Can Liquid Biopsy Based on ctDNA/cfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC?
Abstract
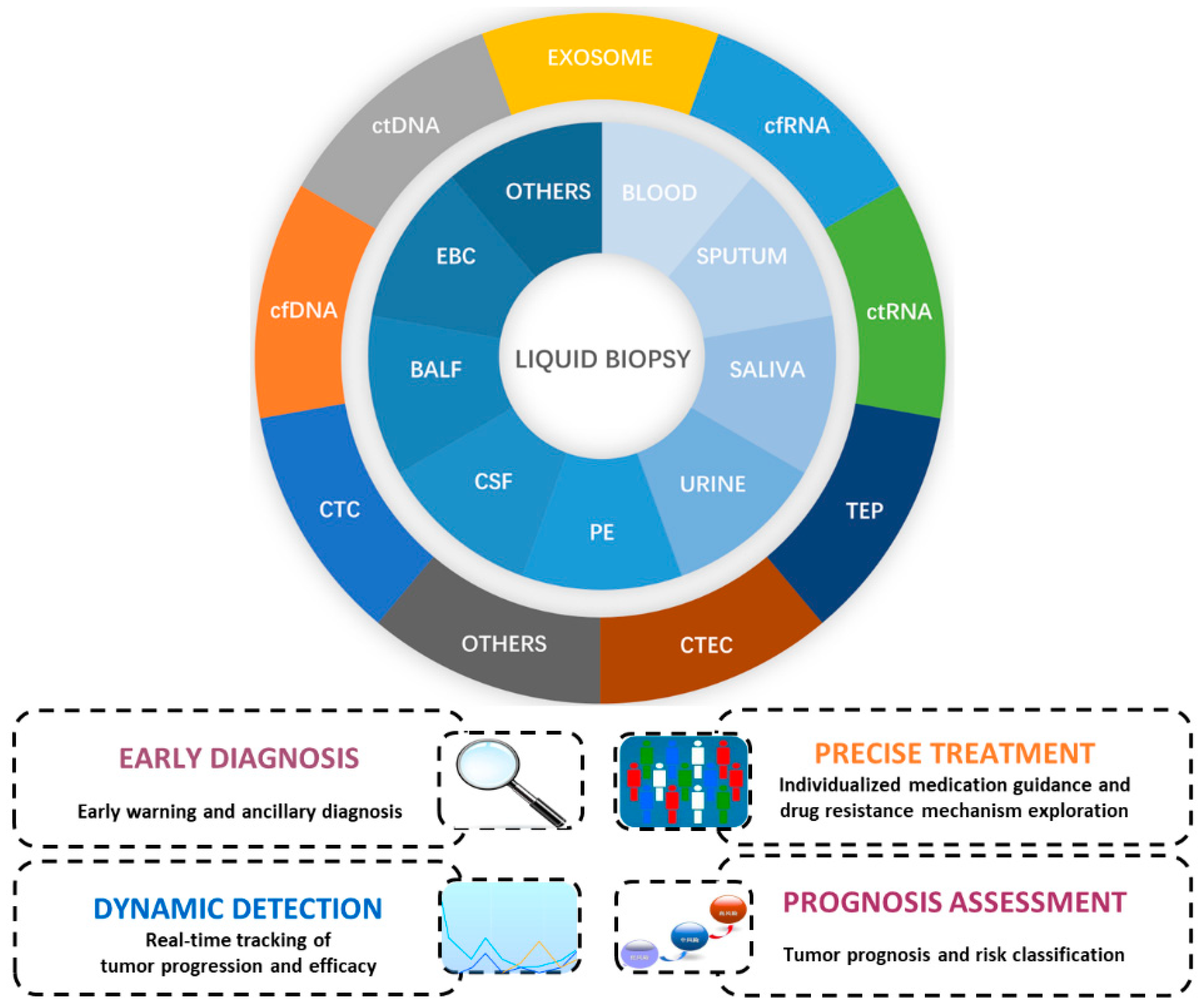
:1. Introduction
2. Plasma ctDNA-Based EGFR Mutations Can Guide Targeted Therapy for Advanced NSCLC
3. Evaluation of Resistance to EGFR-TKI Therapy Based on Plasma ctDNA Detection in NSCLC Patients
4. A Marker for the New Era of NSCLC Treatment-Minimal or Molecular Residual Disease (MRD)
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yoneda, K.; Imanishi, N.; Ichiki, Y.; Tanaka, F. Treatment of Non-small Cell Lung Cancer with EGFR-mutations. J. UOEH 2019, 41, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Mielgo-Rubio, X.; Martín, M.; Remon, J.; Higuera, O.; Calvo, V.; Jarabo, J.R.; Conde, E.; Luna, J.; Provencio, M.; De, Castro, J.; et al. Targeted therapy moves to earlier stages of non-small-cell lung cancer: Emerging evidence, controversies and future challenges. Future Oncol. 2021, 17, 4011–4025. [Google Scholar] [CrossRef] [PubMed]
- Chaft, J.E.; Shyr, Y.; Sepesi, B.; Forde, P.M. Preoperative and Postoperative Systemic Therapy for Operable Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2022, 40, 546–555. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Pisapia, P.; Malapelle, U.; Troncone, G. Liquid Biopsy and Lung Cancer. Acta. Cytol. 2019, 63, 489–496. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [CrossRef]
- Jiang, N.; Meng, X.; Mi, H.; Chi, Y.; Li, S.; Jin, Z.; Tian, H.; He, J.; Shen, W.; Tian, H.; et al. Circulating lncRNA XLOC_009167 serves as a diagnostic biomarker to predict lung cancer. Clin. Chim. Acta 2018, 486, 26–33. [Google Scholar] [CrossRef]
- Xing, S.; Zeng, T.; Xue, N.; He, Y.; Lai, Y.Z.; Li, H.L.; Huang, Q.; Chen, S.L.; Liu, W.L. Development and Validation of Tumor-educated Blood Platelets Integrin Alpha 2b (ITGA2B) RNA for Diagnosis and Prognosis of Non-small-cell Lung Cancer through RNA-seq. Int. J. Biol. Sci. 2019, 15, 1977–1992. [Google Scholar] [CrossRef]
- Lei, Y.; Sun, N.; Zhang, G.; Liu, C.; Lu, Z.; Huang, J.; Zhang, C.; Zang, R.; Che, Y.; Mao, S.; et al. Combined detection of aneuploid circulating tumor-derived endothelial cells and circulating tumor cells may improve diagnosis of early stage non-small-cell lung cancer. Clin. Transl. Med. 2020, 10, e128. [Google Scholar] [CrossRef]
- Kim, Y.J.; Ji, W.J.; Lee, J.C.; Chun, S.M.; Choi, C.M. Assessment of Anti-tumor Efficacy of Osimertinib in Non-Small Cell Lung Cancer Patients by Liquid Biopsy Using Bronchoalveolar Lavage Fluid, Plasma, or Pleural Effusion. Cancer Res. Treat. 2022, 54, 985–995. [Google Scholar] [CrossRef]
- Chiang, C.L.; Huang, H.C.; Luo, Y.H.; Chiu, C.H. Cerebrospinal fluid as a medium of liquid biopsy in the management of patients with non-small-cell lung cancer having central nervous system metastasis. Front. Biosci. (Landmark Ed). 2021, 26, 1679–1688. [Google Scholar] [CrossRef]
- Ryan, D.J.; Toomey, S.; Smyth, R.; Madden, S.F.; Workman, J.; Cummins, R.; Sheehan, K.; Fay, J.; Naidoo, J.; Breathnach, O.S.; et al. Exhaled Breath Condensate (EBC) analysis of circulating tumour DNA (ctDNA) using a lung cancer specific UltraSEEK oncogene panel. Lung Cancer. 2022, 168, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Yong, E. Cancer biomarkers: Written in blood. Nature 2014, 511, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Lo, Y.M.D. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016, 32, 360–371. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Robert, B.; Arnau Peyrotte, E.; Del Rio, M.; Ychou, M.; Molina, F.; Gongora, C.; Thierry, A.R. High fragmentation characterizes tumour-derived circulating DNA. PLoS ONE 2011, 6, e23418. [Google Scholar] [CrossRef]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Jiang, P.; Zheng, Y.W.; Liao, G.J.; Sun, H.; Wong, J.; Siu, S.S.; Chan, W.C.; Chan, S.L.; Chan, A.T.; et al. Cancer genome scanning in plasma: Detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin. Chem. 2013, 59, 211–224. [Google Scholar] [CrossRef]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef]
- Wong, I.H.; Lo, Y.M.; Zhang, J.; Liew, C.T.; Ng, M.H.; Wong, N.; Lai, P.B.; Lau, W.Y.; Hjelm, N.M.; Johnson, P.J. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999, 59, 71–73. [Google Scholar]
- Chan, K.C.; Lai, P.B.; Mok, T.S.; Chan, H.L.; Ding, C.; Yeung, S.W.; Lo, Y.M. Quantitative analysis of circulating methylated DNA as a biomarker for hepatocellular carcinoma. Clin. Chem. 2008, 54, 1528–1536. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.C.; Jiang, P.; Chan, C.W.; Sun, K.; Wong, J.; Hui, E.P.; Chan, S.L.; Chan, W.C.; Hui, D.S.; Ng, S.S.; et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 18761–18768. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- To, E.W.; Chan, K.C.; Leung, S.F.; Chan, L.Y.; To, K.F.; Chan, A.T.; Johnson, P.J.; Lo, Y.M. Rapid clearance of plasma Epstein-Barr virus DNA after surgical treatment of nasopharyngeal carcinoma. Clin. Cancer Res. 2003, 9, 3254–3259. [Google Scholar]
- Yao, W.; Mei, C.; Nan, X.; Hui, L. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016, 590, 142–148. [Google Scholar] [CrossRef]
- Douillard, J.Y.; Ostoros, G.; Cobo, M.; Ciuleanu, T.; Cole, R.; McWalter, G.; Walker, J.; Dearden, S.; Webster, A.; Milenkova, T.; et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: Circulating-free tumor dna as a surrogate for determination of EGFR status. J. Thorac. Oncol. 2014, 9, 1345–1353. [Google Scholar] [CrossRef] [PubMed]
- Mok, T.; Wu, Y.L.; Lee, J.S.; Yu, C.J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef]
- Karachaliou, N.; Mayo-de las Casas, C.; Queralt, C.; de Aguirre, I.; Melloni, B.; Cardenal, F.; Garcia-Gomez, R.; Massuti, B.; Sánchez, J.M.; Porta, R.; et al. Spanish Lung Cancer Group. Association of EGFR L858R Mutation in Circulating Free DNA With Survival in the EURTAC Trial. JAMA Oncol. 2015, 1, 149–157. [Google Scholar] [CrossRef]
