH17 cells in bronchoalveolar lavage fluid characterizes a pop-ulation of patients with severe asthma. J. Allergy Clin. Immunol. 2014, 134, 1175–1186. [Google Scholar] [CrossRef]
- Wakashin, H.; Hirose, K.; Maezawa, Y.; Kagami, S.I.; Suto, A.; Watanabe, N.; Saito, Y.; Hatano, M.; Tokuhisa, T.; Iwakura, Y.; et al. IL-23 and Th17 cells enhance Th2-cell-mediated eosinophilic airway inflammation in mice. Am. J. Respir. Crit. Care Med. 2008, 178, 1023–1032. [Google Scholar] [CrossRef]
- Hasegawa, T.; Uga, H.; Mori, A.; Kurata, H. Increased serum IL-17A and Th2 cytokine levels in patients with severe uncontrolled asthma. Eur. Cytokine Netw. 2017, 28, 8–18. [Google Scholar] [CrossRef]
- Wang, Y.H.; Voo, K.S.; Liu, B.; Chen, C.Y.; Uygungil, B.; Spoede, W.; Bernstein, J.A.; Huston, D.P.; Liu, Y.J. A novel subset of CD4(+) T(H)2memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J. Exp. Med. 2010, 207, 2479–2491. [Google Scholar] [CrossRef]
- Pouliquen, I.J.; Kornmann, O.; Barton, S.V.; Price, J.A.; Ortega, H.G. Characterization of the relationship between dose and blood eosinophil response following subcutaneous administration of mepolizumab. Int. J. Clin. Pharmacol. Ther. 2015, 53, 1015–1027. [Google Scholar] [CrossRef]
- Tsukamoto, N.; Takahashi, N.; Itoh, H.; Pouliquen, I. Pharmacokinetics and pharmacodynamics of mepolizumab, an anti-interleukin 5 monoclonal antibody, in healthy Japanese male subjects. Clin. Pharmacol. Drug Dev. 2016, 5, 102–108. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.H.; Damera, G.; Newbold, P.; Ranade, K. Reductions in eosinophil biomarkers by benralizumab in patients with asthma. Respir. Med. 2016, 111, 21–29. [Google Scholar] [CrossRef]
