AI Algorithms for Modeling the Risk, Progression, and Treatment of Sepsis, Including Early-Onset Sepsis—A Systematic Review
Abstract
1. Introduction
1.1. AI in Medicine
1.2. Sepsis
1.3. Objectives
- To review the research related to AI models for clinical decision support systems (CDSS) in neonatal sepsis, particularly early-onset sepsis (EOS).
- To assess the importance of medical data inputs utilized in the construction of CDSSs, including their potential to develop trustworthy models for medicine.
- To explore the feasibility of modeling the most effective antibiotic therapy through medical data analysis in sepsis to reduce empiric or untargeted antimicrobial treatment.
- To evaluate the models employed by CDSSs and their effectiveness in classifying and predicting neonatal sepsis over time.
2. Materials and Methods
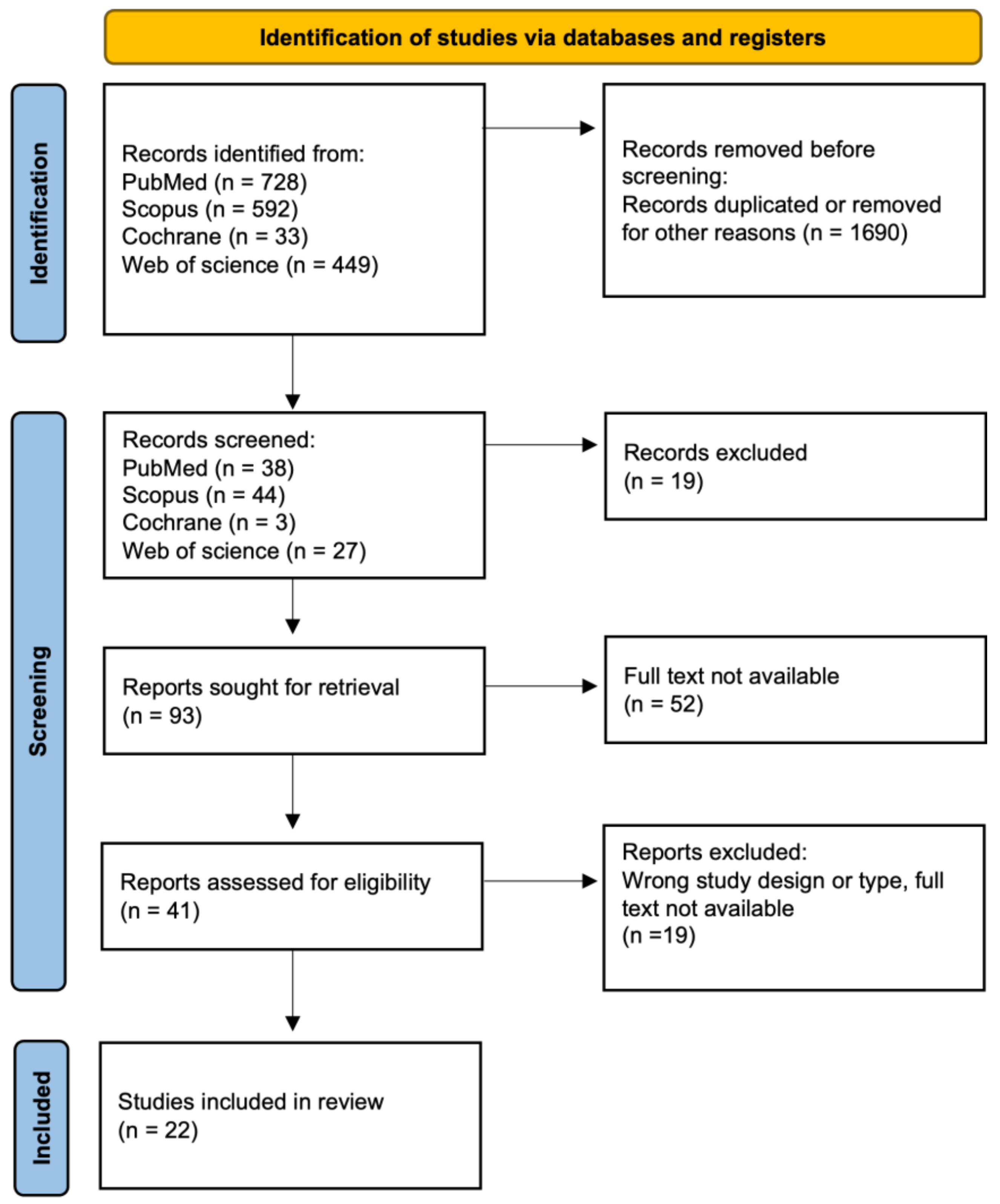
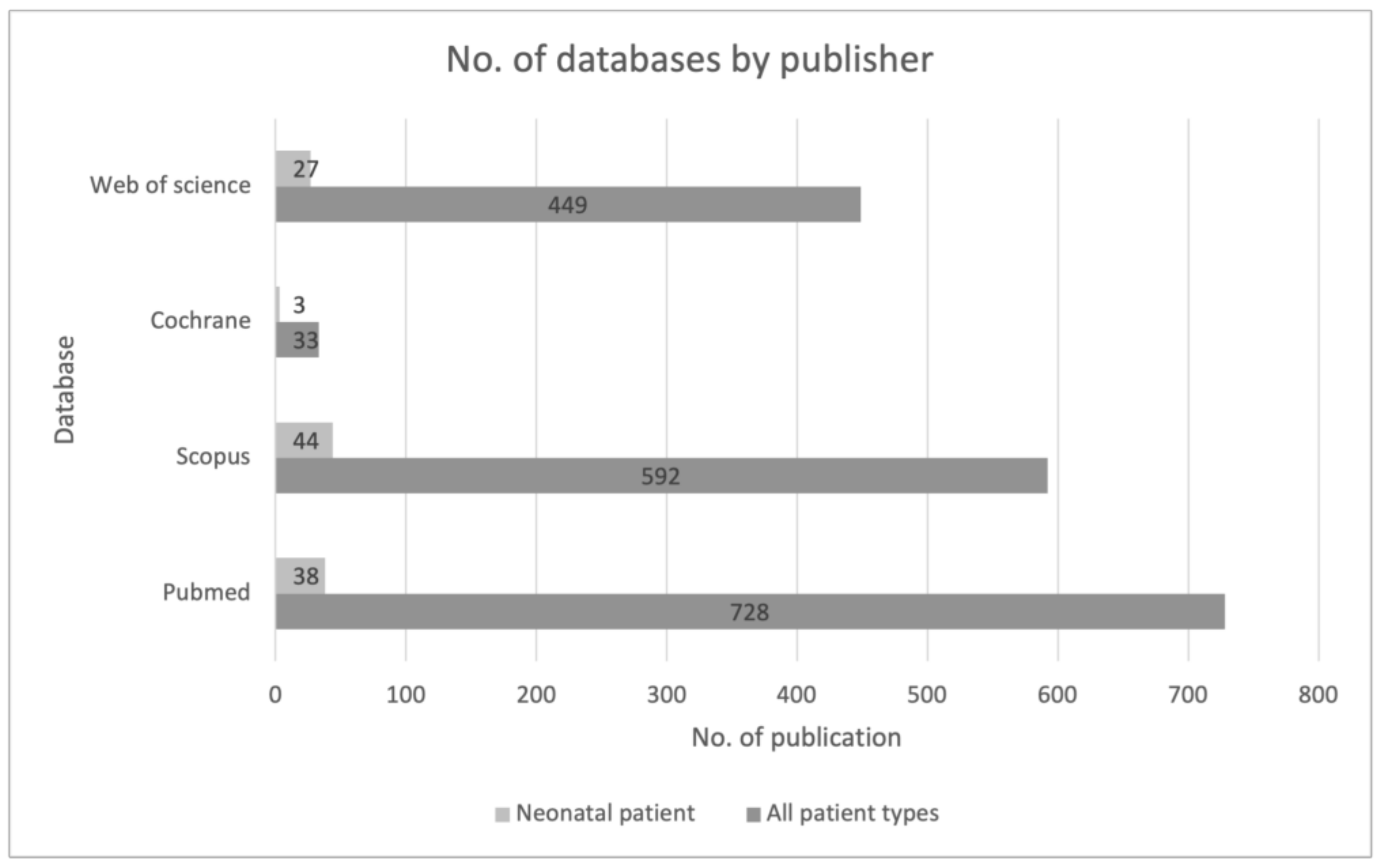
2.1. Research Strategy
2.2. Inclusion Criteria
2.3. Data Extraction
- Identification: Titles, keywords, and abstracts of all identified publications were scrutinized for alignment with the study’s objectives.