- Mayo-de-Las-Casas, C.; Jordana-Ariza, N.; Garzón-Ibañez, M.; Balada-Bel, A.; Bertrán-Alamillo, J.; Viteri-Ramírez, S.; Reguart, N.; Muñoz-Quintana, M.A.; Lianes-Barragan, P.; Camps, C.; et al. Large scale, prospective screening of EGFR mutations in the blood of advanced NSCLC patients to guide treatment decisions. Ann. Oncol. 2017, 28, 2248–2255. [Google Scholar] [CrossRef]
- Xu, H.; Baidoo, A.A.H.; Su, S.; Ye, J.; Chen, C.; Xie, Y.; Bertolaccini, L.; Ismail, M.; Ricciuti, B.; Ng, C.S.H.; et al. A comparison of EGFR mutation status in tissue and plasma cell-free DNA detected by ADx-ARMS in advanced lung adenocarcinoma patients. Transl. Lung Cancer Res. 2019, 8, 135–143. [Google Scholar] [CrossRef]
- Wan, R.; Wang, Z.; Lee, J.J.; Wang, S.; Li, Q.; Tang, F.; Wang, J.; Sun, Y.; Bai, H.; Wang, D.; et al. Comprehensive Analysis of the Discordance of EGFR Mutation Status between Tumor Tissues and Matched Circulating Tumor DNA in Advanced Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cheng, Y.; An, T.; Gao, H.; Wang, K.; Zhou, Q.; Hu, Y.; Song, Y.; Ding, C.; Peng, F.; et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): A phase 2, single-arm, multicentre clinical trial. Lancet Respir. Med. 2018, 6, 681–690. [Google Scholar] [CrossRef]
- Behel, V.; Chougule, A.; Noronha, V.; Patil, V.M.; Menon, N.; Singh, A.; Chopade, S.; Kumar, R.; Shah, S.; More, S.; et al. Clinical Utility of Liquid Biopsy (Cell-free DNA) Based EGFR Mutation Detection Post treatment Initiation as a Disease Monitoring Tool in Patients With Advanced EGFR-mutant NSCLC. Clin. Lung Cancer 2022, 23, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.F.; Lei, L.; Wu, L.X.; Li, X.F.; Zhang, Q.X.; Pan, W.W.; Min, Y.H.; Zhu, Y.C.; Du, K.Q.; Wang, M.; et al. Effect of icotinib on advanced lung adenocarcinoma patients with sensitive EGFR mutation detected in ctDNA by ddPCR. Transl. Cancer Res. 2019, 8, 2858–2863. [Google Scholar] [CrossRef] [PubMed]
- Park, C.K.; Lee, S.Y.; Lee, J.C.; Choi, C.M.; Lee, S.Y.; Jang, T.W.; Oh, I.J.; Kim, Y.C. Phase II open-label multicenter study to assess the antitumor activity of afatinib in lung cancer patients with activating epidermal growth factor receptor mutation from circulating tumor DNA: Liquid-Lung-A. Thorac. Cancer 2021, 12, 444–452. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.C.; Scagliotti, G.V.; Baas, P.; Barlesi, F.; Bivona, T.G.; Herbst, R.S.; Mok, T.S.; Peled, N.; Pirker, R.; et al. Liquid Biopsy for Advanced Non-Small Cell Lung Cancer (NSCLC): A Statement Paper from the IASLC. J. Thorac. Oncol. 2018, 13, 1248–1268. [Google Scholar] [CrossRef]
- Thompson, J.C.; Yee, S.S.; Troxel, A.B.; Savitch, S.L.; Fan, R.; Balli, D.; Lieberman, D.; Morrissette, J.D.; Evans, T.L.; Bauml, J.; et al. Detection of Therapeutically Targetable Driver and Resistance Mutations in Lung Cancer Patients by Next-Generation Sequencing of Cell-Free Circulating Tumor DNA. Clin. Cancer Res. 2016, 22, 5772–5782. [Google Scholar] [CrossRef]
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.L.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping with the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180. [Google Scholar] [CrossRef]
- Park, S.; Olsen, S.; Ku, B.M.; Lee, M.S.; Jung, H.A.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; Choi, Y.L.; et al. High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program. Cancer 2021, 127, 3019–3028. [Google Scholar] [CrossRef]
- Sehayek, O.; Kian, W.; Onn, A.; Stoff, R.; Sorotsky, H.G.; Zemel, M.; Bar, J.; Dudnik, Y.; Nechushtan, H.; Rottenberg, Y.; et al. Liquid First Is “Solid” in Naïve Non-Small Cell Lung Cancer Patients: Faster Turnaround Time With High Concordance to Solid Next-Generation Sequencing. Front. Oncol. 2022, 12, 912801. [Google Scholar] [CrossRef]
- Palmero, R.; Taus, A.; Viteri, S.; Majem, M.; Carcereny, E.; Garde-Noguera, J.; Felip, E.; Nadal, E.; Malfettone, A.; Sampayo, M.; et al. Biomarker Discovery and Outcomes for Comprehensive Cell-Free Circulating Tumor DNA Versus Standard-of-Care Tissue Testing in Advanced Non–Small-Cell Lung Cancer. JCO Precis. Oncol. 2021, 5, 93–102. [Google Scholar] [CrossRef]
- Kok, P.S.; Lee, K.; Lord, S.; Yang, J.C.; Rosell, R.; Goto, K.; John, T.; Wu, Y.L.; Mok, T.S.K.; Lee, C.K. Clinical utility of plasma EGFR mutation detection with quantitative PCR in advanced lung cancer: A meta-analysis. Lung Cancer 2021, 154, 113–117. [Google Scholar] [CrossRef]
- Selvarajah, S.; Plante, S.; Speevak, M.; Vaags, A.; Hamelinck, D.; Butcher, M.; McCready, E.; Grafodatskaya, D.; Blais, N.; Tran-Thanh, D.; et al. A Pan-Canadian Validation Study for the Detection of EGFR T790M Mutation Using Circulating Tumor DNA From Peripheral Blood. JTO Clin. Res. Rep. 2021, 2, 100212. [Google Scholar] [CrossRef] [PubMed]
- Park, C.K.; Cho, H.J.; Choi, Y.D.; Oh, I.J.; Kim, Y.C. A Phase II Trial of Osimertinib in the Second-Line Treatment of Non-small Cell Lung Cancer with the EGFR T790M Mutation, Detected from Circulating Tumor DNA: LiquidLung-O-Cohort 2. Cancer Res. Treat. 2019, 51, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Park, S.; Kim, W.S.; Lee, J.C.; Jang, S.J.; Choi, J.; Choi, C.M. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stage EGFR-mutated non-small cell lung cancer. Thorac. Cancer 2018, 9, 1104–1110. [Google Scholar] [CrossRef]
- Romero, A.; Serna-Blasco, R.; Alfaro, C.; Sánchez-Herrero, E.; Barquín, M.; Turpin, M.C.; Chico, S.; Sanz-Moreno, S.; Rodrigez-Festa, A.; Laza-Briviesca, R.; et al. ctDNA analysis reveals different molecular patterns upon disease progression in patients treated with osimertinib. Transl. Lung Cancer Res. 2020, 9, 532–540. [Google Scholar] [CrossRef]
