CT-Derived Body Composition Is a Predictor of Survival after Esophagectomy
Abstract
1. Introduction
2. Patients and Methods
2.1. Study Population
2.2. Image Acquisition
2.3. Image Features
- (1)
- Basic tumor features: (a) volume (mL), (b) density (HU), (c) mean diameter (mm), (d) maximum length (mm), and density based on average HU value.
- (2)
- CT-based tumor radiomic features: High-dimensional radiomic features (n = 500) were automatically extracted from the segmented regions on the CT images, which included summary, first order, shape, and gray-level co-occurrence features [19].
- (3)
- Tumor PET features: PET features were quantified by mapping tumor ROIs from the CT images onto PET images and then extracting functional characteristics, including maximum SUV, minimum SUV, average SUV, SUV entropy, SUV P75, PET metabolic tumor volume (MTV)/mL, and PET total lesion glycolysis (TLG).
2.4. Clinical Features
2.5. Statistical Analysis
3. Results
3.1. Body Composition
3.2. Cox Regression Analysis
3.3. One-Year Survival
3.4. Three-Year Survival
3.5. Five-Year Survival
4. Comment
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abate, E.; DeMeester, S.R.; Zehetner, J.; Oezcelik, A.; Ayazi, S.; Costales, J.; Banki, F.; Lipham, J.C.; Hagen, J.A.; DeMeester, T.R. Recurrence after esophagectomy for adenocarcinoma: Defining optimal follow-up intervals and testing. J. Am. Coll. Surg. 2010, 210, 428–435. [Google Scholar] [CrossRef]
- Glare, P.; Sinclair, C.; Downing, M.; Stone, P. Predicting survival in patients with advanced disease. Eur. J. Cancer 2008, 44, 1146–1156. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Yan, H.J.; Shiiya, H.; Sato, M.; Shinozaki-Ushiku, A.; Nakajima, J. Machine learning-based radiomic computed tomography phenotyping of thymic epithelial tumors: Predicting pathological and survival outcomes. J. Thorac. Cardiovasc. Surg. 2022, 165, 502–516.e9. [Google Scholar] [CrossRef]
