Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics
Abstract
1. Introduction
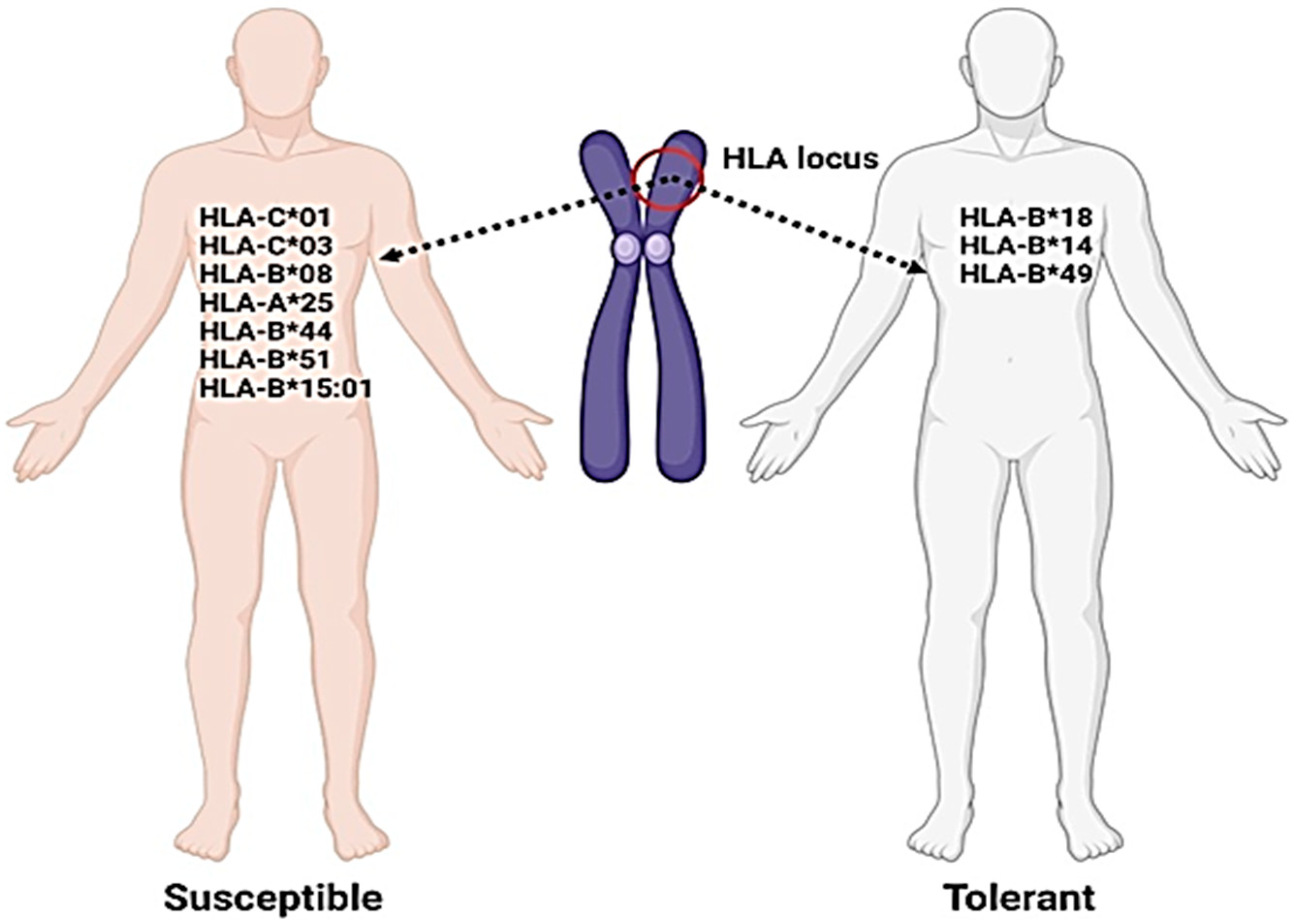
2. HLA Variants in COVID-19 Disease
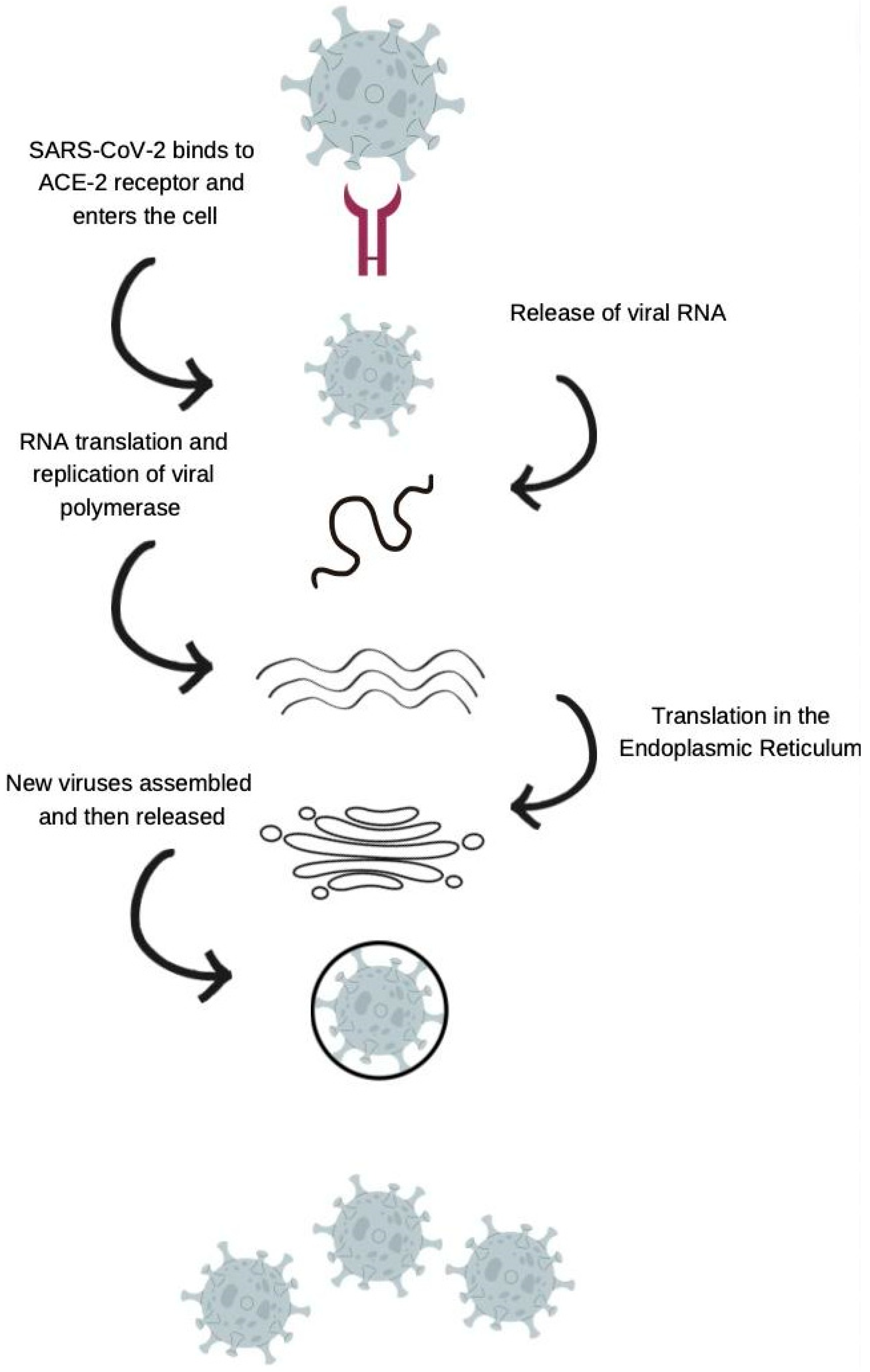
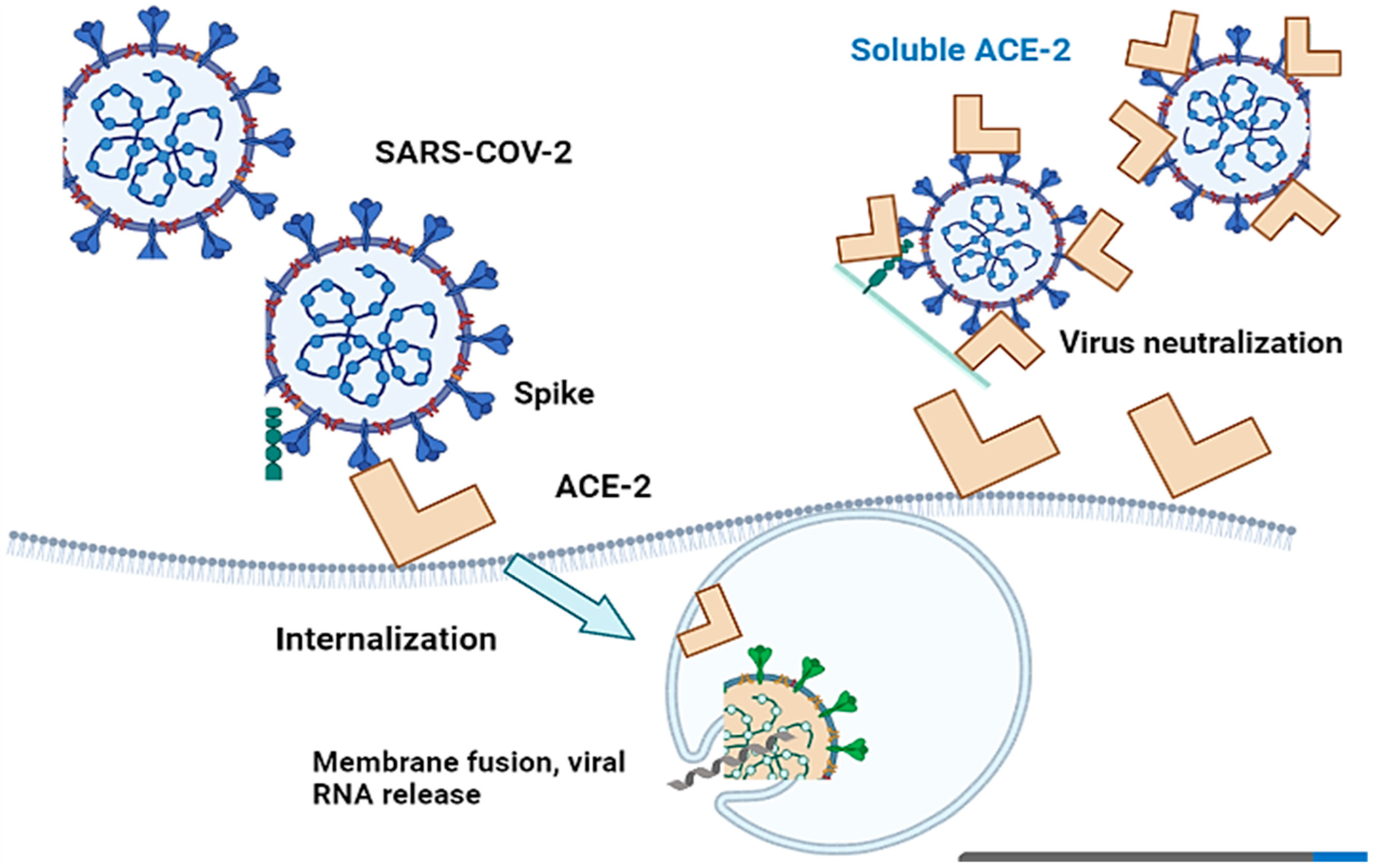
3. Angiotensin-Converting Enzyme 2 (ACE-2) in COVID-19
4. Genetic Polymorphism of ACE-2
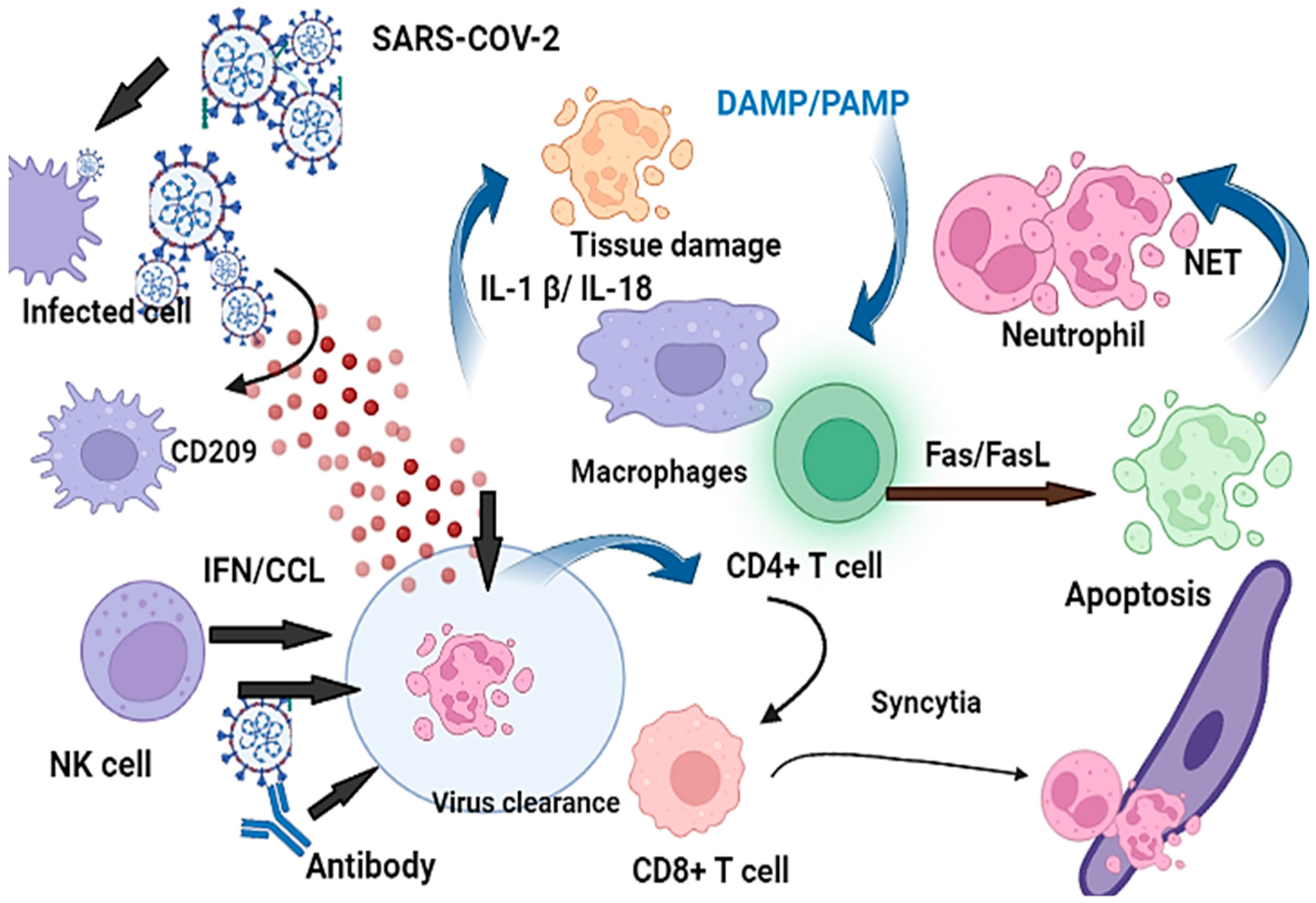
5. The Association of Antiviral Immune Responses to COVID-19 Severity
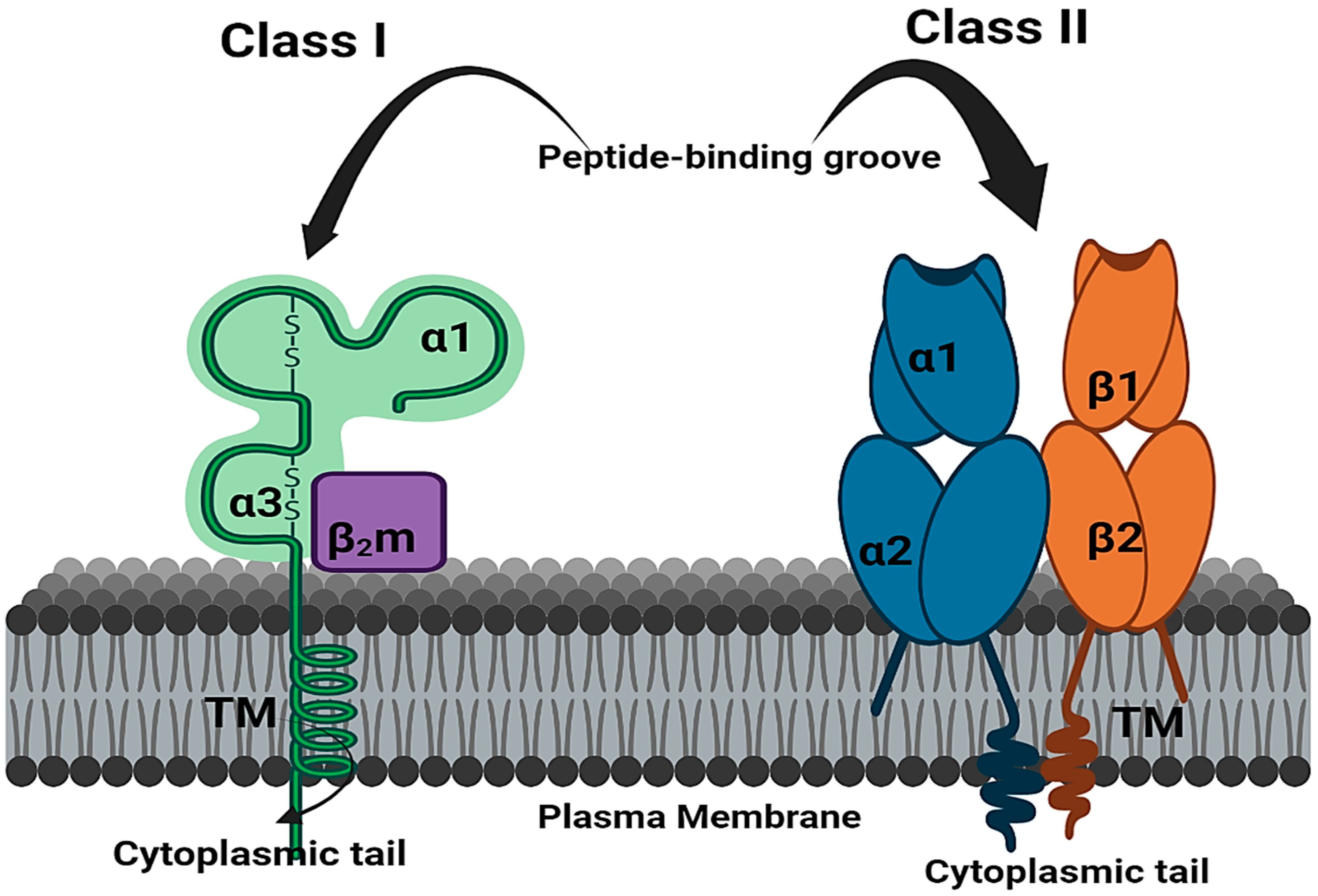
6. The Role of HLA in Antiviral Immunity against SARS-CoV-2
7. Mechanism of HLA Alleles
8. The Regional Analysis of HLA
9. HLA Variants in the Genetic Susceptibility
10. HLA-Based Vaccines
| Human Leukocyte Antigens | Association with SARS-CoV Infections | References |
|---|---|---|
| HLA-B*07:03, HLA-Cw*08:01, HLADRB1*12:02, HLA-B*46:01 | Increase the susceptibility | [86,87] |
| HLA-A*02:01, HLA-DR*03:01, and HLA-Cw*15:02 | Provide protection against SARS-CoV-2 | [24] |
| HLA-C*01 and/or HLA-B*44 | Have association with inflammatory autoimmune diseases in SARS-CoV-2 infection and inappropriate immunological reaction | [88] |