- Takaku, Y.; Soma, T.; Nishihara, F.; Nakagome, K.; Kobayashi, T.; Hagiwara, K.; Kanazawa, M.; Nagata, M. Omalizumab attenuates airway inflammation and interleukin-5 production by mononuclear cells in patients with severe allergic asthma. Int. Arch. Allergy Immunol. 2013, 161 (Suppl. S2), 107–117. [Google Scholar] [CrossRef] [PubMed]
- Caminati, M.; Marcon, A.; Guarnieri, G.; Miotti, J.; Bagnasco, D.; Carpagnano, G.E.; Pelaia, G.; Vaia, R.; Maule, M.; Vianello, A.; et al. Benralizumab Efficacy in Late Non-Responders to Mepolizumab and Variables Associated with Occurrence of Switching: A Real-Word Perspective. J. Clin. Med. 2023, 12, 1836. [Google Scholar] [CrossRef] [PubMed]
- Van Veen, I.H.; Ten Brinke, A.; Gauw, S.A.; Sterk, P.J.; Rabe, K.F.; Bel, E.H. Consistency of sputum eosinophilia in difficult-to-treat asthma: A 5-year follow-up study. J. Allergy Clin. Immunol. 2009, 124, 615–617. [Google Scholar] [CrossRef] [PubMed]
- Green, R.H.; Pavord, I. Stability of inflammatory phenotypes in asthma. Thorax 2012, 67, 665–667. [Google Scholar] [CrossRef][Green Version]
- Hancox, R.J.; Cowan, D.C.; Aldridge, R.E.; Cowan, J.O.; Palmay, R.; Williamson, A.; Town, G.I.; Taylor, D.R. Asthma phenotypes: Consistency of classification using induced sputum. Respirology 2012, 17, 461–466. [Google Scholar] [CrossRef]
- Betancor, D.; Olaguibel, J.M.; Rodrigo-Muñoz, J.M.; Arismendi, E.; Barranco, P.; Barroso, B.; Bobolea, I.; Cárdaba, B.; Cruz, M.J.; Curto, E.; et al. How reliably can algorithms identify eosinophilic asthma phenotypes using non-invasive biomarkers? Clin. Transl. Allergy 2022, 12, e12182. [Google Scholar] [CrossRef]

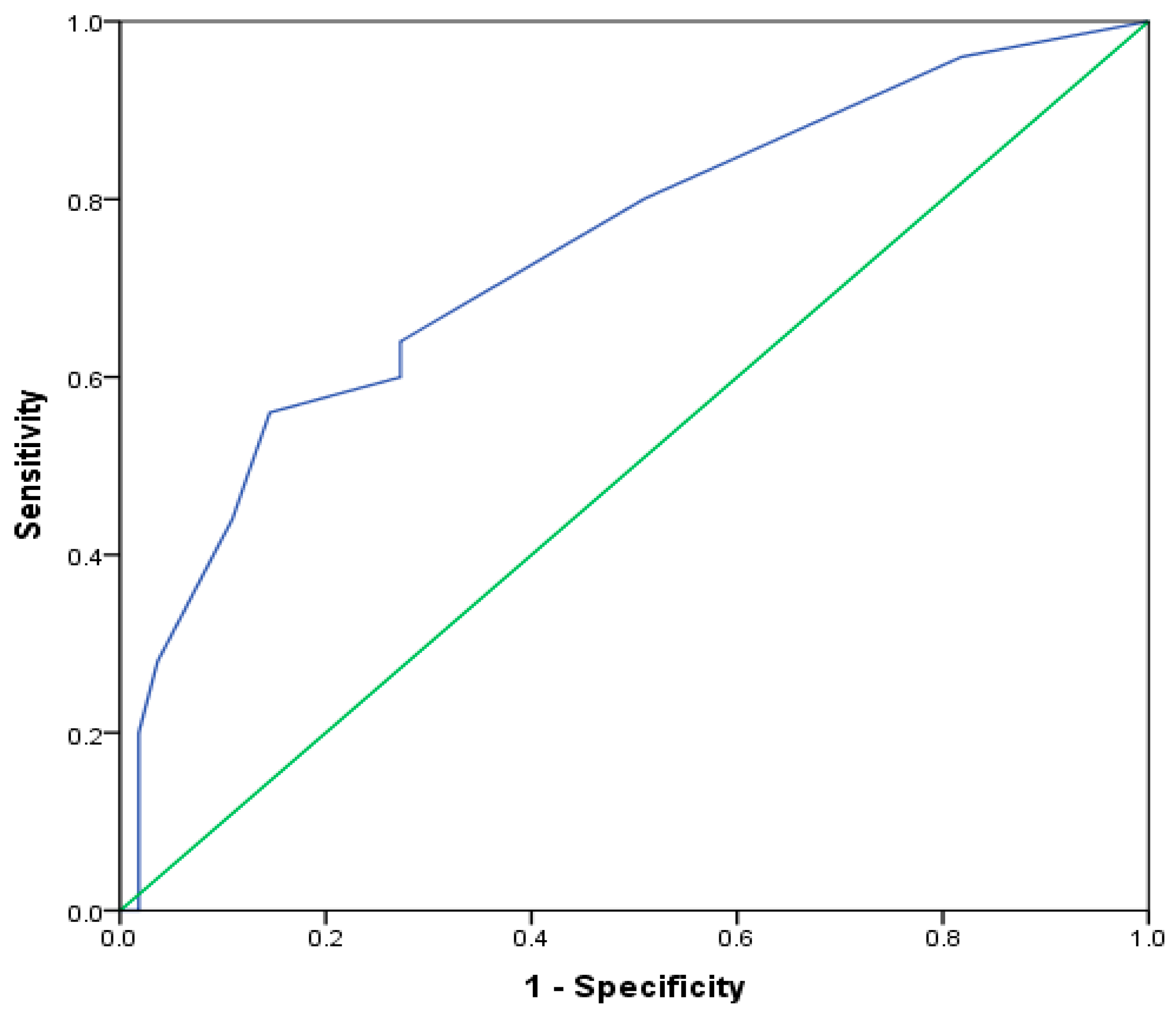