- Shimada, Y.; Kudo, Y.; Maehara, S.; Amemiya, R.; Masuno, R.; Park, J.; Ikeda, N. Radiomics with Artificial Intelligence for the Prediction of Early Recurrence in Patients with Clinical Stage IA Lung Cancer. Ann. Surg. Oncol. 2022, 29, 8185–8193. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Hu, Y.; Han, L.; Fu, J.; Vardhanabhuti, V.; Yang, H. Prediction of Individual Lymph Node Metastatic Status in Esophageal Squamous Cell Carcinoma Using Routine Computed Tomography Imaging: Comparison of Size-Based Measurements and Radiomics-Based Models. Ann. Surg. Oncol. 2022, 29, 8117–8126. [Google Scholar] [CrossRef]
- Ashraf, S.F.; Yin, K.; Meng, C.X.; Wang, Q.; Wang, Q.; Pu, J.; Dhupar, R. Predicting benign, preinvasive, and invasive lung nodules on computed tomography scans using machine learning. J. Thorac. Cardiovasc. Surg. 2022, 163, 1496–1505.e10. [Google Scholar] [CrossRef]
- Cao, B.; Province, R.O.K.L.O.S.; Mi, K.; Dai, W.; Liu, T.; Xie, T.; Li, Q.; Lang, J.; Han, Y.; Peng, L.; et al. Prognostic and incremental value of computed tomography-based radiomics from tumor and nodal regions in esophageal squamous cell carcinoma. Chin. J. Cancer Res. 2022, 34, 71–82. [Google Scholar] [CrossRef]
- Lagarde, S.M.; Reitsma, J.B.; de Castro, S.M.; ten Kate, F.J.; Busch, O.R.; Van Lanschot, J.J. Prognostic nomogram for patients undergoing oesophagectomy for adenocarcinoma of the oesophagus or gastro-oesophageal junction. Br. J. Surg. 2007, 94, 1361–1368. [Google Scholar] [CrossRef]
- Shapiro, J.; van Klaveren, D.; Lagarde, S.M.; Toxopeus, E.L.A.; van der Gaast, A.; Hulshof, M.C.C.M.; Wijnhoven, B.P.L.; Henegouwen, M.I.V.B.; Steyerberg, E.W.; van Lanschot, J.J.B. Prediction of survival in patients with oesophageal or junctional cancer receiving neoadjuvant chemoradiotherapy and surgery. Br. J. Surg. 2016, 103, 1039–1047. [Google Scholar] [CrossRef]
- Bates, D.D.B.; Pickhardt, P.J. CT-Derived Body Composition Assessment as a Prognostic Tool in Oncologic Patients: From Opportunistic Research to Artificial Intelligence-Based Clinical Implementation. AJR Am. J. Roentgenol. 2022, 219, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Park, Y.S.; Na, K.J.; Park, S.; Park, I.K.; Kang, C.H.; Kim, Y.T.; Goo, J.M.; Yoon, S.H. Association of Adipopenia at Preoperative PET/CT with Mortality in Stage I Non-Small Cell Lung Cancer. Radiology 2021, 301, 645–653. [Google Scholar] [CrossRef]