- Deng, Q.; Fang, Q.; Sun, H.; Singh, A.P.; Alexander, M.; Li, S.; Cheng, H.; Zhou, S. Detection of plasma EGFR mutations for personalized treatment of lung cancer patients without pathologic diagnosis. Cancer Med. 2020, 9, 2085–2095. [Google Scholar] [CrossRef] [PubMed]
- Papadimitrakopoulou, V.A.; Han, J.Y.; Ahn, M.J.; Ramalingam, S.S.; Delmonte, A.; Hsia, T.C.; Laskin, J.; Kim, S.W.; He, Y.; Tsai, C.M.; et al. Epidermal growth factor receptor mutation analysis in tissue and plasma from the AURA3 trial: Osimertinib versus platinum-pemetrexed for T790M mutation-positive advanced non-small cell lung cancer. Cancer 2020, 126, 373–380. [Google Scholar] [CrossRef]
- Xing, L.; Pan, Y.; Shi, Y.; Shu, Y.; Feng, J.; Li, W.; Cao, L.; Wang, L.; Gu, W.; Song, Y.; et al. Biomarkers of Osimertinib Response in Patients with Refractory, EGFR-T790M-positive Non-Small Cell Lung Cancer and Central Nervous System Metastases: The APOLLO Study. Clin. Cancer Res. 2020, 26, 6168–6175. [Google Scholar] [CrossRef]
- Yu, H.A.; Schoenfeld, A.J.; Makhnin, A.; Kim, R.; Rizvi, H.; Tsui, D.; Falcon, C.; Houck-Loomis, B.; Meng, F.; Yang, J.L.; et al. Effect of Osimertinib and Bevacizumab on Progression-Free Survival for Patients with Metastatic EGFR-Mutant Lung Cancers: A Phase 1/2 Single-Group Open-Label Trial. JAMA Oncol. 2020, 6, 1048–1054. [Google Scholar] [CrossRef] [PubMed]
- Takahama, T.; Azuma, K.; Shimokawa, M.; Takeda, M.; Ishii, H.; Kato, T.; Saito, H.; Daga, H.; Tsuboguchi, Y.; Okamoto, I.; et al. Plasma screening for the T790M mutation of EGFR and phase 2 study of osimertinib efficacy in plasma T790M-positive non-small cell lung cancer: West Japan Oncology Group 8815L/LPS study. Cancer 2020, 126, 1940–1948. [Google Scholar] [CrossRef]
- Sakai, K.; Takahama, T.; Shimokawa, M.; Azuma, K.; Takeda, M.; Kato, T.; Daga, H.; Okamoto, I.; Akamatsu, H.; Teraoka, S.; et al. Predicting osimertinib-treatment outcomes through EGFR mutant-fraction monitoring in the circulating tumor DNA of EGFR T790M-positive patients with non-small cell lung cancer (WJOG8815L). Mol. Oncol. 2021, 15, 126–137. [Google Scholar] [CrossRef]
- Park, C.K.; Cho, H.J.; Choi, Y.D.; Oh, I.J.; Kim, Y.C. A Phase II Trial of Osimertinib as the First-Line Treatment of Non-Small Cell Lung Cancer Harboring Activating EGFR Mutations in Circulating Tumor DNA: LiquidLung-O-Cohort 1. Cancer Res. Treat. 2021, 53, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, Y.; Meng, Z.; Wu, Y.; Wang, S.; Jin, G.; Qin, Y.; Wang, F.; Wang, J.; Zhou, H.; et al. Plasma EGFR mutation abundance affects clinical response to first-line EGFR-TKIs in patients with advanced non-small cell lung cancer. Ann. Transl. Med. 2021, 9, 635. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Bai, R.L.; Chen, X.; Zhao, Y.G.; Wang, X.; Ma, K.W.; Tian, H.M.; Han, F.J.; Liu, Z.L.; Yang, L.; et al. Correlation of circulating tumor DNA EGFR mutation levels with clinical outcomes in patients with advanced lung adenocarcinoma. Chin. Med. J. (Engl.) 2021, 134, 2430–2437. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Z.; Wang, S.; Zhu, Y.; Ma, D.; Mu, Y.; Ying, J.; Li, J.; Xing, P. Disease monitoring of epidermal growth factor receptor (EGFR)-mutated non-small-cell lung cancer patients treated with tyrosine kinase inhibitors via EGFR status in circulating tumor DNA. Thorac. Cancer 2022, 13, 2201–2209. [Google Scholar] [CrossRef]
- Xu, J.; Liu, Z.; Bai, H.; Dong, G.; Zhong, J.; Wan, R.; Zang, A.; Li, X.; Li, Q.; Guo, J.; et al. Evaluation of Clinical Outcomes of Icotinib in Patients With Clinically Diagnosed Advanced Lung Cancer With EGFR-Sensitizing Variants Assessed by Circulating Tumor DNA Testing: A Phase 2 Nonrandomized Clinical Trial. JAMA Oncol. 2022, 8, 1328–1332. [Google Scholar] [CrossRef]
- Mok, T.S.; Wu, Y.L.; Ahn, M.J.; Garassino, M.C.; Kim, H.R.; Ramalingam, S.S.; Shepherd, F.A.; He, Y.; Akamatsu, H.; Theelen, W.S.; et al. AURA3 Investigators. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N. Engl. J. Med. 2017, 376, 629–640. [Google Scholar] [CrossRef]
- Gray, J.E.; Okamoto, I.; Sriuranpong, V.; Vansteenkiste, J.; Imamura, F.; Lee, J.S.; Pang, Y.K.; Cobo, M.; Kasahara, K.; Cheng, Y.; et al. Tissue and Plasma EGFR Mutation Analysis in the FLAURA Trial: Osimertinib versus Comparator EGFR Tyrosine Kinase Inhibitor as First-Line Treatment in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 6644–6652. [Google Scholar] [CrossRef]
- Morgensztern, D.; Waqar, S.; Subramanian, J.; Gao, F.; Trinkaus, K.; Govindan, R. Prognostic significance of tumor size in patients with stage III non-small-cell lung cancer: A surveillance, epidemiology, and end results (SEER) survey from 1998 to 2003. J. Thorac. Oncol. 2012, 7, 1479–1484. [Google Scholar] [CrossRef]
- Zhao, M.; Zhang, Y.; Cai, W.; Li, J.; Zhou, F.; Cheng, N.; Ren, R.; Zhao, C.; Li, X.; Ren, S.; et al. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer 2014, 120, 2299–2307. [Google Scholar] [CrossRef] [PubMed]
- Canale, M.; Petracci, E.; Delmonte, A.; Chiadini, E.; Dazzi, C.; Papi, M.; Capelli, L.; Casanova, C.; De Luigi, N.; Mariotti, M.; et al. Impact of TP53 Mutations on Outcome in EGFR-Mutated Patients Treated with First-Line Tyrosine Kinase Inhibitors. Clin. Cancer Res. 2017, 23, 2195–2202. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Zhang, X.C.; Chen, Z.H.; Yin, X.L.; Yang, J.J.; Xu, C.R.; Yan, H.H.; Chen, H.J.; Su, J.; Zhong, W.Z.; et al. Relative abundance of EGFR mutations predicts benefit from gefitinib treatment for advanced non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 3316–3321. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.R.; Wang, J.F.; Lin, Y.B.; Wang, F.; Fu, S.; Zhang, S.L.; Su, X.D.; Jiang, L.; Zhang, Y.G.; Shao, J.Y.; et al. Mutation abundance affects the efficacy of EGFR tyrosine kinase inhibitor readministration in non-small-cell lung cancer with acquired resistance. Med. Oncol. 2014, 31, 810. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Tang, W.; Ma, J.; Wei, B.; Niu, Y.; Zhang, G.; Li, P.; Yan, X.; Ma, Z. Mutation abundance affects the therapeutic efficacy of EGFR-TKI in patients with advanced lung adenocarcinoma: A retrospective analysis. Cancer Biol. Ther. 2018, 19, 687–694. [Google Scholar] [CrossRef]