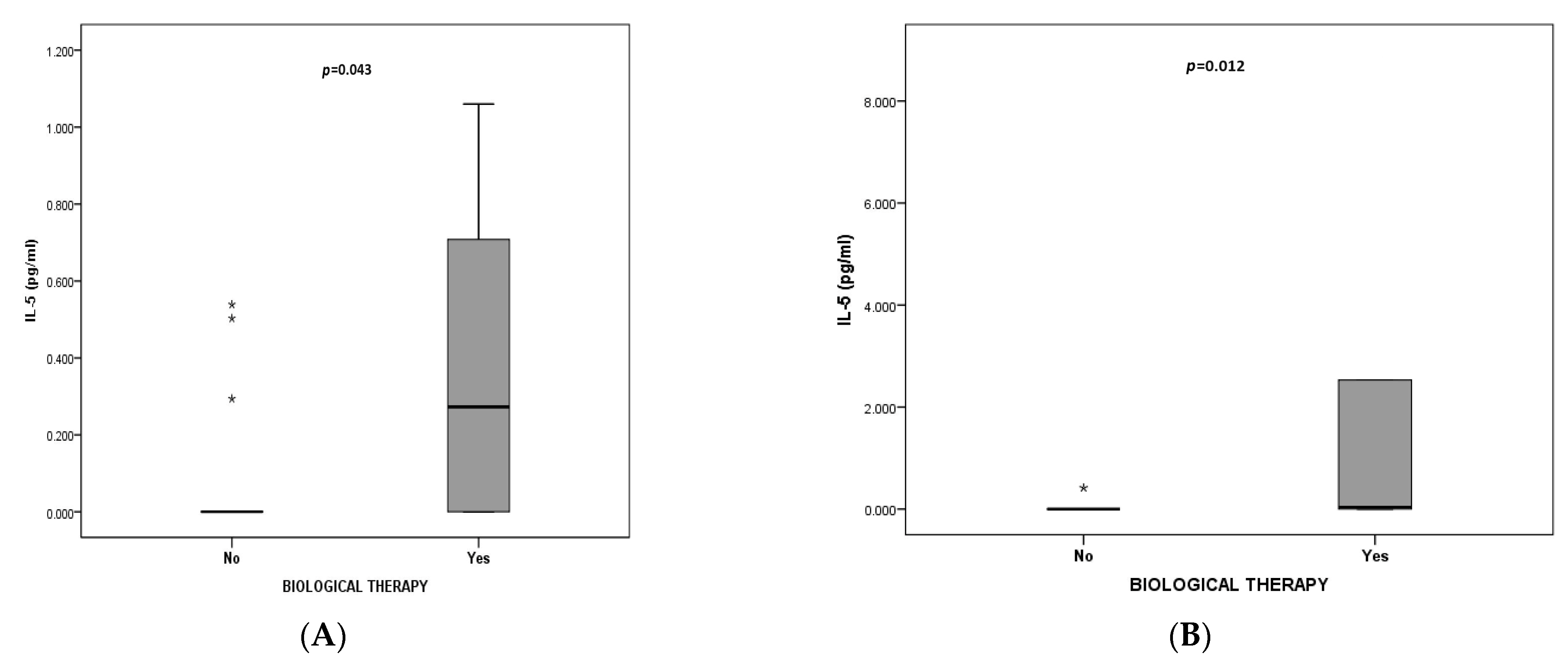
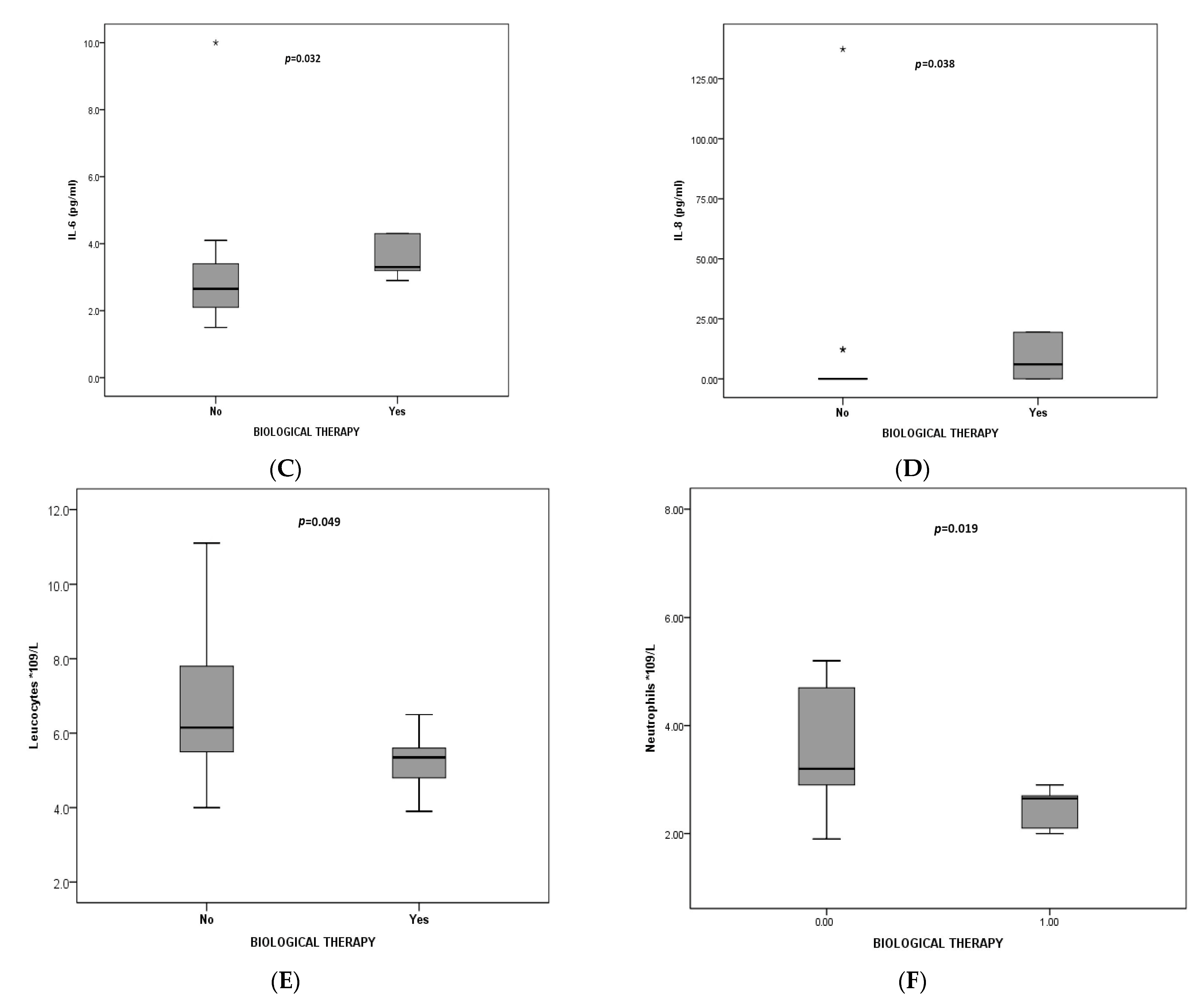
| Characteristic | Total | EA n = 25 | NA n = 12 | MGA n = 24 | PGA n = 19 | p * |
|---|---|---|---|---|---|---|
| Age (years), mean ± SD | 43.65 ± 12.72 | 45.44 ± 13.62 | 41.67 ± 12.46 | 41.71 ± 13.78 | 45.00 ± 10.56 | 0.677 |
| Male | 23 (28.8) | 8 (32.0) | 3 (25.0) | 7 (29.2) | 5 (26.3) | 0.966 |
| Female | 57 (71.3) | 17 (68.0) | 9 (75.0) | 17 (70.8) | 14 (73.7) | |
| Smoking habit, n (%) | ||||||
| Active smoker | 20 (25.0) | 6 (24.0) | 2 (16.7) | 6 (25.0) | 6 (31.6) | 0.960 |
| Ex-smoker | 15 (18.8) | 4 (16.0) | 2 (16.7) | 5 (20.8) | 4 (21.1) | |
| Non-smoker | 55 (56.3) | 15 (60.0) | 8 (66.7) | 13 (54.2) | 9 (47.4) | |
| Smoking duration (years), med (min–max) | 14 (1–50) | 15 (3–27) | 15 (10–20) | 10 (5–50) | 20 (1–30) | 0.817 |
| Asthma duration (years), med (min–max) | 7 (0–52) | 10 (1–45) | 3.5 (0–23) | 6.5 (0–50) | 7 (1–52) | 0.155 |
| BMI, mean ± SD | 26.43 ± 4.64 | 26.52 ± 2.80 | 24.67 ± 4.46 | 27.26 ± 6.24 | 26.39 ± 4.36 | 0.483 |
| Number of severe exacerbations during last year, med (min–max) | 0 (0–10) | 1 (0–8) | 0 (0–7) | 0 (0–4) | 0 (0–10) | 0.134 |
| Allergic rhinitis, n (%) | 63 (78.8%) | 18 (72.2) | 10 (83.3) | 20 (83.3) | 15 (78.9) | 0.769 |
| Nonallergic rhinitis, n (%) | 15 (18.8) | 6 (16.7) | 42(16.7) | 4 (16.7) | 3 (15.8) | 0.882 |
| Drug allergy, n (%) | 22 (27.5) | 6 (24.0) | 4 (33.3) | 7 (29.2) | 5 (26.3) | 0.939 |
| CRSwNP, n (%) | 12 (15.0) | 5 (20.0) | 2 (16.7) | 3 (12.5) | 2 (10.5) | N/A |
| CRSsNP, n (%) | 10 (12.5) | 2 (8.0) | 1 (8.3) | 3 (12.5) | 4 (21.1) | N/A |
| GERB, n (%) | 10 (12.5) | 4 (16.0) | 0 (0.0) | 3 (12.5) | 3 (15.8) | N/A |