- Murnane, L.C.; Forsyth, A.K.; Koukounaras, J.; Pilgrim, C.H.; Shaw, K.; Brown, W.A.; Mourtzakis, M.; Tierney, A.C.; Burton, P.R. Myosteatosis predicts higher complications and reduced overall survival following radical oesophageal and gastric cancer surgery. Eur. J. Surg. Oncol. 2021, 47, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Srpcic, M.; Jordan, T.; Popuri, K.; Sok, M. Sarcopenia and myosteatosis at presentation adversely affect survival after esophagectomy for esophageal cancer. Radiol. Oncol. 2020, 54, 237–246. [Google Scholar] [CrossRef]
- Anconina, R.; Ortega, C.; Metser, U.; Liu, Z.A.; Elimova, E.; Allen, M.; Darling, G.E.; Wong, R.M.; Taylor, K.; Yeung, J.; et al. Combined 18 F-FDG PET/CT Radiomics and Sarcopenia Score in Predicting Relapse-Free Survival and Overall Survival in Patients With Esophagogastric Cancer. Clin. Nucl. Med. 2022, 47, 684–691. [Google Scholar] [CrossRef]
- Pu, L.; Gezer, N.S.; Ashraf, S.F.; Ocak, I.; Dresser, D.E.; Dhupar, R. Automated segmentation of five different body tissues on computed tomography using deep learning. Med. Phys. 2023, 50, 178–191. [Google Scholar] [CrossRef]
- Pu, L.; Ashraf, S.F.; Gezer, N.S.; Ocak, I.; Dresser, D.E.; Leader, J.K.; Dhupar, R. Estimating 3-D whole-body composition from a chest CT scan. Med. Phys. 2022, 49, 7108–7177. [Google Scholar] [CrossRef] [PubMed]
- García, S.; Schmidt, J.; Ploeg, H. A validation study: Using CT scans to calculate volume, weight, and density. In Proceedings of the ASB 29th Annual Meeting, Cleveland, OH, USA, 31 July–5 August 2005; p. 850. [Google Scholar]