| HLA-C*01 and B*44 alleles | Can significantly help in the understanding of disease transmission and physiopathogenesis | [89] |
11. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ul Haq, I.; Krukiewicz, K.; Yahya, G.; Haq, M.U.; Maryam, S.; Mosbah, R.A.; Saber, S.; Alrouji, M. The breadth of bacteriophages contributing to the development of the phage-based vaccines for COVID-19: An ideal platform to design the multiplex vaccine. Int. J. Mol. Sci. 2023, 24, 1536. [Google Scholar] [CrossRef] [PubMed]
- Egbuna, C.; Chandra, S.; Awuchi, C.G.; Aklani, S.; Ulhaq, I.; Akram, M.; Khan, J. Myth surrounding the FDA disapproval ofhydroxychloroquine sulfate and chloroquine phosphate as drugs for coronavirus disease 2019. In Coronavirus Drug Discovery; Elsevier: Amsterdam, The Netherlands, 2022; pp. 153–168. [Google Scholar]
- Maryam, S.; Ul Haq, I.; Yahya, G.; Ul Haq, M.; Algammal, A.M.; Saber, S.; Cavalu, S. COVID19 surveillance in wastewater: An epidemiological tool for the monitoring of SARS-CoV-2. Front. Cell. Infect. Microbiol. 2023, 12, 978643. [Google Scholar] [CrossRef] [PubMed]