| Allergic rhinitis, family, n (%) | 39 (48.8) | 9 (36.0) | 4 (33.3) | 16 (66.7) | 10 (52.6) | 0.113 |
| Asthma, family, n (%) | 31 (38.8) | 9 (36.0) | 6 (50.0) | 9 (37.5) | 7 (36.8) | 0.858 |
| FEV1, % predicted, mean ± SD | 92.60 ± 14.54 | 90.56 ± 17.09 | 91.42 ± 14.58 | 95.86 ± 11.98 | 91.92 ± 14.30 | 0.619 |
| FEV1/FVC %, mean ± SD | 71.36 ± 8.54 | 69.78 ± 6.45 | 71.46 ± 9.25 | 74.59 ± 10.68 | 69.28 ± 6.67 | 0.142 |
| MMEF, %, mean ± SD | 67.15 ± 21.55 | 60.20 ± 21.89 | 67.92 ± 22.02 | 74.71 ± 21.10 | 66.26 ± 19.72 | 0.170 |
| MEF25, %, med (IQR) | 50.0 (37.2–62.7) | 45.0 (30.9–55.5) | 48.0 (40.5–72.22) | 52.5 (36.0–63.0) | 56.0 (38.0–68.0) | 0.230 |
| MEF50, %, med (IQR) | 70 (56.0–87.7) | 68.0 (37.4–77) | 69.0 (53.5–69.0) | 79.0 (61.0–95.0) | 74.7 (53.0–82.0) | 0.185 |
| MEF75, %, med (IQR) | 93.0 (76.2–109.0) | 98.0 (55.8–109.0) | 87.5 (73.5–97.7) | 105.0 (81.7–111) | 86.9 (71.0–99.0 | 0.042 |
| SAD, n (%) | 36 (45.0) | 14 (56.0) | 3 (25.0) | 11 (45.8) | 8 (42.1) | 0.357 |
| COVID 19 infection, n (%) | 62 (78.5) | 18 (75.0) | 9 (75.0) | 17 (70.8) | 18 (94.7) | 0.255 |
| Characteristic | Total | EA n = 25 | NA n = 12 | MGA n = 24 | PGA n = 19 | p * |
|---|---|---|---|---|---|---|
| GINA symptom control, n (%) | ||||||
| Well-controlled | 17(21.3) | 4 (16) | 2 (16.7) | 7 (29.2) | 4 (21.1) | |
| Partly controlled | 30 (37.5) | 10 (40) | 4 (33.3) | 8(33.4) | 8(42.1) | 0.928 |
| Uncontrolled | 33 (41.3) | 11 (44) | 6 (50) | 9 (37.5) | 7 (36.7) | |
| ACT, med (IQR) | 16.5 (12.0–22.0) | 16.0 (11.0–20.5) | 16.5 (12.2–24.7) | 18.5 (11.5–24.7) | 16.0 (12.0–21.0) | 0.572 |
| ACQ, med (IQR) | 1.5 (0.5–2.6) | 2.0 (0.7–3.0) | 1.2 (0.3–2.4) | 1.2 (0.4–3.0) | 1.7 (0.8–2.5) | 0.581 |
| Therapy | ||||||