- Verification of full texts: The complete texts of all publications identified in the preceding step were independently assessed for inclusion in the review and data extraction. This process adhered rigorously to the defined inclusion/exclusion criteria and study objectives.
- Based on the collated materials, a presentation of the results was crafted, and conclusions with recommendations were formulated.
2.4. Research Questions
- Are AI algorithms suitable for diagnosing sepsis as a support for medical professionals?
- Can AI algorithms accurately assess the risk of sepsis and its rapid progression?
- To what extent can AI algorithms contribute to enhancing treatment strategies for neonatal patients and decreasing antibiotic usage in the future?
- What technological and clinical challenges arise in the utilization of AI for assessing sepsis progression and treatment, including early-onset sepsis (EOS)?
3. Results
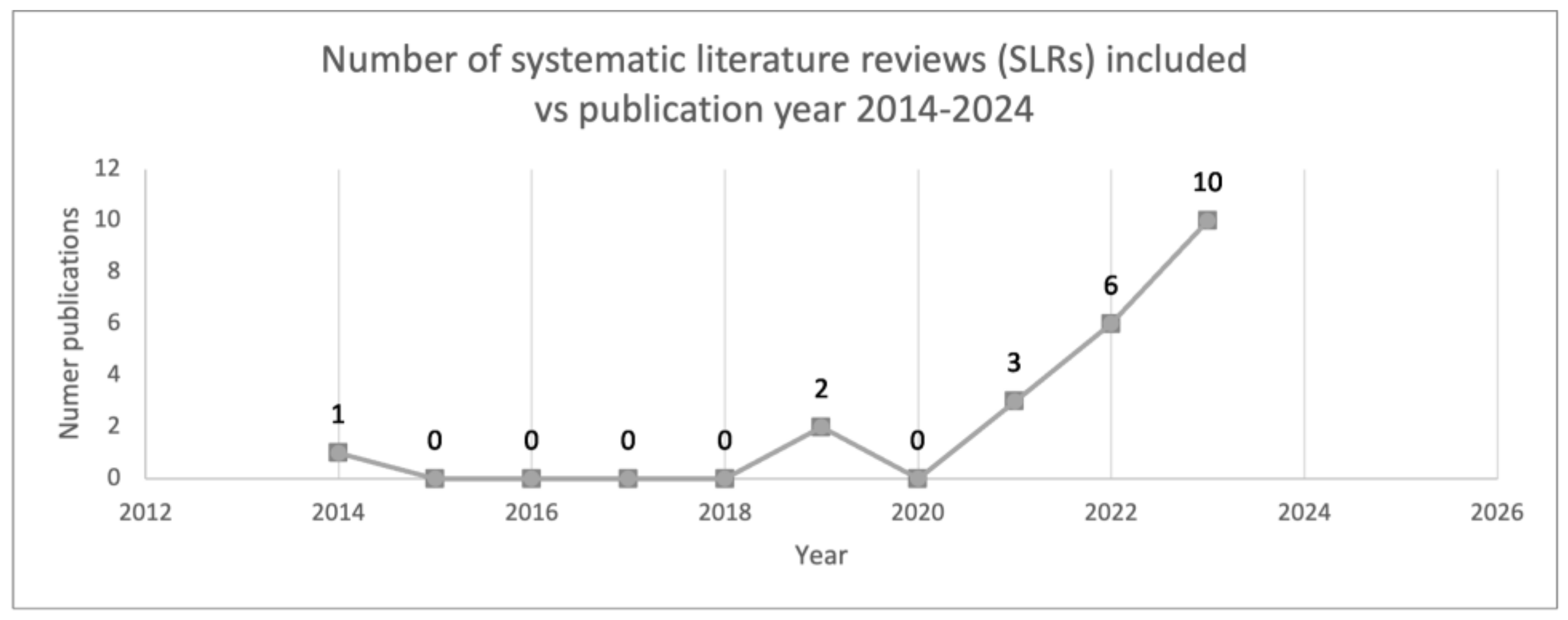
3.1. Study Characteristics
3.2. Quality Assessment
4. Discussion
4.1. Strengths
4.2. Limitations
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Slawomirski, L.; Lindner, L.; de Bienassis, K.; Haywood, P.; Hashiguchi, T.C.O.; Steentjes, M.; Oderkirk, J. Progress on Implementing and Using Electronic Health Record Systems: Developments in OECD Countries as of 2021; OECD Health Working Papers, No. 160; OECD Publishing: Paris, France, 2023. [Google Scholar] [CrossRef]
- Holzinger, A.; Langs, G.; Denk, H.; Zatloukal, K.; Müller, H. Causability and explainability of artificial intelligence in medicine. WIREs Data Min. Knowl. Discov. 2019, 9, e1312. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Shafi, S.; Parwani, A.V. Artificial intelligence in diagnostic pathology. Diagn. Pathol. 2023, 18, 109. [Google Scholar] [CrossRef]
- Iyngkaran, P.; Usmani, W.; Hanna, F.; de Courten, M. Challenges of Health Data Use in Multidisciplinary Chronic Disease Care: Perspective from Heart Failure Care. J. Cardiovasc. Dev. Dis. 2023, 10, 486. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Rajpurkar, P.; Chen, E.; Banerjee, O.; Topol, E.J. AI in health and medicine. Nat. Med. 2022, 28, 31–38. [Google Scholar] [CrossRef] [PubMed]
- U.S Food and Drug Administration. Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices (accessed on 17 March 2024).
- Zuhair, V.; Babar, A.; Ali, R.; Oduoye, M.O.; Noor, Z.; Chris, K.; Okon, I.I.; Rehman, L.U. Exploring the Impact of Artificial Intelligence on Global Health and Enhancing Healthcare in Developing Nations. J. Prim. Care Community Health 2024, 15, 21501319241245847. [Google Scholar] [CrossRef]
- Amisha; Malik, P.; Pathania, M.; Rathaur, V.K. Overview of artificial intelligence in medicine. J. Fam. Med. Prim. Care 2019, 8, 2328–2331. [Google Scholar] [CrossRef]