- Ulhaq, I.; Basit, A.; Ali, I.; Hussain, F.; Ali, Z.; Siddique, F.; Ahmed, H.; Aqib, A.I.; Rahim, K. Coronavirus Disease-2019 (COVID-19) Epidemiology. In COVID-19: Epidemiology, Biochemistry, and Diagnostics; Coronavirus Disease-19(COVID-19): A Perspective of New Scenario Series; Bentham Science: Oak Park, IL, USA, 2021; Volume 1, pp. 1–41. [Google Scholar] [CrossRef]
- Basit, A.; Ulhaq, I.; Hussain, F.; Ud-Din, Z.; Rahim, K. Nucleic Acid Based Detection of COVID-19. In COVID-19: Epidemiology, Biochemistry, and Diagnostics; Coronavirus Disease-19(COVID-19): A Perspective of New Scenario Series; Bentham Science: Oak Park, IL, USA, 2021; Volume 1, pp. 405–419. [Google Scholar] [CrossRef]
- Ayaz, M.M.; Ulhaq, I.; Rahim, k. Histopathologic Evaluation and Scoring of SARS-CoV-2 Infection. In COVID-19: Different Models and Treatment Strategies; Coronavirus Disease-19(COVID-19): A Perspective of New Scenario Series; Bentham Science: Oak Park, IL, USA, 2021; Volume 2, pp. 52–71. [Google Scholar] [CrossRef]