| Antihistamine, n (%) | 58 (72.5) | 21 (84) | 9 (75) | 17 (70.8) | 11 (57.9) | 0.288 |
| Nasal glucocorticoids, n (%) | 47 (58.8) | 19 (76) | 6 (50.0) | 15 (62.5) | 7 (36.8) | 0.062 |
| Allergic immunotherapy | 12 (15) | 3 (12) | 0 (0) | 7 (29.2) | 2 (10.5) | N/A |
| ICS, n (%) | 10 (12.5) | 4 (16) | 1 (8.3) | 4 (16.7) | 1 (5.3) | N/A |
| SABA n (%), | 33 (41.3) | 8 (32) | 6 (50) | 11 (45.8) | 8 (42.1) | 0.688 |
| LAMA, n (%) | 9 (11.3) | 3 (12) | 1 (8.3) | 4 (16.7) | 1 (5.3) | N/A |
| LTRA, n (%) | 46 (57.5) | 18 (72.0) | 7 (58.3) | 12 (50.0) | 9 (47.4) | 0.320 |
| Methylxanthines, n(%) | 3 (3.8) | 2 (8) | 0 (0) | 0 (0) | 1 (5.3) | N/A |
| ICS/LABA n (%), | 29 (36.3) | 9 (36.0) | 4 (33.3) | 8 (33.3) | 8 (42.1) | 0.937 |
| ICS/LABA MART protocol, n (%) | 30 (37.5) | 11 (44.0) | 4 (33.3) | 9 (37.5) | 6 (31.6) | 0.844 |
| Biologic, n (%) | 17 (21.3) | 7 (41.2) | 2 (11.8) | 6 (35.2) | 2 (11.8) | N/A |
| Omalizumab, n (%) | 10 (58.8) | 6 (85.7) | 1 (50) | 2 (33.3) | 1 (50) | N/A |
| Benralizumab, n (%) | 7 (41.2) | 1 (14.3) | 1 (50) | 4 (66.7) | 1(50) | N/A |
| Marker | Univariate Logistic Regression | Multivariate Logistic Regression | ||||
|---|---|---|---|---|---|---|
| OR | 95%CI OR | p | OR | 95%CI OR | p | |
| EA | Step 4 | |||||
| Number of exacerbations during last year | 1.235 | 1.01–1.52 | 0.044 | |||
| Smoke as trigger | 3.857 | 1.24–12.04 | 0.020 | 5.966 | 1.53–23.30 | 0.010 |
| Nasal glucocorticoids | 3.054 | 1.06–8.81 | 0.039 | |||
| MEF25 | 0.970 | 0.94–0.99 | 0.033 | 0.964 | 0.94–0.99 | 0.019 |
| MEF50 | 0.976 | 0.95–0.99 | 0.030 | |||
| CRP | 1.198 | 1.01–1.43 | 0.049 | 1.246 | 1.01–1.54 | 0.043 |
| Periostin | 1.001 | 1.00–1.01 | 0.050 | 1.001 | 1.00–1.01 | 0.057 |