- Pu, J.; Leader, J.K.; Sechrist, J.; Beeche, C.A.; Singh, J.P.; Ocak, I.K.; Risbano, M.G. Automated identification of pulmonary arteries and veins depicted in non-contrast chest CT scans. Med. Image Anal. 2022, 77, 102367. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Subasi, A. Chapter 3—Machine learning techniques. In Practical Machine Learning for Data Analysis Using Python; Subasi, A., Ed.; Academic Press: Cambridge, MA, USA, 2020; pp. 91–202. [Google Scholar]
- Ghantasala, G.P.; Kumari, N.V.; Patan, R. Chapter 9—Cancer prediction and diagnosis hinged on HCML in IOMT environment. In Machine Learning and the Internet of Medical Things in Healthcare; Singh, K.K., Elhoseny, M., Singh, A., Elngar, A.A., Eds.; Academic Press: Cambridge, MA, USA, 2021; pp. 179–207. [Google Scholar]
- Suthaharan, S. Chapter 6—A Cognitive Random Forest: An Intra- and Intercognitive Computing for Big Data Classification under Cune Condition. In Handbook of Statistics; Gudivada, V.N., Raghavan, V.V., Govindaraju, V., Rao, C., Eds.; Elsevier: Hoboken, NJ, USA, 2016; pp. 207–227. [Google Scholar]
- Gholami, R.; Fakhari, N. Chapter 27—Support Vector Machine: Principles, Parameters, and Applications. In Handbook of Neural Computation; Samui, P., Sekhar, S., Balas, V., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 515–535. [Google Scholar]
- Pisner, D.A.; Schnyer, D.M. Chapter 6—Support vector machine. In Machine Learning; Mechelli, A., Vieira, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 101–121. [Google Scholar]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Xiong, J.; Yu, W.; Ma, J.; Ren, Y.; Fu, X.; Zhao, J. The Role of PET-Based Radiomic Features in Predicting Local Control of Esophageal Cancer Treated with Concurrent Chemoradiotherapy. Sci. Rep. 2018, 8, 9902. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Ou, J.; Wu, Y.P.; Li, R.; Chen, T.W.; Zhang, X.M. Contrast-enhanced CT radiomics features to predict recurrence of locally advanced oesophageal squamous cell cancer within 2 years after trimodal therapy: A case-control study. Medicine 2021, 100, e26557. [Google Scholar] [CrossRef] [PubMed]