- Ali, I.; Rasheed, M.A.; Cavalu, S.; Rahim, K.; Ijaz, S.; Yahya, G.; Goh, L.P.W.; Popoviciu, M.S. Identification of Natural Lead Compounds against Hemagglutinin-Esterase Surface Glycoprotein in Human Coronaviruses Investigated via MD Simulation, Principal Component Analysis, Cross-Correlation, H-Bond Plot and MMGBSA. Biomedicines 2023, 11, 793. [Google Scholar] [CrossRef] [PubMed]
- Haq, I.U.; Khan, Z.I.; Aziz, I.; Basit, A.; Hussain, F.; Bibi, A.; Aqib, A.I.; Younas, U.; Rahim, K. Chapter 12—Phages and SARS-CoV-2. In Application of Natural Products in SARS-CoV-2; Elsevier: Amsterdam, The Netherlands, 2023; pp. 273–292. ISBN 9780323950473. [Google Scholar] [CrossRef]
- Metwally, K.; Abo-Dya, N.E.; Alahmdi, M.I.; Albalawi, M.Z.; Yahya, G.; Aljoundi, A.; Salifu, E.Y.; Elamin, G.; Ibrahim, M.A.A.; Sayed, Y. The Unusual Architecture of RNA-Dependent RNA Polymerase (RdRp)’s Catalytic Chamber Provides a Potential Strategy for Combination Therapy against COVID-19. Molecules 2023, 28, 2806. [Google Scholar] [CrossRef]
- Available online: http://covid19.who.int/ (accessed on 21 February 2023).