| NA | Step 2 | |||||
| Eosinophils | 0.006 | 0.00–0.77 | 0.039 | 0.008 | 0.00–1.30 | 0.063 |
| IL-8 | 1.010 | 1.01–1.02 | 0.009 | 1.009 | 1.00–1.02 | 0.025 |
| IgE | 0.958 | 0.92–0.99 | 0.026 | |||
| MGA | Step 4 | |||||
| Allergic rhinitis in the family | 2.870 | 1.05–7.8 | 0.039 | |||
| Physical activity as a trigger | 7.105 | 1.27–39.72 | 0.026 | 5.011 | 0.80–31.38 | 0.085 |
| Allergic immunotherapy | 4.200 | 1.18–15.00 | 0.027 | 3.812 | 0.96–15.13 | 0.057 |
| FEV1/FVC | 1.072 | 1.01–1.14 | 0.036 | |||
| MEF50 | 1.027 | 1.01–1.05 | 0.021 | 1.030 | 1.01–1.06 | 0.023 |
| MEF75 | 1.032 | 1.01–1.06 | 0.023 | |||
| PGA | ||||||
| Nasal glucocorticoids | 0.306 | 0.10–0.89 | 0.030 | |||
| Biomarker, Med (IQR) | EA n = 18 | NA n = 10 | MGA n = 18 | PGA n = 17 | p * |
|---|---|---|---|---|---|
| ESR (mm/h) | 12 (8–22.5) | 9 (7.5–18.5) | 13 (9.5–18) | 12 (8–17) | 0.825 |
| CRP (mg/L) | 1.9 (1.5–4.2) | 1.1 (1.0–2.3) | 1.25 (1.0–3.4) | 1.2 (1.0–3.0) | 0.131 |
| Leucocytes(109/L) | 6.6 (5.9–8.2) | 6.4 (5.1–7.0) | 6.1 (5.5–8.0) | 6.0 (5.5–7.5) | 0.729 |
| Neutrophils(109/L) | 3.6 (2.8–4.5) | 3.3 (3.0–4.6) | 3.2 (2.8–4.7) | 3.3 (2.6–4.0) | 0.790 |
| Lymphocytes(109/L) | 2.2 (1.9–2.6) | 1.7 (1.2–2.6) | 2.2 (1.8–2.8) | 2.1 (1.7–2.2) | 0.157 |
| Eosinophils(109/L) | 0.3 (0.2–0.6) | 0.1 (0.1–0.2) | 0.2 (0.1–0.3) | 0.2 (0.1–0.3) | 0.034 |
| IgE (IU/mL) | 182.5 (75.7–363.5) | 87.0 (60.7–288.7) | 81.5 (20.2–309.5) | 78.0 (18.0–205.0) | 0.212 |
| FeNO (ppb) | 24.2 (14.1–39.4) | 16.3 (11.6–26.3) | 16.2 (7.8–32.9) | 20.9 (11.6–31.6) | 0.420 |
| IL-6 (pg/mL) | 2.8 (1.9–3.2) | 2.4 (1.7–5.3) | 2.6 (2.1–3.4) | 3.1 (1.8–3.3) | 0.969 |