- Hak, F.; Guimarães, T.; Santos, M. Towards effective clinical decision support systems: A systematic review. PLoS ONE 2022, 17, e0272846. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lee, T.C.; Shah, N.U.; Haack, A.; Baxter, S.L. Clinical Implementation of Predictive Models Embedded within Electronic Health Record Systems: A Systematic Review. Informatics 2020, 7, 25. [Google Scholar] [CrossRef]
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Georgiou, P.; Lescure, F.-X.; Birgand, G.; Holmes, A.H. Machine learning for clinical decision support in infectious diseases: A narrative review of current applications. Clin. Microbiol. Infect. 2020, 26, 584–595, Erratum in: Clin. Microbiol. Infect. 2020, 26, 1118. [Google Scholar] [CrossRef] [PubMed]
- Weston, E.J.; Pondo, T.M.; Lewis, M.M.; Martell-Cleary, P.M.; Morin, C.; Jewell, B.; Daily, P.; Apostol, M.; Petit, S.; Farley, M.; et al. The burden of invasive early-onset neonatal sepsis in the United States, 2005–2008. Pediatr. Infect. Dis. J. 2011, 30, 937–941. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Simonsen, K.A.; Anderson-Berry, A.L.; Delair, S.F.; Dele Davies, H. Early-Onset Neonatal Sepsis. Clin. Microbiol. Rev. 2014, 27, 21–47. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Fleischmann-Struzek, C.; Goldfarb, D.M.; Schlattmann, P.; Schlapbach, L.J.; Reinhart, K.; Kissoon, N. The global burden of paediatric and neonatal sepsis: A systematic review. Lancet Respir. Med. 2018, 6, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Johnston, N.; de Waal, K. Clinical and haemodynamic characteristics of preterm infants with early onset sepsis. J. Paediatr. Child Health 2022, 58, 2267–2272. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, A.; Bielicki, J.; Mathur, S.; Sharland, M.; Van Den Anker, J.N. Reviewing the WHO guidelines for antibiotic use for sepsis in neonates and children. Paediatr. Int. Child Health 2018, 38, S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Boscarino, G.; Romano, R.; Iotti, C.; Tegoni, F.; Perrone, S.; Esposito, S. An Overview of Antibiotic Therapy for Early- and Late-Onset Neonatal Sepsis: Current Strategies and Future Prospects. Antibiotics 2024, 13, 250. [Google Scholar] [CrossRef]
- Prisma. Available online: http://www.prisma-statement.org/ (accessed on 2 October 2020).
- Lyra, S.; Jin, J.; Leonhardt, S.; Lüken, M. Early Prediction of Neonatal Sepsis from Synthetic Clinical Data Using Machine Learning. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; pp. 1–4. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wang, R.; Masino, A.J.; Obstfeld, A.E. Aiding clinical assessment of neonatal sepsis using hematological analyzer data with machine learning techniques. Int. J. Lab. Hematol. 2021, 43, 1341–1356. [Google Scholar] [CrossRef]
- Masino, A.J.; Harris, M.C.; Forsyth, D.; Ostapenko, S.; Srinivasan, L.; Bonafide, C.P.; Balamuth, F.; Schmatz, M.; Grundmeier, R.W. Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data. PLoS ONE 2019, 14, e0212665. [Google Scholar] [CrossRef]