- El-Sokkary, M.M.; El-Baz, A.M.; El-Morsi, R.M.; Keuper, K.; El-Hawary, S.; Shata, A.; Yahya, G. Early forecasting of COVID-19 case progression with hematological and biochemical parameters of patients in Egypt. Paki. J. Pharma. Sci. 2022, 35, 401–408. [Google Scholar]
- Al Naggar, Y.; Giesy, J.P.; Abdel-Daim, M.M.; Javed Ansari, M.; Al-Kahtani, S.N.; Yahya, G. Fighting against the second wave of COVID-19: Can honeybee products help protect against the pandemic? Saudi. J. Biol. Sci. 2021, 28, 1519–1527. [Google Scholar] [CrossRef]
- Elmorsy, M.A.; El-Baz, A.M.; Mohamed, N.H.; Almeer, R.; Abdel-Daim, M.M.; Yahya, G. In silico screening of potent inhibitors against COVID-19 key targets from a library of FDA-approved drugs. Environ. Sci. Pollut. Res. 2022, 29, 12336–12346. [Google Scholar] [CrossRef]
- Mostafa, I.; Mohamed, N.H.; Mohamed, B.; Almeer, R.; Abulmeaty, M.M.A.; Bungau, S.G.; El-Shazly, A.M.; Yahya, G. In-silico screening of naturally derived phytochemicals against SARS-CoV Main protease. Environ. Sci. Pollut. Res. 2022, 29, 26775–26791. [Google Scholar] [CrossRef]
- Shaldam, M.A.; Yahya, G.; Mohamed, N.H.; Abdel-Daim, M.M.; Al Naggar, Y. In silico screening of potent bioactive compounds from honeybee products against COVID-19 target enzymes. Environ. Sci. Pollut. Res. 2021, 28, 40507–40514. [Google Scholar] [CrossRef]
- Yahya, G.; Mansour, B.; Keuper, K.; Shaldam, M.; El-Baz, A.M. Virtual Screening Attributes Male Biased COVID-19 Mortality to Predicted Antiviral Activity of Female Sex Hormones. Let. Drug Des. Discov. 2021, 18, 872–883. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Soltane, R.; Abo Elmaaty, A.; Tantawy, M.A.; Antar, S.A.; Yahya, G.; Chrouda, A.; Pashameah, R.A.; Mustafa, M.; Abu Mraheil, M.; et al. Coronavirus Disease (COVID-19) Control between Drug Repurposing and Vaccination: A Comprehensive Overview. Vaccines 2021, 9, 1317. [Google Scholar] [CrossRef]
- Lan, N.T.P.; Kikuchi, M.; Huong, V.T.Q.; Ha, D.Q.; Thuy, T.T.; Tham, V.D.; Tuan, H.M.; Van Tuong, V.; Nga, C.T.P.; Van Dat, T.; et al. Protective and Enhancing HLA Alleles, HLA-DRB1*0901 and HLA-A*24, for Severe Forms of Dengue Virus Infection, Dengue Hemorrhagic Fever and Dengue Shock Syndrome. PLoS Negl. Trop. Dis. 2008, 2, e304. [Google Scholar] [CrossRef]
- Lin, M.; Tseng, H.-K.; Trejaut, J.A.; Lee, H.-L.; Loo, J.-H.; Chun-Hsiung, H.; Chen, P.-J.; Su, Y.-W.; Lim, K.H.; Tsai, Z.-U.; et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC. Med. Gen. 2003, 4, 9. [Google Scholar] [CrossRef]
- Hajeer, A.H.; Balkhy, H.; Johani, H.; Yousef, M.; Arabi, Y. Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome—Coronavirus infection. Ann. Thorac. Med. 2016, 11, 211–213. [Google Scholar] [CrossRef]
- Luckey, D.; Weaver, E.A.; Osborne, D.G.; Billadeau, D.D.; Taneja, V. Immunity to Influenza is dependent on MHC II polymorphism: Study with 2 HLA transgenic strains. Sci. Rep. 2019, 9, 19061. [Google Scholar] [CrossRef]
- Nishida, N.; Ohashi, J.; Khor, S.-S.; Sugiyama, M.; Tsuchiura, T.; Sawai, H.; Hino, K.; Honda, M.; Kaneko, S.; Yatsuhashi, H.; et al. Understanding of HLA-conferred susceptibility to chronic hepatitis B infection requires HLA genotyping-based association analysis. Sci. Rep. 2016, 6, 24767. [Google Scholar] [CrossRef]
- Shiina, T.; Hosomichi, K.; Inoko, H.; Kulski, J.K. The HLA genomic loci map: Expression, interaction, diversity and disease. J. Hum. Genet. 2009, 54, 15–39. [Google Scholar] [CrossRef]
- Nguyen, A.; David, J.K.; Maden, S.K.; Wood, M.A.; Weeder, B.R.; Nellore, A.; Thompson, R.F. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome coronavirus 2. J. Virol. 2020, 94, e00510–e00520. [Google Scholar] [CrossRef]
- Savage, S.A.; Viard, M.; O’Huigin, C.; Zhou, W.; Yeager, M.; Li, S.A.; Wang, T.; Ramsuran, V.; Vince, N.; Vogt, A.; et al. Genome-wide association study identifies HLA-DPB1 as a significant risk factor for severe aplastic anemia. Am. J. Hum. Gen. 2020, 106, 264–271. [Google Scholar] [CrossRef]
- Patel, N.; Sethi, Y.; Kaka, N.; Kaiwan, O.; Gupta, I.; Shaheen, R.S.; Sapoor, S.; Chopra, H.; Popoviciu, M.S.; Emran, T.B.; et al. Acute Hepatitis of Unknown Origin in Pediatric Age Group: Recent Outbreaks and Approach to Management. J. Clin. Med. 2023, 12, 9. [Google Scholar] [CrossRef]
- Ni, W.; Yang, X.; Yang, D.; Bao, J.; Li, R.; Xiao, Y.; Hou, C.; Wang, H.; Liu, J.; Yang, D.; et al. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care 2020, 24, 422. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.; de Oliveira, S.C. The Impact of Angiotensin-Converting Enzyme 2 (ACE2) Expression Levels in Patients with Comorbidities on COVID-19 Severity: A Comprehensive Review. Microorganisms 2021, 9, 1692. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Wang, J.; Bachtiar, M.; Chong, S.S.; Lee, C.G.L. Architecture of polymorphisms in the human genome reveals functionally important and positively selected variants in immune response and drug transporter genes. Hum. Genom. 2018, 12, 43. [Google Scholar] [CrossRef] [PubMed]