- Linardou, H.; Dahabreh, I.J.; Kanaloupiti, D.; Siannis, F.; Bafaloukos, D.; Kosmidis, P.; Papadimitriou, C.A.; Murray, S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: A systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol. 2008, 9, 962–972. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Zhang, C.; Bollag, G.; Shokat, K.M.; Rosen, N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature 2010, 464, 427–430. [Google Scholar] [CrossRef]
- Sos, M.L.; Koker, M.; Weir, B.A.; Heynck, S.; Rabinovsky, R.; Zander, T.; Seeger, J.M.; Weiss, J.; Fischer, F.; Frommolt, P.; et al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009, 69, 3256–3261. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Azizian, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef]
- Sequist, L.V.; Waltman, B.A.; Dias-Santagata, D.; Digumarthy, S.; Turke, A.B.; Fidias, P.; Bergethon, K.; Shaw, A.T.; Gettinger, S.; Cosper, A.K.; et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci. Transl. Med. 2011, 3, 75ra26. [Google Scholar] [CrossRef] [PubMed]
- Campo, M.; Gerber, D.; Gainor, J.F.; Heist, R.S.; Temel, J.S.; Shaw, A.T.; Fidias, P.; Muzikansky, A.; Engelman, J.A.; Sequist, L.V. Acquired Resistance to First-Line Afatinib and the Challenges of Prearranged Progression Biopsies. J. Thorac. Oncol. 2016, 11, 2022–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonetti, A.; Sharma, S.; Minari, R.; Perego, P.; Giovannetti, E.; Tiseo, M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br. J. Cancer 2019, 121, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Yu, D.; Tian, W.; Wu, F. Resistance mechanisms to osimertinib and emerging therapeutic strategies in nonsmall cell lung cancer. Curr. Opin. Oncol. 2022, 34, 54–65. [Google Scholar] [CrossRef]
- Niederst, M.J.; Hu, H.; Mulvey, H.E.; Lockerman, E.L.; Garcia, A.R.; Piotrowska, Z.; Sequist, L.V.; Engelman, J.A. The Allelic Context of the C797S Mutation Acquired upon Treatment with Third-Generation EGFR Inhibitors Impacts Sensitivity to Subsequent Treatment Strategies. Clin. Cancer Res. 2015, 21, 3924–3933. [Google Scholar] [CrossRef]
- Arulananda, S.; Do, H.; Musafer, A.; Mitchell, P.; Dobrovic, A.; John, T. Combination Osimertinib and Gefitinib in C797S and T790M EGFR-Mutated Non-Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1728–1732. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, J.J.; Huang, J.; Ye, J.Y.; Zhang, X.C.; Tu, H.Y.; Han, Z.H.; Wu, Y.L. Lung Adenocarcinoma Harboring EGFR T790M and In Trans C797S Responds to Combination Therapy of First- and Third-Generation EGFR TKIs and Shifts Allelic Configuration at Resistance. J. Thorac. Oncol. 2017, 12, 1723–1727. [Google Scholar] [CrossRef]
- Eng, J.; Woo, K.M.; Sima, C.S.; Plodkowski, A.; Hellmann, M.D.; Chaft, J.E.; Kris, M.G.; Arcila, M.E.; Ladanyi, M.; Drilon, A. Impact of Concurrent PIK3CA Mutations on Response to EGFR Tyrosine Kinase Inhibition in EGFR-Mutant Lung Cancers and on Prognosis in Oncogene-Driven Lung Adenocarcinomas. J. Thorac. Oncol. 2015, 10, 1713–1719. [Google Scholar] [CrossRef]
- Bean, J.; Riely, G.J.; Balak, M.; Marks, J.L.; Ladanyi, M.; Miller, V.A.; Pao, W. Acquired resistance to epidermal growth factor receptor kinase inhibitors associated with a novel T854A mutation in a patient with EGFR-mutant lung adenocarcinoma. Clin. Cancer Res. 2008, 14, 7519–7525. [Google Scholar] [CrossRef]
- Costa, D.B.; Schumer, S.T.; Tenen, D.G.; Kobayashi, S. Differential responses to erlotinib in epidermal growth factor receptor (EGFR)-mutated lung cancers with acquired resistance to gefitinib carrying the L747S or T790M secondary mutations. J. Clin. Oncol. 2008, 26, 1182–1184. [Google Scholar] [CrossRef]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.M.; Zhao, X.; Christensen, J.; et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef]
- Yu, H.A.; Arcila, M.E.; Rekhtman, N.; Sima, C.S.; Zakowski, M.F.; Pao, W.; Kris, M.G.; Miller, V.A.; Ladanyi, M.; Riely, G.J. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin. Cancer Res. 2013, 19, 2240–2247. [Google Scholar] [CrossRef] [Green Version]
- Patil, T.; Mushtaq, R.; Marsh, S.; Azelby, C.; Pujara, M.; Davies, K.D.; Aisner, D.L.; Purcell, W.T.; Schenk, E.L.; Pacheco, J.M.; et al. Clinicopathologic Characteristics, Treatment Outcomes, and Acquired Resistance Patterns of Atypical EGFR Mutations and HER2 Alterations in Stage IV Non-Small-Cell Lung Cancer. Clin. Lung Cancer 2020, 21, e191–e204. [Google Scholar] [CrossRef]
- Yonesaka, K.; Takegawa, N.; Watanabe, S.; Haratani, K.; Kawakami, H.; Sakai, K.; Chiba, Y.; Maeda, N.; Kagari, T.; Hirotani, K.; et al. An HER3-targeting antibody-drug conjugate incorporating a DNA topoisomerase I inhibitor U3-1402 conquers EGFR tyrosine kinase inhibitor-resistant NSCLC. Oncogene 2019, 38, 1398–1409. [Google Scholar] [CrossRef]
- Lin, M.W.; Su, K.Y.; Su, T.J.; Chang, C.C.; Lin, J.W.; Lee, Y.H.; Yu, S.L.; Chen, J.S.; Hsieh, M.S. Clinicopathological and genomic comparisons between different histologic components in combined small cell lung cancer and non-small cell lung cancer. Lung Cancer 2018, 125, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.C.; Liao, W.Y.; Lin, C.A.; Shih, J.Y.; Yu, C.J.; Yang, J.C. Acquired BRAF V600E Mutation as Resistant Mechanism after Treatment with Osimertinib. J. Thorac. Oncol. 2017, 12, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Offin, M.; Somwar, R.; Rekhtman, N.; Benayed, R.; Chang, J.C.; Plodkowski, A.; Lui, A.J.W.; Eng, J.; Rosenblum, M.; Li, B.T.; et al. Acquired ALK and RET Gene Fusions as Mechanisms of Resistance to Osimertinib in EGFR-Mutant Lung Cancers. JCO Precis. Oncol. 2018, 2, PO.18.00126. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Zhu, V.W.; Hsieh, W.S.; Madison, R.; Creelan, B.; Silberberg, J.; Costin, D.; Bharne, A.; Bonta, I.; Bosemani, T.; et al. Receptor Tyrosine Kinase Fusions and BRAF Kinase Fusions are Rare but Actionable Resistance Mechanisms to EGFR Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2018, 13, 1312–1323. [Google Scholar] [CrossRef]