- Demler, O.V.; Pencina, M.J.; D’Agostino, R.B., Sr. Misuse of DeLong test to compare AUCs for nested models. Stat. Med. 2012, 31, 2577–2587. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, J.; Lysaght, J.; Donohoe, C.L.; Reynolds, J.V. Obesity and gastrointestinal cancer: The interrelationship of adipose and tumour microenvironments. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 699–714. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, D.P.; Krull, J.L.; Lockwood, C.M. Equivalence of the mediation, confounding and suppression effect. Prev. Sci. 2000, 1, 173–181. [Google Scholar] [CrossRef] [PubMed]
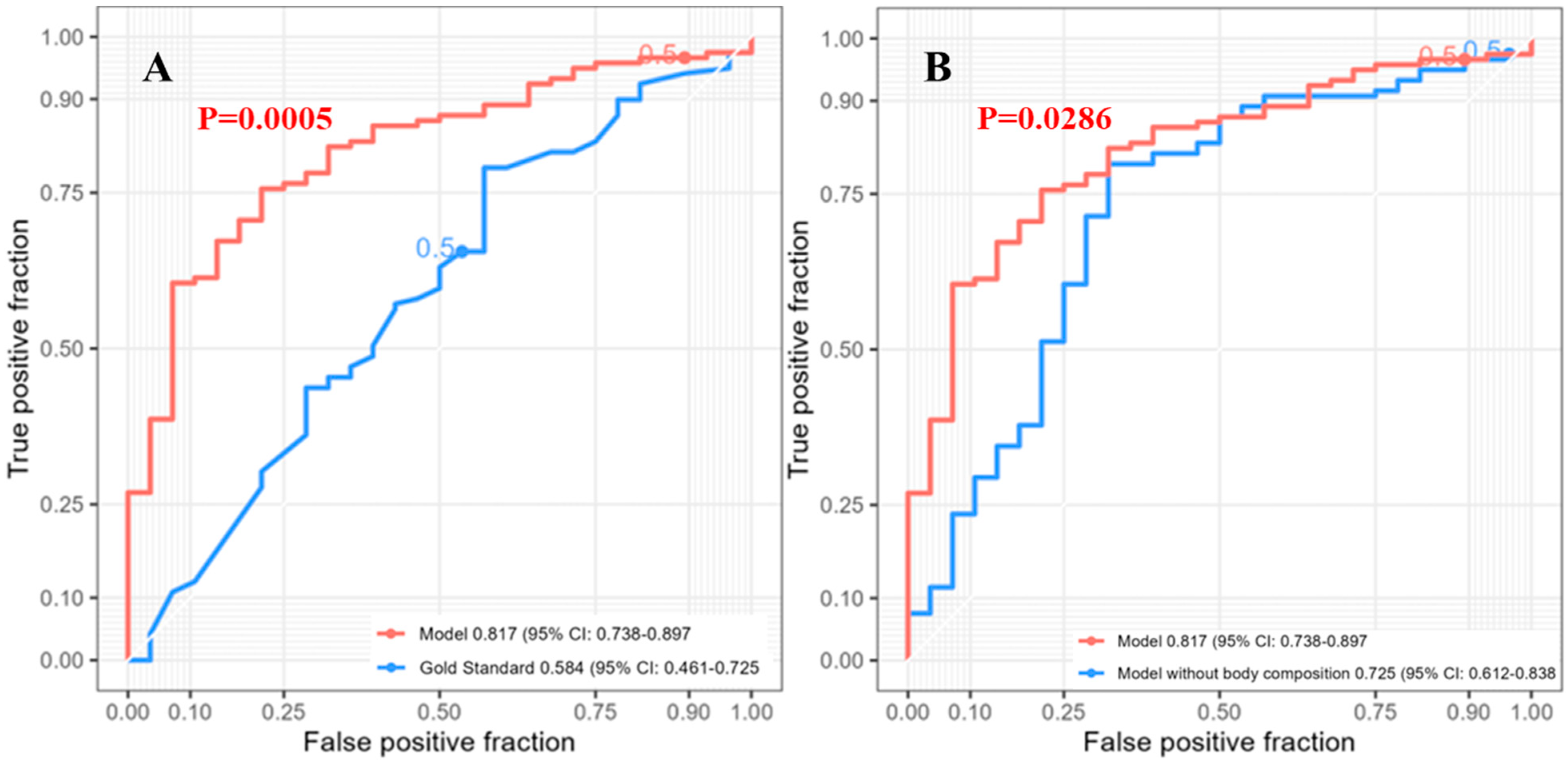
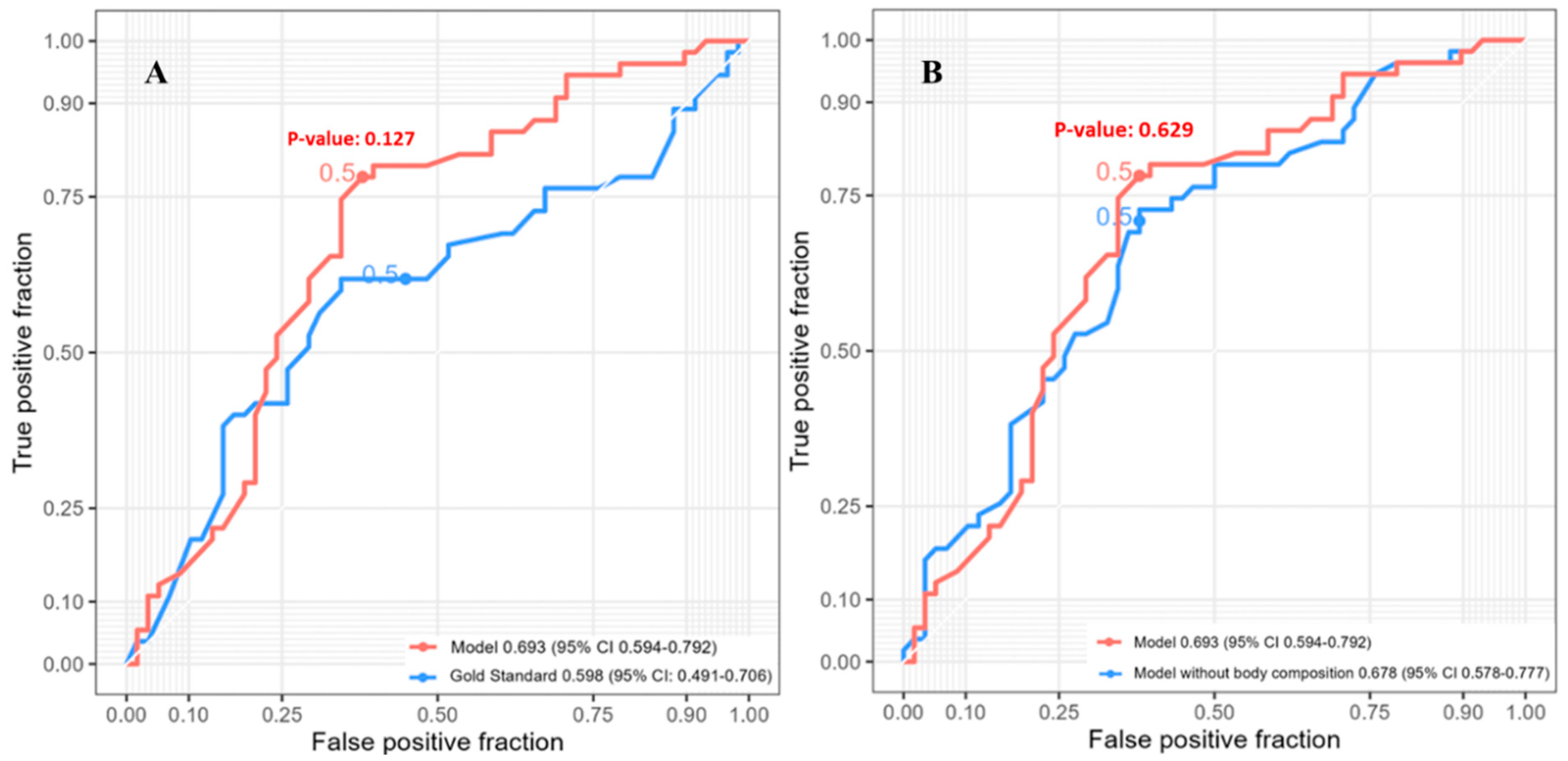
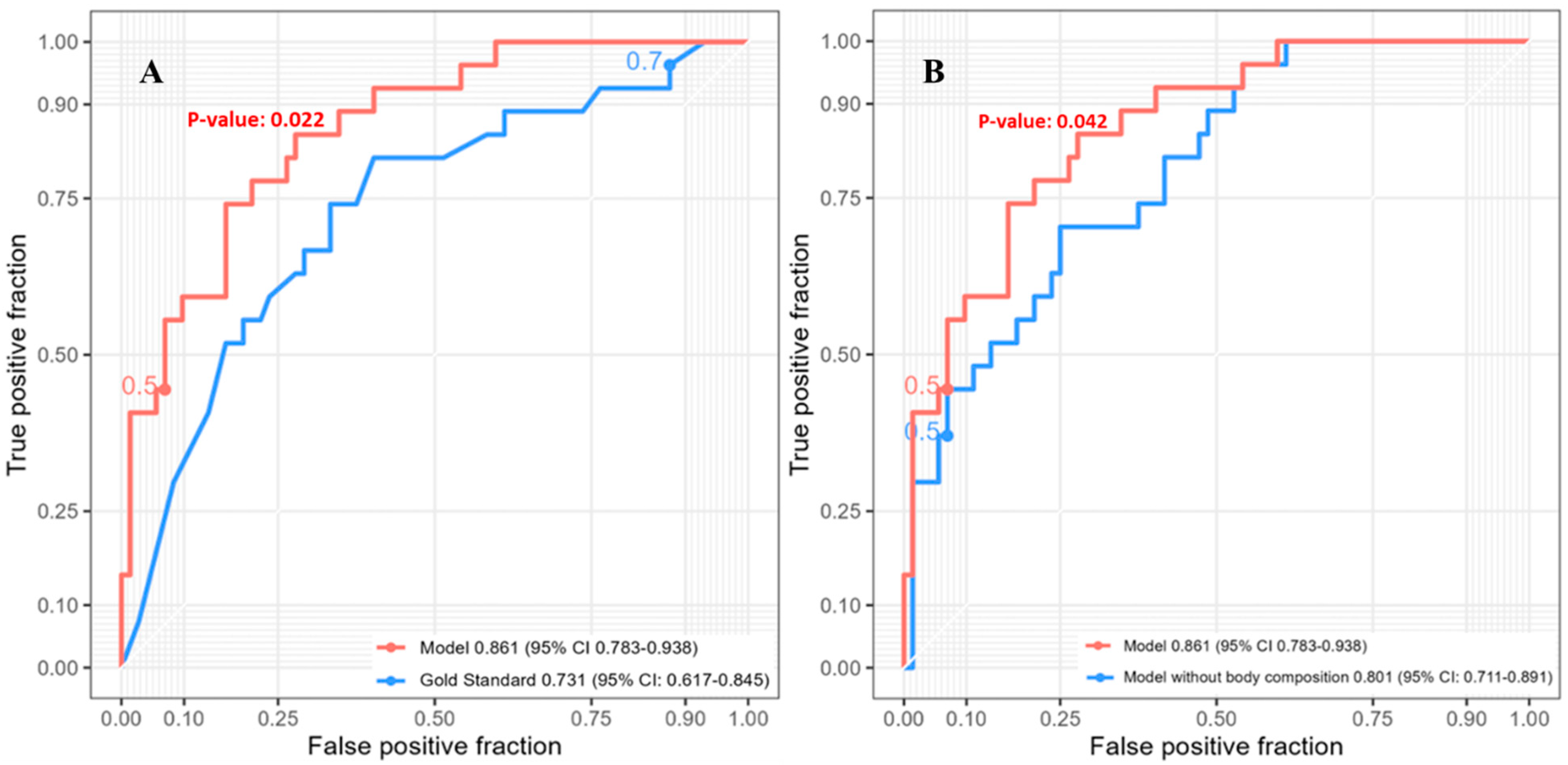
| Characteristic | Value |
|---|---|
| Age | 6.7 ± 10.48 |
| Height (cm) | 171 ± 9.97 |
| Weight (kg) | 85 ± 21.20 |
| Male | 138 (75.41%) |
| Race | |
| White | 179 (97.81%) |
| Black | 3 (1.64%) |
| Asian | 1 (0.05%) |
| Smoking | |
| Former-smoker | 120 (65.57%) |
| Never-smoker | 51 (27.87%) |
| Current-smoker | 12 (6.56%) |
| Cancer Stage | |
| I | 65 (35.52%) |
| II | 31 (16.94%) |
| III | 59 (32.79%) |
| IV | 27 (14.75%) |
| Postoperative Complications | |
| Atrial Fibrillation | 42 (22.95%) |
| Pneumonia | 41 (22.40%) |
| Effusion | 37 (20.22%) |
| Mean ± (SD); n (%) |
| Covariate | Coefficient | Hazard Ratio | Lower 95% | Upper 95% | p-Value |
|---|---|---|---|---|---|
| Age | 0.024 | 1.024 | 0.004 | 0.044 | 0.021 |
| BMI | −0.049 | 0.952 | −0.089 | −0.01 | 0.014 |
| Height(cm) | 0.009 | 1.009 | −0.013 | 0.032 | 0.422 |
| Weight(kg) | −0.009 | 0.991 | −0.02 | 0.002 | 0.1 |
| Sex | |||||
| Female | −0.081 | 0.922 | −0.621 | 0.459 | 0.768 |
| Male | 0.081 | 1.084 | −0.459 | 0.621 | 0.768 |
| Race | |||||
| White | −0.484 | 0.617 | −1.32 | 0.353 | 0.257 |
| Black | 1.476 | 4.373 | −0.533 | 3.484 | 0.15 |
| Asian | −21.885 | 0 | −83.864 | 40.095 | 0.489 |
| Smoking | |||||
| Never-smoker | 0.03 | 1.031 | −0.455 | 0.515 | 0.903 |
| Current-smoker | −0.138 | 0.871 | −0.614 | 0.337 | 0.569 |
| Former-smoker | 2.47 | 11.826 | 0.974 | 3.967 | 0.001 |
| Pathological T-stage | |||||
| T1 | −0.877 | 0.416 | −1.398 | −0.355 | 0.001 |