- Robi, Y.G.; Sitote, T.M. Neonatal Disease Prediction Using Machine Learning Techniques. J. Health Eng. 2023, 2023, 3567194. [Google Scholar] [CrossRef]
- Mani, S.; Ozdas, A.; Aliferis, C.; Varol, H.A.; Chen, Q.; Carnevale, R.; Chen, Y.; Romano-Keeler, J.; Nian, H.; Weitkamp, J.-H. Medical decision support using machine learning for early detection of late-onset neonatal sepsis. J. Am. Med. Inform. Assoc. 2014, 21, 326–336. [Google Scholar] [CrossRef]
- Honoré, A.; Forsberg, D.; Adolphson, K.; Chatterjee, S.; Jost, K.; Herlenius, E. Vital sign-based detection of sepsis in neonates using machine learning. Acta Paediatr. 2023, 112, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Stocker, M.; Daunhawer, I.; van Herk, W.; el Helou, S.; Dutta, S.; Schuerman, F.A.B.A.; Groot, R.K.v.D.T.-D.; Wieringa, J.W.; Janota, J.; van der Meer-Kappelle, L.H.; et al. Machine Learning Used to Compare the Diagnostic Accuracy of Risk Factors, Clinical Signs and Biomarkers and to Develop a New Prediction Model for Neonatal Early-onset Sepsis. Pediatr. Infect. Dis. J. 2022, 41, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, M.A.; O’Jay, O.A.G.; Benders, M.M.; Bartels, R.R.; Vijlbrief, D.D. Development and clinical impact assessment of a machine-learning model for early prediction of late-onset sepsis. Comput. Biol. Med. 2023, 163, 107156. [Google Scholar] [CrossRef] [PubMed]
- Hasan, B.S.; Hoodbhoy, Z.; Khan, A.; Nogueira, M.; Bijnens, B.; Chowdhury, D. Can machine learning methods be used for identification of at-risk neonates in low-resource settings? A prospective cohort study. BMJ Paediatr. Open 2023, 7, e002134. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.A.; Beam, K.; Vesoulis, Z.A.; Aziz, K.B.; Husain, A.N.; Knake, L.A.; Moreira, A.G.; Hooven, T.A.; Weiss, E.M.; Carr, N.R.; et al. Transforming neonatal care with artificial intelligence: Challenges, ethical consideration, and opportunities. J. Perinatol. 2024, 44, 1–11. [Google Scholar] [CrossRef]
- Peng, Z.; Varisco, G.; Long, X.; Liang, R.H.; Kommers, D.; Cottaar, W.; Andriessen, P.; van Pul, C. A Continuous Late-Onset Sepsis Prediction Algorithm for Preterm Infants using Multimodal Physiological Signals from a Patient Monitor. IEEE J. Biomed. Health Inform. 2022, 27, 550–561. [Google Scholar] [CrossRef]
- O’sullivan, C.; Tsai, D.H.-T.; Wu, I.C.-Y.; Boselli, E.; Hughes, C.; Padmanabhan, D.; Hsia, Y. Machine learning applications on neonatal sepsis treatment: A scoping review. BMC Infect. Dis. 2023, 23, 441. [Google Scholar] [CrossRef]
- Khalilzad, Z.; Hasasneh, A.; Tadj, C. Newborn Cry-Based Diagnostic System to Distinguish between Sepsis and Respiratory Distress Syndrome Using Combined Acoustic Features. Diagnostics 2022, 12, 2802. [Google Scholar] [CrossRef]
- Gojak, D.; Gvožđar, K.; Hećimović, Z.; Smajović, A.; Bečić, E.; Deumić, A.; Bećirović, L.S.; Pokvić, L.G.; Badnjević, A. The use of artificial intelligence in the diagnosis of neonatal sepsis. IFAC-Pap. 2022, 55, 62–67. [Google Scholar] [CrossRef]