- Helissey, C.; Favre, L.; Nguyen, A.T.; Mamou, E.; Lamboley, J.L. What management for epidermal growth factor receptor-mutated non-small-cell lung cancer, with squamous cell transformation and T790M-acquired resistance mechanisms? A Case report and review of literature. Anticancer Drugs 2022, 33, e720–e723. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, L.; Liu, L.; Niu, X. EMT-Mediated Acquired EGFR-TKI Resistance in NSCLC: Mechanisms and Strategies. Front. Oncol. 2019, 9, 1044. [Google Scholar] [CrossRef]
- Deng, Q.; Xie, B.; Wu, L.; Ji, X.; Li, C.; Feng, L.; Fang, Q.; Bao, Y.; Li, J.; Jin, S.; et al. Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients. Heliyon 2018, 4, e01031. [Google Scholar] [CrossRef]
- Olsen, S.; Liao, J.; Hayashi, H. Real-World Clinical Outcomes after Genomic Profiling of Circulating Tumor DNA in Patients with Previously Treated Advanced Non-Small Cell Lung Cancer. Curr. Oncol. 2022, 29, 382. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.S.; Yang, J.C.; Lee, C.K.; Kurata, T.; Kim, D.W.; John, T.; Nogami, N.; Ohe, Y.; Mann, H.; Rukazenkov, Y.; et al. Osimertinib as First-Line Treatment of EGFR Mutation-Positive Advanced Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Iwama, E.; Sakai, K.; Azuma, K.; Harada, D.; Nosaki, K.; Hotta, K.; Nishio, M.; Kurata, T.; Fukuhara, T.; Akamatsu, H.; et al. Exploration of resistance mechanisms for epidermal growth factor receptor-tyrosine kinase inhibitors based on plasma analysis by digital polymerase chain reaction and next-generation sequencing. Cancer Sci. 2018, 109, 3921–3933. [Google Scholar] [CrossRef]
- Usui, K.; Yokoyama, T.; Naka, G.; Ishida, H.; Kishi, K.; Uemura, K.; Ohashi, Y.; Kunitoh, H. Plasma ctDNA monitoring during epidermal growth factor receptor (EGFR)-tyrosine kinase inhibitor treatment in patients with EGFR-mutant non-small cell lung cancer (JP-CLEAR trial). Jpn. J. Clin. Oncol. 2019, 49, 554–558. [Google Scholar] [CrossRef]
- Lei, L.; Wang, W.X.; Zhu, Y.C.; Li, J.L.; Fang, Y.; Wang, H.; Zhuang, W.; Zhang, Y.B.; Wang, L.P.; Fang, M.Y.; et al. Real-world efficacy and potential mechanism of resistance of icotinib in Asian advanced non-small cell lung cancer with EGFR uncommon mutations: A multi-center study. Cancer Med. 2020, 9, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Francaviglia, I.; Magliacane, G.; Lazzari, C.; Grassini, G.; Brunetto, E.; Dal Cin, E.; Girlando, S.; Medicina, D.; Smart, C.E.; Bulotta, A.; et al. Identification and monitoring of somatic mutations in circulating cell-free tumor DNA in lung cancer patients. Lung Cancer 2019, 134, 225–232. [Google Scholar] [CrossRef]
- Hochmair, M.J.; Buder, A.; Schwab, S.; Burghuber, O.C.; Prosch, H.; Hilbe, W.; Cseh, A.; Fritz, R.; Filipits, M. Liquid-Biopsy-Based Identification of EGFR T790M Mutation-Mediated Resistance to Afatinib Treatment in Patients with Advanced EGFR Mutation-Positive NSCLC, and Subsequent Response to Osimertinib. Target Oncol. 2019, 14, 75–83. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, C.; Zhao, J.; Wang, Q.; Chu, X.; Li, J.; Zhou, F.; Ren, S.; Li, X.; Su, C.; et al. Re-biopsy and liquid biopsy for patients with non-small cell lung cancer after EGFR-tyrosine kinase inhibitor failure. Thorac. Cancer 2019, 10, 957–965. [Google Scholar] [CrossRef]
- Ariyasu, R.; Uchibori, K.; Sasaki, T.; Tsukahara, M.; Kiyotani, K.; Yoshida, R.; Ono, Y.; Kitazono, S.; Ninomiya, H.; Ishikawa, Y.; et al. Monitoring epidermal growth factor receptor C797S mutation in Japanese non-small cell lung cancer patients with serial cell-free DNA evaluation using digital droplet PCR. Cancer Sci. 2021, 112, 2371–2380. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiong, L.; Xie, F.; Zheng, X.; Li, Y.; Zhu, L.; Sun, J. Next-generation sequencing of tissue and circulating tumor DNA: Resistance mechanisms to EGFR targeted therapy in a cohort of patients with advanced non-small cell lung cancer. Cancer Med. 2021, 10, 4697–4709. [Google Scholar] [CrossRef]
- Choi, W.; Cho, Y.; Park, S.Y.; Hwang, K.H.; Han, J.Y.; Lee, Y. A nanowire-based liquid biopsy method using cerebrospinal fluid cell-free DNA for targeted management of leptomeningeal carcinomatosis. J. Cancer Res. Clin. Oncol. 2021, 147, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Minari, R.; Mazzaschi, G.; Bordi, P.; Gnetti, L.; Alberti, G.; Altimari, A.; Gruppioni, E.; Sperandi, F.; Parisi, C.; Guaitoli, G.; et al. DETECTION Study Group. Detection of EGFR-Activating and T790M Mutations Using Liquid Biopsy in Patients With EGFR-Mutated Non-Small-Cell Lung Cancer Whose Disease Has Progressed During Treatment with First- and Second-Generation Tyrosine Kinase Inhibitors: A Multicenter Real-Life Retrospective Study. Clin. Lung Cancer 2020, 21, e464–e473. [Google Scholar] [PubMed]
- Cavic, M.; Krivokuca, A.; Pavlovic, M.; Boljevic, I.; Rakobradovic, J.; Mihajlovic, M.; Tanic, M.; Damjanovic, A.; Malisic, E.; Jankovic, R. EGFR mutation testing from liquid biopsy of non-small cell lung cancer at the Institute for Oncology and Radiology of Serbia. J. BUON 2020, 25, 2635–2642. [Google Scholar]
- Li, Y.; Zhang, F.; Yuan, P.; Guo, L.; Jianming, Y.; He, J. High MAF of EGFR mutations and high ratio of T790M sensitizing mutations in ctDNA predict better third-generation TKI outcomes. Thorac. Cancer 2020, 11, 1503–1511. [Google Scholar] [CrossRef]