- Keicho, N.; Itoyama, S.; Kashiwase, K.; Phi, N.C.; Long, H.T.; Van Ban, V.; Hoa, B.K.; Le Hang, N.T.; Hijikata, M.; Sakurada, S.; et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009, 70, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Reusch, N.; De Domenico, E.; Bonaguro, L.; Schulte-Schrepping, J.; Babler, K.; Schultze, J.L.; Aschenbrenner, A.C. Neutrophils in COVID-19. Front. Immunol. 2021, 12, 652470. [Google Scholar] [CrossRef]
- Tavasolian, F.; Rashidi, M.; Hatam, G.R.; Jeddi, M.; Hosseini, A.Z.; Mosawi, S.H.; Abdollahi, E.; Inman, R.D. HLA, Immune Response, and Susceptibility to COVID-19. Front. Imunol. 2021, 11, 601886. [Google Scholar] [CrossRef]
- Augusto, D.G.; Hollenbach, J.A. HLA variation and antigen presentation in COVID-19 and SARS-CoV-2 infection. Curr. Opin. Immunol. 2022, 76, 102178. [Google Scholar] [CrossRef]
- Abbasifard, M.; Khorramdelazad, H. The bio-mission of interleukin-6 in the pathogenesis of COVID-19: A brief look at potential therapeutic tactics. Life Sci. 2020, 257, 118097. [Google Scholar] [CrossRef]
- Wang, W.; Zhang, W.; Zhang, J.; He, J.; Zhu, F. Distribution of Allele Frequencies in 82 Chinese Individuals with Coronavirus Disease-2019 (COVID-19). HLA 2020, 96, 194–196. [Google Scholar] [CrossRef]
- Khor, S.-S.; Omae, Y.; Nishida, N.; Sugiyama, M.; Kinoshita, N.; Suzuki, T.; Suzuki, M.; Suzuki, S.; Izumi, S.; Hojo, M.; et al. HLA-A*11:01:01:01, HLA-C*12:02:02:01-HLA-B*52:01:02:02, Age and Sex Are Associated with Severity of Japanese COVID-19 with Respiratory Failure. Front. Immunol. 2021, 12, 658570. [Google Scholar] [CrossRef]
- Zahn, L.M. HLA Genetics and COVID-19. Science 2020, 368, 841–2841. [Google Scholar] [CrossRef]
- Available online: http://hla.alleles.org/nomenclature/naming.html (accessed on 15 December 2022).
- Douillard, V.; Castelli, E.C.; Mack, S.J.; Hollenbach, J.A.; Gourraud, P.A.; Vince, N.; Limou, S.; Covid-19|HLA & Immunogenetics Consortium and the SNP-HLA Reference Consortium. Current HLA Investigations on SARS-CoV-2 and Perspectives. Front. Genet. 2021, 12, 774922. [Google Scholar] [CrossRef]
- Astbury, S.; Reynolds, C.J.; Butler, D.K.; Muñoz-Sandoval, D.C.; Lin, K.M.; Pieper, F.P.; Otter, A.; Kouraki, A.; Cusin, L.; Nightingale, J.; et al. HLA-DR polymorphism in SARS-CoV-2 infection and susceptibility to symptomatic COVID-19. Immunology 2022, 166, 68–77. [Google Scholar] [CrossRef]
- Li, M.; Li, L.; Zhang, Y.; Wang, X.S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 2020, 9, 23–29. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Resuction and functional exhaustion of T cells in patients with Coronavirus Disease COVID-19. Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Xu, J.; Chu, M.; Zhong, F.; Tan, X.; Tang, G.; Mai, J.; Lai, N.; Guan, C.; Liang, Y.; Liao, G. Digestive symptoms of COVID-19 and expression of ACE2 in digestive tract organs. Cell Death. Discov. 2020, 6, 76. [Google Scholar] [CrossRef]
- Suryamohan, K.; Diwanji, D.; Stawiski, E.W.; Gupta, R.; Miersch, S.; Liu, J.; Chen, C.; Jiang, Y.-P.; Fellouse, F.A.; Sathirapongsasuti, J.F.; et al. Human ACE2 receptor polymorphisms and altered susceptibility to SARS-CoV-2. Commun. Biol. 2021, 4, 475. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Beyerstedt, S.; Casaro, E.B.; Rangel, É.B. COVID-19: Angiotensin-converting enzyme 2 (ACE2) expression and tissue susceptibility to SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 905–919. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. J. Biol. Chem. 2000, 275, 33238–33324. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; She, Z.-G.; Cheng, X.; Qin, J.-J.; Zhang, X.-J.; Cai, J.; Lei, F.; Wang, H.; Xie, J.; Wang, W.; et al. Association of blood glucose control and outcomes in patients with COVID-19 and pre-existing type 2 diabetes. Cell Metab. 2020, 31, 1068–1077.e3. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses 2015, 1282, 1–23. [Google Scholar] [CrossRef]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar]
- Zost, S.J.; Gilchuk, P.; Case, J.B.; Binshtein, E.; Chen, R.E.; Nkolola, J.P.; Schäfer, A.; Reidy, J.X.; Trivette, A.; Nargi, R.S.; et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 2020, 584, 443–449. [Google Scholar] [CrossRef]
- Carter-timofte, M.E.; Jørgensen, S.E.; Freytag, M.R. Deciphering the Role of Host Genetics in Susceptibility to Severe COVID-19. Front. Immunol. 2020, 11, 1606. [Google Scholar] [CrossRef]
- Rahimi, P.; Tarharoudi, R.; Rahimpour, A.; Amroabadi, J.M.; Ahmadi, I.; Anvari, E.; Siadat, S.D. The association between interferon lambda 3 and 4 gene single-nucleotide polymorphisms and the recovery of COVID-19 patients. Virol. J. 2021, 18, 221. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, M.; Yang, C.; Zhang, N.; Wang, X.C.; Yang, X.P.; Dong, X.Q. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef]