- Sahu, P.; Stanly, E.A.R.; Lewis, L.E.S.; Prabhu, K.; Rao, M.; Kunhikatta, V. Prediction modelling in the early detection of neonatal sepsis. World J. Pediatr. 2022, 18, 160–175. [Google Scholar] [CrossRef]
- Iqbal, F.; Chandra, P.; Khan, A.A.; Lewis, L.E.S.; Acharya, D.; Vandana, K.; Jayashree, P.; Shenoy, P.A. Prediction of mortality among neonates with sepsis in the neonatal intensive care unit: A machine learning approach. Clin. Epidemiol. Glob. Health 2023, 24, 101414. [Google Scholar] [CrossRef]
- McAdams, R.M.; Kaur, R.; Sun, Y.; Bindra, H.; Cho, S.J.; Singh, H. Predicting clinical outcomes using artificial intelligence and machine learning in neonatal intensive care units: A systematic review. J. Perinatol. 2022, 42, 1561–1575. [Google Scholar] [CrossRef]
- Hsu, J.-F.; Chang, Y.-F.; Cheng, H.-J.; Yang, C.; Lin, C.-Y.; Chu, S.-M.; Huang, H.-R.; Chiang, M.-C.; Wang, H.-C.; Tsai, M.-H. Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit. J. Pers. Med. 2021, 11, 695. [Google Scholar] [CrossRef] [PubMed]
- Mangold, C.; Zoretic, S.; Thallapureddy, K.; Moreira, A.; Chorath, K.; Moreira, A. Machine Learning Models for Predicting Neonatal Mortality: A Systematic Review. Neonatology 2021, 118, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Singh, C.; Kumar, Y. Diagnosis and Detection of Congenital Diseases in New-Borns or Fetuses Using Artificial Intelligence Techniques: A Systematic Review. Arch. Comput. Methods Eng. 2023, 30, 3031–3058. [Google Scholar] [CrossRef]
- Moorman, J.R. The principles of whole-hospital predictive analytics monitoring for clinical medicine originated in the neonatal ICU. npj Digit. Med. 2022, 5, 41. [Google Scholar] [CrossRef]
- Shirwaikar, R.D.; Acharya, D.; Makkithaya, K.; Surulivelrajan, M.; Srivastava, S. Optimizing neural networks for medical data sets: A case study on neonatal apnea prediction. Artif. Intell. Med. 2019, 98, 59–76. [Google Scholar] [CrossRef] [PubMed]
- De Camargo, J.F.; Caldas, J.P.d.S.; Marba, S.T.M. Early neonatal sepsis: Prevalence, complications and outcomes in newborns with 35 weeks of gestational age or more. Rev. Paul. Pediatr. 2022, 40, e2020388. [Google Scholar] [CrossRef]
| Title | Author | Year | Country | Number of Patients in the Study | AI and Statistic Models | Scope of Research | Top Significant Parameters for the Models |
|---|---|---|---|---|---|---|---|
| Medical decision support using machine learning for early detection of late-onset neonatal sepsis | Subramani Mani et al. [24] | 2014 | USA | 299 (90 positive) | Support Vector Machine (SVM), the naive Bayes (NB) classifier, tree augmented naive Bayes (TAN), averaged one dependence estimators (AODE), K-nearest neighbor (KNN), classification and regression trees (CART), andom forests (RF), logistic regression (LR), lazy Bayesian rules (LBR) | sepsis, antibiotic treatment | chorioamnionitis, white blood cells count, anemia of prematurity, maternal age and resuscitation at birth to be strong predictive factors for LOS |
| Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data | Aaron J. Masino et al. [22] | 2019 | USA | 1188 (375 positive) | logistic regression (LR), naïve Bayes (NB), SVM with a radial basis function kernel, K-nearest neighbors (KNN), Gaussian process, RF, AdaBoost, and gradient boosting | sepsis, antibiotic treatment | heart rate difference, systemic and diastolic blood pressure, platelet count, I/T ratio, mean arterial pressure, gestational age |
| Optimizing neural networks for medical data sets: A case study on neonatal apnea prediction | Rudresh Deepak Shirwaikar et al. [41] | 2019 | India | 367 | KNN, SVM, RF, MPL decision trees, deep autoencoders | sepsis | birth weight, heart rate, desaturation, gestation age |
| Aiding clinical assessment of neonatal sepsis using hematological analyzer data with machine learning techniques | Huang, B et al. [21] | 2021 | USA | 2900 (2032 sepsis positive) | extreme gradient boosting (XGB), Random Forest (RF), and Support Vector Machine (SVM) | sepsis | neutrophil fluorescence intensity (NE_SFL), neutrophil cell size (NE_FSC), neutrophil dispersion width (NE_WY), gestational age, and monocyte fluorescence intensity (MO_Y) |
| Machine Learning Approaches to Predict In-Hospital Mortality among Neonates with Clinically Suspected Sepsis in the Neonatal Intensive Care Unit | Jen-Fu Hsu et al. [37] | 2021 | Taiwan | 2472 (1095 posotove) | DNN model, k-nearest neighbors (k-NN), vector machine (SVM), random forest (RF), extreme gradient boost (XGB), Glmnet, regression tree algorithm (Treebag) | sepsis | requirement of ventilator support, feeding conditions intravascular volume expansion |
| Machine Learning Used to Compare the Diagnostic Accuracy of Risk Factors, Clinical Signs and Biomarkers and to Develop a New Prediction Model for Neonatal Early-onset Sepsis | Stocker, Martin et al. [26] | 2022 | Netherlands, Canada, Czech Republic and Switzerland | 1685 (28 positive) | CSs, Random Forest (RF) | early-onset sepsis (EOS) | C-reactive protein, leukocyte count, platelet count, birth weight, gestational age |
| A Continuous Late-Onset Sepsis Prediction Algorithm for Preterm Infants Using Multi-Channel Physiological Signals From a Patient Monitor | Zheng Peng et al. [30] | 2022 | Netherlands | 127 | extreme gradient boosting (XGB), k-nearest neighbors (KNN), logistic regression (LR), support vector machine (SVM) | LOS | HRV features, breathing features, movement features, combination of HRV and breathing features and combination of all features |
| Newborn Cry-Based Diagnostic System to Distinguish between Sepsis and Respiratory Distress Syndrome Using Combined Acoustic Features | Zahra Khalilzad et al. [32] | 2022 | Canada, Lebanon | 50 (17 positive) | deep feedforward neural network (DFNN), vector machine (SVM) model, convolutional neural network (CNN), Multilayer Perceptron (MLP) | sepsis | recording newborn cry-based |