- Fu, Y.; Wang, A.; Zhou, J.; Feng, W.; Shi, M.; Xu, X.; Zhao, H.; Cai, L.; Feng, J.; Lv, X. Advanced NSCLC Patients with EGFR T790M Harboring TP53 R273C or KRAS G12V Cannot Benefit from Osimertinib Based on a Clinical Multicentre Study by Tissue and Liquid Biopsy. Front. Oncol. 2021, 11, 621992. [Google Scholar] [CrossRef] [PubMed]
- Jóri, B.; Schatz, S.; Kaller, L.; Kah, B.; Roeper, J.; Ramdani, H.O.; Diehl, L.; Hoffknecht, P.; Grohé, C.; Griesinger, F.; et al. Comparison of Resistance Spectra after First and Second Line Osimertinib Treatment Detected by Liquid Biopsy. Cancers 2021, 13, 2861. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Cao, N.T.; Van Nguyen, T.C.; Le, K.N.D.; Nguyen, D.T.; Nguyen, Q.T.; Nguyen, T.T.; Van Nguyen, C.; Le, H.T.; Nguyen, M.T.; et al. Liquid biopsy uncovers distinct patterns of DNA methylation and copy number changes in NSCLC patients with different EGFR-TKI resistant mutations. Sci. Rep. 2021, 11, 16436. [Google Scholar] [CrossRef]
- Fan, Y.; Sun, R.; Wang, Z.; Zhang, Y.; Xiao, X.; Liu, Y.; Xin, B.; Xiong, H.; Lu, D.; Ma, J. Detection of MET amplification by droplet digital PCR in peripheral blood samples of non-small cell lung cancer. J. Cancer Res. Clin. Oncol. 2022, 18, 1–11. [Google Scholar] [CrossRef]
- Gelatti, A.C.Z.; Drilon, A.; Santini, F.C. Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC). Lung Cancer 2019, 137, 113–122. [Google Scholar] [CrossRef]
- Lei, L.; Wang, W.X.; Zhu, Y.C.; Li, J.L.; Fang, Y.; Wang, H.; Zhuang, W.; Zhang, Y.B.; Wang, L.P.; Fang, M.Y.; et al. Potential mechanism of primary resistance to icotinib in patients with advanced non-small cell lung cancer harboring uncommon mutant epidermal growth factor receptor: A multi-center study. Cancer Sci. 2020, 111, 679–686. [Google Scholar] [CrossRef]
- Wang, Y.; He, Y.; Tian, P.; Wang, W.; Wang, K.; Chuai, S.; Li, Y.; Zhao, S.; Wang, Y.; Li, W. Low T790M relative allele frequency indicates concurrent resistance mechanisms and poor responsiveness to osimertinib. Transl. Lung Cancer Res. 2020, 9, 1952–1962. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Lee, V.H.F.; Chen, L.; Ma, L.; Wang, D.D.; Yan, H. Deciphering mechanisms of acquired T790M mutation after EGFR inhibitors for NSCLC by computational simulations. Sci. Rep. 2017, 7, 6595. [Google Scholar] [CrossRef]
- Verzè, M.; Minari, R.; Gnetti, L.; Bordi, P.; Leonetti, A.; Cosenza, A.; Ferri, L.; Majori, M.; De Filippo, M.; Buti, S.; et al. Monitoring cfDNA in Plasma and in Other Liquid Biopsies of Advanced EGFR Mutated NSCLC Patients: A Pilot Study and a Review of the Literature. Cancers 2021, 13, 5403. [Google Scholar] [CrossRef]
- Minari, R.; Bordi, P.; Del Re, M.; Facchinetti, F.; Mazzoni, F.; Barbieri, F.; Camerini, A.; Comin, C.E.; Gnetti, L.; Azzoni, C.; et al. Primary resistance to osimertinib due to SCLC transformation: Issue of T790M determination on liquid re-biopsy. Lung Cancer 2018, 115, 21–27. [Google Scholar] [CrossRef]
- Tsui, D.W.Y.; Murtaza, M.; Wong, A.S.C.; Rueda, O.M.; Smith, C.G.; Chandrananda, D.; Soo, R.A.; Lim, H.L.; Goh, B.C.; Caldas, C.; et al. Dynamics of multiple resistance mechanisms in plasma DNA during EGFR-targeted therapies in non-small cell lung cancer. EMBO. Mol. Med. 2018, 10, e7945. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.; Stewart, E.L.; Martins-Filho, S.N.; Cabanero, M.; Wang, A.; Bao, H.; Wu, X.; Patel, D.; Chen, Z.; Law, J.H.; et al. Early Detection of Multiple Resistance Mechanisms by ctDNA Profiling in a Patient With EGFR-mutant Lung Adenocarcinoma Treated with Osimertinib. Clin. Lung Cancer 2020, 21, e488–e492. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Hu, Y.; Mileham, K.F.; Husain, H.; Costa, D.B.; Tracy, P.; Feeney, N.; Sholl, L.M.; Dahlberg, S.E.; Redig, A.J.; et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients with EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018, 4, 1527–1534. [Google Scholar] [CrossRef]
- Kato, R.; Hayashi, H.; Sakai, K.; Suzuki, S.; Haratani, K.; Takahama, T.; Tanizaki, J.; Nonagase, Y.; Tanaka, K.; Yoshida, T.; et al. CAPP-seq analysis of circulating tumor DNA from patients with EGFR T790M-positive lung cancer after osimertinib. Int. J. Clin. Oncol. 2021, 26, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- De Carlo, E.; Schiappacassi, M.; Pelizzari, G.; Baresic, T.; Del Conte, A.; Stanzione, B.; Da Ros, V.; Doliana, R.; Baldassarre, G.; Bearz, A. Acquired EGFR C797G Mutation Detected by Liquid Biopsy as Resistance Mechanism After Treatment with Osimertinib: A Case Report. In Vivo 2021, 35, 2941–2945. [Google Scholar] [CrossRef]
- Shen, F.; Liang, N.; Fan, Z.; Zhao, M.; Kang, J.; Wang, X.; Hu, Q.; Mu, Y.; Wang, K.; Yuan, M.; et al. Genomic Alterations Identification and Resistance Mechanisms Exploration of NSCLC With Central Nervous System Metastases Using Liquid Biopsy of Cerebrospinal Fluid: A Real-World Study. Front. Oncol. 2022, 12, 889591. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xing, P.; Shi, M.; Guo, W.; Zhao, F.; Zhu, H.; Xiao, J.; Wan, J.; Li, J. Cerebrospinal Fluid Cell-Free DNA-Based Detection of High Level of Genomic Instability Is Associated with Poor Prognosis in NSCLC Patients with Leptomeningeal Metastases. Front. Oncol. 2022, 12, 664420. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.M.; Li, Y.S.; Tu, H.Y.; Jiang, B.Y.; Yang, J.J.; Zhou, Q.; Xu, C.R.; Yang, X.R.; Wu, Y.L. Genotyping of Cerebrospinal Fluid Associated with Osimertinib Response and Resistance for Leptomeningeal Metastases in EGFR-Mutated NSCLC. J. Thorac. Oncol. 2021, 16, 250–258. [Google Scholar] [CrossRef]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef]