| Periostin (pmol/L), med | 865.7 (697.6–1851.8) | 661.4 (521.7–1004.1) | 925.3 (713.2–1125.8) | 1049.6 (836.4–1355.4) | 0.256 |
| IL-5 (pg/mL) | 0.0 (0.0–0.0) | 0.0 (0.0–1.0) | 0.0 (0.0–0.0) | 0.0 (0.0–0.0) | 0.081 |
| IL-8 (pg/mL) | 0.0 (0.0–2.8) | 0.2 (0.0–166.5) | 0.0 (0.0–0.0) | 0.0 (0.0–7.9) | 0.271 |
| IL-33 (pg/mL) | 41.6 (10.8–315.0) | 42.2 (3.7–406.1) | 9.4 (0.2–89.7) | 22.9 (4.3–194.9) | 0.346 |
| IL-17A (pg/mL) | 0 (0.0–0.6) | 0 (0.0–0.0) | 0 (0.0–0.1) | 0 (0.0–0.0) | 0.004 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Plavsic, A.; Nikolic, B.B.; Milenkovic, B.; Miskovic, R.; Kusic, N.; Dimitrijevic, M.; Arandjelovic, S.; Milosevic, K.; Buha, I.; Spiric, V.T. Asthma Inflammatory Phenotypes: How Can We Distinguish Them? J. Clin. Med. 2024, 13, 526. https://doi.org/10.3390/jcm13020526
Plavsic A, Nikolic BB, Milenkovic B, Miskovic R, Kusic N, Dimitrijevic M, Arandjelovic S, Milosevic K, Buha I, Spiric VT. Asthma Inflammatory Phenotypes: How Can We Distinguish Them? Journal of Clinical Medicine. 2024; 13(2):526. https://doi.org/10.3390/jcm13020526
Chicago/Turabian StylePlavsic, Aleksandra, Branka Bonaci Nikolic, Branislava Milenkovic, Rada Miskovic, Natasa Kusic, Milan Dimitrijevic, Snezana Arandjelovic, Katarina Milosevic, Ivana Buha, and Vesna Tomic Spiric. 2024. "Asthma Inflammatory Phenotypes: How Can We Distinguish Them?" Journal of Clinical Medicine 13, no. 2: 526. https://doi.org/10.3390/jcm13020526
APA StylePlavsic, A., Nikolic, B. B., Milenkovic, B., Miskovic, R., Kusic, N., Dimitrijevic, M., Arandjelovic, S., Milosevic, K., Buha, I., & Spiric, V. T. (2024). Asthma Inflammatory Phenotypes: How Can We Distinguish Them? Journal of Clinical Medicine, 13(2), 526. https://doi.org/10.3390/jcm13020526