| The use of artificial intelligence in the diagnosis of neonatal sepsis | Dž. Gojak et al. [33] | 2022 | Bosnia and Herzegovina | 1000 | artificial neural network (ANN) | sepsis | Body temperature, C-reactive protein, leukocyte count, Platelet count |
| Neonatal Disease Prediction Using Machine Learning Techniques | Yohanes Gutema Robi et al. [23] | 2023 | Etiopia | 180 | XGBoost (XGB), Random Forest (RF), and Support Vector Machine (SVM) | late-onset sepsis (LOS) | APGAR, C-reactive protein, resuscitation, low lung volume and bleaching (LLVB), intercostal subcostal retractions (ICSCR), SpO2, gestational age, white blodd cells count, convulsions, respiratory function, weight |
| Vital sign-based detection of sepsis in neonates using machine learning | Antoine Honoré et al. [25] | 2023 | Sweden | 325 (20 positive) | model Hidden Markowa | sepsis | birth weight, gender and postnatal age, heart rate, respiratory characteristics |
| Development and clinical impact assessment of a machine-learning model for early prediction of late-onset sepsis | Merel (A.M.) van den Berg et al. [27] | 2023 | Netherlands | 2519 (389 positive) | logistic regression (LR), GAM i XGBoost | LOS | C-reactive protein, leukocyte count, neutrophil count and thrombocyte counts |
| Prediction of mortality among neonates with sepsis in the neonatal intensive care unit: A machine learning approach | Colleen O’Sullivan et al. [31] | 2023 | India | 388 (184 positive) | naive bayes (base line model), ogistic regression, sequential minimal optimization (SMO), classification and regression tree (CART), Random Forest | EOS, LOS | PROM, absent end diastolic flow, chorioamnionitis-maternal predictors; preterm birth, birthweight (>2500 g), appearance, pulse, grimace, activity, and respiration, APGAR at 5 min, C-reactive protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tądel, K.; Dudek, A.; Bil-Lula, I. AI Algorithms for Modeling the Risk, Progression, and Treatment of Sepsis, Including Early-Onset Sepsis—A Systematic Review. J. Clin. Med. 2024, 13, 5959. https://doi.org/10.3390/jcm13195959
Tądel K, Dudek A, Bil-Lula I. AI Algorithms for Modeling the Risk, Progression, and Treatment of Sepsis, Including Early-Onset Sepsis—A Systematic Review. Journal of Clinical Medicine. 2024; 13(19):5959. https://doi.org/10.3390/jcm13195959
Chicago/Turabian StyleTądel, Karolina, Andrzej Dudek, and Iwona Bil-Lula. 2024. "AI Algorithms for Modeling the Risk, Progression, and Treatment of Sepsis, Including Early-Onset Sepsis—A Systematic Review" Journal of Clinical Medicine 13, no. 19: 5959. https://doi.org/10.3390/jcm13195959
APA StyleTądel, K., Dudek, A., & Bil-Lula, I. (2024). AI Algorithms for Modeling the Risk, Progression, and Treatment of Sepsis, Including Early-Onset Sepsis—A Systematic Review. Journal of Clinical Medicine, 13(19), 5959. https://doi.org/10.3390/jcm13195959