- Li, W.; Guo, L.; Liu, Y.; Dong, L.; Yang, L.; Chen, L.; Liu, K.; Shao, Y.; Ying, J. Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC. J. Thorac. Oncol. 2021, 16, 404–418. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Li, W.; Chen, L.; Ying, J. Intergenic Breakpoints Identified by DNA Sequencing Confound Targetable Kinase Fusion Detection in NSCLC. J. Thorac. Oncol. 2020, 15, 1223–1231. [Google Scholar] [CrossRef] [PubMed]
- Cohen, D.; Hondelink, L.M.; Solleveld-Westerink, N.; Uljee, S.M.; Ruano, D.; Cleton-Jansen, A.M.; von der Thüsen, J.H.; Ramai, S.R.S.; Postmus, P.E.; Graadt van Roggen, J.F.; et al. Optimizing Mutation and Fusion Detection in NSCLC by Sequential DNA and RNA Sequencing. J. Thorac. Oncol. 2020, 15, 1000–1014. [Google Scholar] [CrossRef]
- Wu, C.Y.; Lee, C.L.; Wu, C.F.; Fu, J.Y.; Yang, C.T.; Wen, C.T.; Liu, Y.H.; Liu, H.P.; Hsieh, J.C. Circulating Tumor Cells as a Tool of Minimal Residual Disease Can Predict Lung Cancer Recurrence: A longitudinal, Prospective Trial. Diagnostics 2020, 10, 144. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.A.; Chabon, J.J.; Lovejoy, A.F.; Newman, A.M.; Stehr, H.; Azad, T.D.; Khodadoust, M.S.; Esfahani, M.S.; Liu, C.L.; Zhou, L.; et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017, 7, 1394–1403. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Chen, K.; Zhao, H.; Shi, Y.; Yang, F.; Wang, L.T.; Kang, G.; Nie, Y.; Wang, J. Perioperative Dynamic Changes in Circulating Tumor DNA in Patients with Lung Cancer (DYNAMIC). Clin. Cancer Res. 2019, 25, 7058–7067. [Google Scholar] [CrossRef] [PubMed]
- Ohara, S.; Suda, K.; Sakai, K.; Nishino, M.; Chiba, M.; Shimoji, M.; Takemoto, T.; Fujino, T.; Koga, T.; Hamada, A.; et al. Prognostic implications of preoperative versus postoperative circulating tumor DNA in surgically resected lung cancer patients: A pilot study. Transl. Lung Cancer Res. 2020, 9, 1915–1923. [Google Scholar] [CrossRef] [PubMed]
- Kuang, P.P.; Li, N.; Liu, Z.; Sun, T.Y.; Wang, S.Q.; Hu, J.; Ou, W.; Wang, S.Y. Circulating Tumor DNA Analyses as a Potential Marker of Recurrence and Effectiveness of Adjuvant Chemotherapy for Resected Non-Small-Cell Lung Cancer. Front. Oncol. 2021, 10, 595650. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Shao, C.; Han, L.; Liu, H.; Ma, Z.; Yang, Y.; Feng, Y.; Pan, M.; Santarpia, M.; Carmo-Fonseca, M.; et al. Detection of epidermal growth factor receptor (EGFR) mutations from preoperative circulating tumor DNA (ctDNA) as a prognostic predictor for stage I-III non-small cell lung cancer (NSCLC) patients with baseline tissue EGFR mutations. Transl. Lung Cancer Res. 2021, 10, 3213–3225. [Google Scholar] [CrossRef]
- Qiu, B.; Guo, W.; Zhang, F.; Lv, F.; Ji, Y.; Peng, Y.; Chen, X.; Bao, H.; Xu, Y.; Shao, Y.; et al. Dynamic recurrence risk and adjuvant chemotherapy benefit prediction by ctDNA in resected NSCLC. Nat. Commun. 2021, 12, 6770. [Google Scholar] [CrossRef]
- Peng, M.; Huang, Q.; Yin, W.; Tan, S.; Chen, C.; Liu, W.; Tang, J.; Wang, X.; Zhang, B.; Zou, M.; et al. Circulating Tumor DNA as a Prognostic Biomarker in Localized Non-small Cell Lung Cancer. Front. Oncol. 2020, 10, 561598. [Google Scholar] [CrossRef]
- Li, N.; Wang, B.X.; Li, J.; Shao, Y.; Li, M.T.; Li, J.J.; Kuang, P.P.; Liu, Z.; Sun, T.Y.; Wu, H.Q.; et al. Perioperative circulating tumor DNA as a potential prognostic marker for operable stage I to IIIA non-small cell lung cancer. Cancer 2022, 128, 708–718. [Google Scholar] [CrossRef]
- Waldeck, S.; Mitschke, J.; Wiesemann, S.; Rassner, M.; Andrieux, G.; Deuter, M.; Mutter, J.; Lüchtenborg, A.M.; Kottmann, D.; Titze, L.; et al. Early assessment of circulating tumor DNA after curative-intent resection predicts tumor recurrence in early-stage and locally advanced non-small-cell lung cancer. Mol. Oncol. 2022, 16, 527–537. [Google Scholar] [CrossRef]
- Xia, L.; Mei, J.; Kang, R.; Deng, S.; Chen, Y.; Yang, Y.; Feng, G.; Deng, Y.; Gan, F.; Lin, Y.; et al. Perioperative ctDNA-Based Molecular Residual Disease Detection for Non-Small Cell Lung Cancer: A Prospective Multicenter Cohort Study (LUNGCA-1). Clin. Cancer Res. 2022, 28, 3308–3317. [Google Scholar] [CrossRef]
- Gale, D.; Heider, K.; Ruiz-Valdepenas, A.; Hackinger, S.; Perry, M.; Marsico, G.; Rundell, V.; Wulff, J.; Sharma, G.; Knock, H.; et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 2022, 33, 500–510. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Liu, W.; Chen, C.; Zhang, T.; Ma, Y.; Cui, L.; Gu, Y.; Bei, T.; Zhao, X.; Zhang, B. Circulating tumor DNA predicts neoadjuvant immunotherapy efficacy and recurrence-free survival in surgical non-small cell lung cancer patients. Transl. Lung Cancer Res. 2022, 11, 263–276. [Google Scholar] [CrossRef]
- Guo, H.; Li, W.; Wang, B.; Chen, N.; Qian, L.; Cui, J. Coexisting opportunities and challenges: In which scenarios can minimal/measurable residual disease play a role in advanced non-small cell lung cancer? Chin. J. Cancer Res. 2021, 33, 574–582. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, T.; Wakuda, K.; Kenmotsu, H.; Miyawaki, E.; Mamesaya, N.; Kobayashi, H.; Omori, S.; Ono, A.; Naito, T.; Murakami, H.; et al. Proposing synchronous oligometastatic non-small-cell lung cancer based on progression after first-line systemic therapy. Cancer Sci. 2021, 112, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Spaggiari, L.; Bertolaccini, L.; Facciolo, F.; Gallina, F.T.; Rea, F.; Schiavon, M.; Margaritora, S.; Congedo, M.T.; Lucchi, M.; Ceccarelli, I.; et al. A risk stratification scheme for synchronous oligometastatic non-small cell lung cancer developed by a multicentre analysis. Lung Cancer 2021, 154, 29–35. [Google Scholar] [CrossRef]
- Gauvin, C.; Krishnan, V.; Kaci, I.; Tran-Thanh, D.; Bédard, K.; Albadine, R.; Leduc, C.; Gaboury, L.; Blais, N.; Tehfe, M.; et al. Survival Impact of Aggressive Treatment and PD-L1 Expression in Oligometastatic NSCLC. Curr. Oncol. 2021, 28, 59. [Google Scholar] [CrossRef]