| T2 | 0.028 | 1.029 | −0.551 | 0.608 | 0.924 |
| T3 | 0.579 | 1.784 | 0.118 | 1.04 | 0.014 |
| T4 | 1.569 | 4.801 | 0.613 | 2.524 | 0.001 |
| Pathological N-stage | |||||
| N0 | −0.891 | 0.41 | −1.376 | −0.406 | 0 |
| N1 | 0.391 | 1.479 | −0.083 | 0.866 | 0.106 |
| N2 | 0.796 | 2.217 | 0.119 | 1.474 | 0.021 |
| N3 | 0.51 | 1.665 | −0.112 | 1.132 | 0.108 |
| Pathological M-stage | |||||
| M0 | −0.714 | 0.49 | −1.626 | 0.199 | 0.125 |
| M1 | 0.714 | 2.041 | −0.199 | 1.626 | 0.125 |
| Postoperative Complications | |||||
| Atrial Fibrillation | 0.175 | 1.191 | −0.345 | 0.694 | 0.510 |
| Pneumonia | 0.447 | 1.563 | −0.049 | 0.943 | 0.078 |
| Effusion | 0.432 | 1.540 | −0.099 | 0.962 | 0.111 |
| Covariate | Coefficient | Hazard Ratio | Lower 95% | Upper 95% | p-Value |
|---|---|---|---|---|---|
| Tumor Features | |||||
| Tumor volume | 0.003 | 1.003 | −0.002 | 0.009 | 0.272 |
| Tumor density | 0.004 | 1.004 | −0.002 | 0.010 | 0.192 |
| Tumor mean diameter | 0.025 | 1.026 | −0.001 | 0.052 | 0.061 |
| Tumor maximum length | 0.003 | 1.003 | 0.000 | 0.006 | 0.093 |
| PET Features | |||||
| Average SUV | −0.014 | 0.986 | −0.087 | 0.059 | 0.710 |
| Maximum SUV | −0.013 | 0.987 | −0.068 | 0.041 | 0.633 |
| Minimum SUV | 0.030 | 1.031 | −0.199 | 0.259 | 0.795 |
| PET MTV/ml | 0.000 | 1.000 | 0.000 | 0.001 | 0.135 |
| PET TLG | 0.000 | 1.000 | 0.000 | 0.000 | 0.270 |
| SUV entropy | −0.129 | 0.879 | −0.271 | 0.013 | 0.076 |
| SUV P75 | −0.012 | 0.988 | −0.068 | 0.044 | 0.678 |
| Body Composition Features | |||||
| VAT volume | −0.048 | 0.953 | −0.120 | 0.024 | 0.191 |
| VAT density | 0.034 | 1.034 | 0.002 | 0.065 | 0.037 |
| VAT mass | −0.047 | 0.954 | −0.117 | 0.023 | 0.192 |
| SAT volume | −0.021 | 0.979 | −0.046 | 0.003 | 0.090 |
| SAT density | 0.013 | 1.013 | −0.008 | 0.033 | 0.236 |
| SAT mass | −0.021 | 0.979 | −0.045 | 0.003 | 0.090 |
| IMAT volume | 0.023 | 1.023 | −0.273 | 0.320 | 0.878 |
| IMAT density | 0.032 | 1.033 | −0.006 | 0.071 | 0.096 |
| IMAT mass | 0.022 | 1.022 | −0.261 | 0.305 | 0.878 |