- McCoy, K.; Peterson, A.; Tian, Y.; Sang, Y. Immunogenetic Association Underlying Severe COVID-19. Vaccines 2020, 8, 700. [Google Scholar] [CrossRef]
- Duque, G.A.; Descoteaux, A. Macrophage cytokines: Involvement in immunity and infectious diseases. Front. Immunol. 2014, 5, 491. [Google Scholar]
- Mozafari, N.; Azadi, S.; Mehdi-Alamdarlou, S.; Ashrafi, H.; Azadi, A. Inflammation: A bridge between diabetes and COVID-19, and possible management with sitagliptin. Med. Hypoth. 2020, 143, 110111. [Google Scholar] [CrossRef]
- Ryan, S.O.; Cobb, B.A. Roles for major histocompatibility complex glycosylation in immune function. Semin. Immunopathol. 2012, 34, 425–441. [Google Scholar] [CrossRef]
- Burgner, D.; Jamieson, S.E.; Blackwell, J.M. Genetic susceptibility to infectious diseases: Big is beautiful, but will bigger be even better? Lancet Infect. Dis. 2006, 6, 653–663. [Google Scholar] [CrossRef]
- Loi, E.; Moi, L.; Cabras, P.; Arduino, G.; Costanzo, G.; Del Giacco, S.; Erlich, H.A.; Firinu, D.; Caddori, A.; Zavattari, P. HLA-C dysregulation as a possible mechanism of immune evasion in SARS-CoV-2 and other RNA-virus infections. Front. Immunol. 2022, 13, 1011829. [Google Scholar] [CrossRef]
- Littera, R.; Campagna, M.; Deidda, S.; Angioni, G.; Cipri, S.; Melis, M.; Firinu, D.; Santus, S.; Lai, A.; Porcella, R.; et al. Human Leukocyte Antigen Complex and Other Immunogenetic and Clinical Factors Influence Susceptibility or Protection to SARS-CoV-2 Infection and Severity of the Disease Course. Sard. Exp. 2020, 11, 60568. [Google Scholar] [CrossRef]
- Pisanti, S.; Deelen, J.; Gallina, A.M.; Caputo, M.; Citro, M.; Abate, M.; Sacchi, N.; Vecchione, C.; Martinelli, R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of COVID-19. J. Transl. Med. 2020, 18, 352. [Google Scholar] [CrossRef]
- Migliorini, F.; Torsiello, E.; Spiezia, F.; Oliva, F.; Tingart, M.; Maffuli, M. Association between HLA genotypes and COVID-19 susceptibility, severity and progression: A comprehensive review of the literature. Eur. J. Med. Res. 2021, 26, 84. [Google Scholar] [CrossRef]
- Silvestr, D.; Kourilsk, F.M.; Niccolai, M.G.; Levy, J.P. Presence of HLA antigens on human reticulocytes as demonstrated by electron microscopy. Nature 1970, 228, 67–68. [Google Scholar] [CrossRef]
- Ujvari, B.; Belov, K. Major Histocompatibility Complex (MHC) Markers in Conservation Biology. Int. J. Mol. Sci. 2011, 12, 5168–5186. [Google Scholar] [CrossRef]
- Allard, M.; Oger, R.; Benlalam, H.; Florenceau, L.; Echasserieau, K.; Bernardeau, K.; Labarrière, N.; Lang, F.; Gervois, N. Soluble HLA-I/peptide monomers mediate antigen-specific CD8 T cell activation through passive peptide exchange with cell-bound HLA-I molecules. J Immunol. 2014, 192, 5090–5097. [Google Scholar] [CrossRef] [PubMed]
- Leddon, S.A.; Sant, A.J. Generation of MHC class II-peptide ligands for CD4 T-cell allorecognition of MHC class II molecules. Curr. Opin. Organ Transplant. 2010, 15, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, C.; Furst, D.; Fae, I.; Wenda, S.; Zollikofer, C.; Mytilineos, J.; Fischer, G.F. HLA typing by next-generation sequencing-getting closer to reality. Tissue Antigens 2014, 83, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Lundegaard, C.; Lamberth, K.; Harndahl, M.; Buus, S.; Lund, O.; Nielsen, M. NetMHC-3.0: Accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008, 36, W509–W512. [Google Scholar] [CrossRef] [PubMed]
- Sadegh-Nasseri, S.; Kim, A. Exogenous antigens bind MHC class II first and are processed by cathepsins later. Mol. Immunol. 2015, 68, 81–84. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Cooper, M.A.; Nuovo, G.J.; Cella, M.; Facchetti, F.; Colonna, M.; Caligiuri, M.A. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: A potential new link between adaptive and innate immunity. Blood 2003, 101, 3052–3057. [Google Scholar] [CrossRef]
- Kachuri, L.; Francis, S.S.; Morrison, M.L.; Wendt, G.A.; Bosse, Y.; Cavazos, T.B.; Rashkin, S.R. The landscape of host genetic factors involved in immune response to common viral infections. Genome Med. 2020, 12, 93. [Google Scholar] [CrossRef]
- Grifoni, A.; Sidney, J.; Vita, R.; Peters, B.; Crotty, S.; Weiskopf, D.; Sette, D. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe 2021, 29, 1076–1092. [Google Scholar] [CrossRef]
- Mai, H.; Chen, J.; Chen, H.; Liu, Z.; Huang, G.; Wang, J.; Xiao, Q.; Ren, W.; Zhou, B. Fine Mapping of the MHC Region Identifies Novel Variants Associated with HBV-Related Hepatocellular Carcinoma in Han Chinese. J. Hepatocell. Carcinoma 2021, 8, 951–961. [Google Scholar] [CrossRef]
- Wieczorek, M.; Abualrous, E.T.; Sticht, J.; Álvaro-Benito, M.; Stolzenberg, S.; Noé, F.; Freund, C. Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 2017, 8, 292. [Google Scholar] [CrossRef]
- Sahin Tekin, M.; Yorulmaz, G.; Yantir, E.; Gunduz, E.; Colak, E. A Novel Finding of an HLA Allele’s and a Haplotype’s Relationship with SARS-CoV-2 Vaccine-Associated Subacute Thyroiditis. Vaccines 2022, 10, 1986. [Google Scholar] [CrossRef]
- Kuznetsov, A.; Voronina, A.; Govorun, V.; Arapidi, G. Critical Review of Existing MHC I Immunopeptidome Isolation Methods. Molecules 2020, 25, 5409. [Google Scholar] [CrossRef]
- Lassale, C.; Gaye, B.; Hamer, M.; Galle, C.R.; Batty, G.D. Ethnic disparities in hospitalization for COVID-19 in England: The role of socioeconomic factors, mental health, and inflammatory and pro-inflammatory factors in a community-based cohort study. Brain Behav. Immun. 2020, 88, 44–49. [Google Scholar] [CrossRef]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Quadeer, A.A.; Mckay, M.R. Preliminary Identification of Potential Vaccine Targets for the COVID-19 Coronavirus (SARS-CoV-2 ) Based on SARS-CoV Immunological Studies. Viruses 2020, 12, 254. [Google Scholar] [CrossRef]
- Lim, H.X.; Lim, J.; Jazayeri, S.D.; Popemma, S.; Pohl, C.L. Development of multi-epitope peptide-based vaccines against SARS-CoV-2. Biomed. J. 2021, 44, 18–30. [Google Scholar] [CrossRef]
- Ihtisham, U.H.; Kashif, R.; Muhammad, R.; Tayyaba, A.; Sifa, A.; Kinza, Y. Chapter 18—Polyketides and SARS-CoV-2. In Application of Natural Products in SARS-CoV-2; Elsevier: Amsterdam, The Netherlands, 2023; pp. 423–444. ISBN 9780323950473. [Google Scholar] [CrossRef]
- Ihtisham, U.H.; Fatima, F.; Amna, S.; Abdul, B.; Firasat, H.; Israr, A.; Zarak, I.K.; Amjad, I.A.; Faisal, S.; Umair, Y.; et al. Natural Products and SARS-CoV-2. In Application of Natural Products in SARS-CoV-2; Elsevier: Amsterdam, The Netherlands, 2023; pp. 1–24. ISBN 9780323950473. [Google Scholar]
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagalli, R.; et al. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 232, 1574–1581. [Google Scholar] [CrossRef]
- Wang, S.F.; Chen, K.H.; Chen, M.; Li, W.Y.; Chen, Y.J.; Tsao, C.H.; Yen, M.Y.; Huang, J.C.; Chen, Y.M. Human-leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral. Immunol. 2011, 24, 421–426. [Google Scholar] [CrossRef]
- Correale, P.; Mutti, L.; Pentimalli, F.; Baglio, G.; Saladino, R.E.; Sileri, P.; Giordan, A. HLA-B*44 and C*01 prevalence correlates with Covid19 spreading across Italy. Int. J. Mol. Sci. 2020, 21, 5205. [Google Scholar] [CrossRef]
- Li, C.; Zhao, C.; Bao, J.; Tang, B.; Wang, Y.; Gu, B. Laboratory diagnosis of coronavirus disease-2019 (COVID-19). Clin. Chim. Acta 2020, 510, 35–46. [Google Scholar] [CrossRef]



| Alleles | Description | Reference |
|---|---|---|
| HLA | Refers to the HLA region and prefix for an HLA gene | [38] |
| HLA-DRB1*13 | Refers to all alleles in the DR13 serologic group | |
| HLA-DRB1*13:01 | Refers to a specific HLA allele | |
| HLA-DRB1*13:01:02 | Refers to an allele that differs by a synonymous mutation from DRB1*13:01:01 | |
| HLA-DRB1*13:01:01:02 | Refers to an allele that contains a mutation outside the coding region from DRB1*13:01:01:01 | |
| HLA-DRB1 | Refers to a particular HLA locus, i.e., DRB1 | |
| HLA-A*24:02:01:02L | Refers to an allele encoding a protein with significantly reduced or ‘low’ cell surface expression, where the mutation is found outside the coding region | |
| HLA-B*44:02:01:02S | Refers to an allele encoding a protein that is expressed as a ‘secreted’ molecule only | |
| HLA-A*32:11Q | Refers to an allele that has a mutation that has previously been shown to have a significant effect on cell surface expression, although this has not been confirmed and its expression remains ‘questionable’ | |
| HLA-A*24:09N | Refers to a ‘null’ allele, an allele that is not expressed | |
| HLA-A*30:14L | Refers to an allele encoding a protein with significantly reduced or ‘low’ cell surface expression. |
| Locus | Antigen Specificities | Alleles | Reference |
|---|---|---|---|
| HLA-A | 24 | 303 | [38] |
| HLA-B | 55 | 559 | |
| HLA-C | 9 | 150 | |
| HLADRB1 | 17 | 362 | |
| Total | 105 | 1374 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haq, I.U.; Krukiewicz, K.; Tayyab, H.; Khan, I.; Khan, M.; Yahya, G.; Cavalu, S. Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics. J. Clin. Med. 2023, 12, 2645. https://doi.org/10.3390/jcm12072645
Haq IU, Krukiewicz K, Tayyab H, Khan I, Khan M, Yahya G, Cavalu S. Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics. Journal of Clinical Medicine. 2023; 12(7):2645. https://doi.org/10.3390/jcm12072645
Chicago/Turabian StyleHaq, Ihtisham Ul, Katarzyna Krukiewicz, Hamnah Tayyab, Imran Khan, Mehtab Khan, Galal Yahya, and Simona Cavalu. 2023. "Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics" Journal of Clinical Medicine 12, no. 7: 2645. https://doi.org/10.3390/jcm12072645
APA StyleHaq, I. U., Krukiewicz, K., Tayyab, H., Khan, I., Khan, M., Yahya, G., & Cavalu, S. (2023). Molecular Understanding of ACE-2 and HLA-Conferred Differential Susceptibility to COVID-19: Host-Directed Insights Opening New Windows in COVID-19 Therapeutics. Journal of Clinical Medicine, 12(7), 2645. https://doi.org/10.3390/jcm12072645