| SM volume | −0.035 | 0.966 | −0.086 | 0.017 | 0.186 |
| SM density | 0.000 | 1.000 | −0.026 | 0.026 | 0.984 |
| SM muscle | −0.029 | 0.971 | −0.073 | 0.014 | 0.183 |
| Bone volume | −0.008 | 0.992 | −0.259 | 0.244 | 0.951 |
| Bone density | −0.004 | 0.996 | −0.009 | 0.001 | 0.108 |
| Bone mass | −0.024 | 0.976 | −0.179 | 0.131 | 0.762 |
| Covariate | Coefficient | Hazard Ratio | Lower 95% | Upper 95% | p-Value |
|---|---|---|---|---|---|
| Race | 0.363 | 1.438 | 0.124 | 0.603 | 0.003 |
| BMI | −0.254 | 0.776 | −0.356 | −0.151 | <0.0001 |
| Staging | |||||
| Pathological N-Stage | 0.533 | 1.705 | 0.315 | 0.752 | <0.0001 |
| Postoperative Complications | |||||
| Pneumonia | 0.684 | 1.982 | 0.161 | 1.207 | 0.010 |
| Body Composition | |||||
| Bone density | −0.006 | 0.994 | −0.011 | −0.001 | 0.031 |
| Muscle density | 0.062 | 1.064 | 0.015 | 0.109 | 0.009 |
| IMAT volume | 1.016 | 2.763 | 0.468 | 1.565 | <0.0001 |
| SAT volume | 0.081 | 1.084 | 0.019 | 0.143 | 0.010 |
| c-index: 0.754 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iyer, K.; Beeche, C.A.; Gezer, N.S.; Leader, J.K.; Ren, S.; Dhupar, R.; Pu, J. CT-Derived Body Composition Is a Predictor of Survival after Esophagectomy. J. Clin. Med. 2023, 12, 2106. https://doi.org/10.3390/jcm12062106
Iyer K, Beeche CA, Gezer NS, Leader JK, Ren S, Dhupar R, Pu J. CT-Derived Body Composition Is a Predictor of Survival after Esophagectomy. Journal of Clinical Medicine. 2023; 12(6):2106. https://doi.org/10.3390/jcm12062106
Chicago/Turabian StyleIyer, Kartik, Cameron A. Beeche, Naciye S. Gezer, Joseph K. Leader, Shangsi Ren, Rajeev Dhupar, and Jiantao Pu. 2023. "CT-Derived Body Composition Is a Predictor of Survival after Esophagectomy" Journal of Clinical Medicine 12, no. 6: 2106. https://doi.org/10.3390/jcm12062106
APA StyleIyer, K., Beeche, C. A., Gezer, N. S., Leader, J. K., Ren, S., Dhupar, R., & Pu, J. (2023). CT-Derived Body Composition Is a Predictor of Survival after Esophagectomy. Journal of Clinical Medicine, 12(6), 2106. https://doi.org/10.3390/jcm12062106