- Pécuchet, N.; Zonta, E.; Didelot, A.; Combe, P.; Thibault, C.; Gibault, L.; Lours, C.; Rozenholc, Y.; Taly, V.; Laurent-Puig, P.; et al. Base-Position Error Rate Analysis of Next-Generation Sequencing Applied to Circulating Tumor DNA in Non-Small Cell Lung Cancer: A Prospective Study. PLoS. Med. 2016, 13, e1002199. [Google Scholar] [CrossRef] [PubMed]
- Moding, E.J.; Liu, Y.; Nabet, B.Y.; Chabon, J.J.; Chaudhuri, A.A.; Hui, A.B.; Bonilla, R.F.; Ko, R.B.; Yoo, C.H.; Gojenola, L.; et al. Circulating Tumor DNA Dynamics Predict Benefit from Consolidation Immunotherapy in Locally Advanced Non-Small Cell Lung Cancer. Nat. Cancer 2020, 1, 176–183. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Nabet, B.Y.; Rizvi, H.; Chaudhuri, A.A.; Wells, D.K.; Dunphy, M.P.S.; Chabon, J.J.; Liu, C.L.; Hui, A.B.; Arbour, K.C.; et al. Circulating Tumor DNA Analysis to Assess Risk of Progression after Long-term Response to PD-(L)1 Blockade in NSCLC. Clin. Cancer Res. 2020, 26, 2849–2858. [Google Scholar] [CrossRef]
- Tang, C.; Lee, W.C.; Reuben, A.; Chang, L.; Tran, H.; Little, L.; Gumbs, C.; Wargo, J.; Futreal, A.; Liao, Z.; et al. Immune and Circulating Tumor DNA Profiling After Radiation Treatment for Oligometastatic Non-Small Cell Lung Cancer: Translational Correlatives from a Mature Randomized Phase II Trial. Int. J. Radiat. Oncol. Biol. Phys. 2020, 106, 349–357. [Google Scholar] [CrossRef]
- Yan, X.; Liu, C. Application of Non-Blood-Derived Fluid Biopsy in Monitoring Minimal Residual Diseases of Lung Cancer. Front. Surg. 2022, 9, 865040. [Google Scholar] [CrossRef]
| Reference (Year) | Characteristics of Subjects | n | ctDNA/cfDNA Detection Method | Prognostic Relevance |
|---|---|---|---|---|
| [28] (2015) |
| 97 | TaqMan assay |
L858R mutation in tissues and cfDNA vs. L858R mutation in tissues but not in cfDNA: 13.7 m (95% CI = 7.1–17.7) vs. 27.7 m (95% CI = 16.1–46.2) (HR = 2.22; 95% CI = 1.09–4.52; p = 0.03) |
| [31] (2016) |
Group B: positive for EGFR mutations in blood only (T−/C+) (n = 28) Group C: positive for EGFR mutations in tissue only (T+/C−) (n = 180)
| 472 | ARMS, ddPCR and NGS |
|
| [45] (2018) |
| 19 | Cobas EGFR Mutaion Test v2 or PANA Mutyper |
|
| [46] (2018) |
| 57 | Qualitative (PANAMutyper) and quantitative (PANAGENE-SQI) |
|
| [32] (2018) |
| 183 | ddPCR |
|
| [47] (2019) |
| 22 | dPCR and NGS |
|
| [30] (2019) |
| 71 | ADx-ARMS |
C+EGFR wild type vs. C+EGFR mutant type: 8.7 m vs. 11.0 m (p = 0.001) |
| [48] (2019) |
| 30 | NGS or ARMS |
|
| [49] (2019) |
| 307 | Cobas EGFR Mutation Test v2 |
T790M-positive plasma: 8.3 m with osimertinib (95% CI = 6.8–10.5) ; 4.2 m with platinum–pemetrexed (95% CI = 4.1–5.4) |
| [34] (2019) |
| 66 | ddPCR |
|
| [50] (2020) |
| 12 | NGS |
Patients with undetectable plasma EGFR mutations at week 6 had better overall PFS compared to those with detectable mutations (NR vs. 4.5 m; 95% CI = 0.0–1.1; p < 0.05). No significant changes in PFS were observed based on the absence of detectable EGFR-sensitizing mutations in CSF at week 6 (p = 0.68) |
| [51] (2020) |
| 30 | ddPCR |
|
| [52] (2020) |
| 53 | Cobas EGFR Mutation Test v2 or ddPCR |
|
| [53] (2020) |
| 52 | Cobas EGFR Mutation Test v2, ddPCR and NGS |
|
| [35] (2021) |
Group B: EGFR mutations in ctDNA and tumor DNA (n = 10)
| 21 | PNA-based RT-PCR |
|
| [54] (2021) |
| 19 | Mutyper and Cobas EGFR Mutation Test v2 |
|
| [55] (2021) |
| 54 | cSMART assay |
60.0% for the >0.1% group vs. 21.4% for the ≤0.1% group (p = 0.028)
|
| [56] (2021) |
| 41 | R-superARMS |
ΔCt >4.89 vs. 4.89: NR vs. 11.0 m (p = 0.014) mPFS: mutation clearance vs. incomplete mutation clearance: NR vs. 27.5 m (p = 0.088) |
| [57] (2022) |
| 28 | NGS |
EGFR clearance vs. EGFR non-clearance (week4): 11.4 m vs. 5.67 m (p = 0.011; HR = 0.23; 95% CI = 0.08–0.72) Non-clearance vs. EGFR clearance only vs. total-clearance (week4): 11.4 m vs. 9.2 m vs. 5.07 m
|
| [58] (2022) |
| 116 | SuperARMS ddPCR NGS |
|
| [33] (2022) |
| 158 | Cobas EGFR Mutation Test v2 |
|
| References (Year) | No.of Patients | Prior Treatment | Detection Method | Genetic Treatment-Resistant Alterations Detected in Plasma (%) |
|---|---|---|---|---|
| [106] (2021) | 50 | First-generation EGFR-TKIs | ddPCR and NGS | T790M: 38% (19/50) |
| [91] (2018) | 53 | First-generation EGFR-TKIs | NGS for 124-genes panel | T790M: 45.28% (24/53) EGFR point mutations: 20.75% (11/53) KRAS/NRAS point mutations: 15.09% (8/53) EGFR amplification: 7.54% (4/53) BRAF amplification: 1.8% (1/53) MET amplification: 3.7% (2/53) ERBB2 amplification: 1.8% (1/53) |
| [101] (2020) | 37 | First-generation EGFR-TKIs | ddPCRand NGS for 223-genes panel | EGFR T790M: 51.35% (19/37) TP53: 67.57% (25/37) KRAS: 8.11% (3/37) c-Met amplification: 5.41% (2/37) STK11: 5.41% (2/37) FANCA: 5.41% (2/37) ERBB2: 5.41% (2/37) PIK3CA: 2.7% (1/37) FGFR1: 2.7% (1/37) BRAF: 2.7% (1/37) |
| [105] (2020) | 147 | First-generation EGFR-TKIs | NGS for 168-genes panel | T790M: 40.13% (59/147) MET and ERBB2 amplification: 2.04% (3/147) TP53: 45.86% (61/133) |
| [96] (2019) | 48 | Icotinib | NGS for 170-genes panel | T790M: 81.2% (39/48) EGFR amplification: 72.9% (35/48) CTNNB1: 2.1% (1/48) PIK3CA: 2.1% (1/48) BRAF: 2.1% (1/48) EML4-ALK: 2.1% (1/48) SLC342-ROS1: 2.1% (1/48) Unknown mutations: 2.1% (1/48) |
| [45] (2018) | 80 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2 or PANA Mutyper | T790M: 26.3% (21/80) |
| [95] (2019) | 66 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2. | T790M: 33.3% (22/66) |
| [97] (2019) | 50 | First- and second-generation EGFR-TKIs | NGS | T790M: 71% (30/42) |
| [99] (2019) | 118 | First- and second-generation EGFR-TKIs | ARMS-PCR or super ARMS-PCR | T790M: 41.5% (49/118) |
| [52] (2020) | 276 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2 or ddPCR | T790M: 26.8% (74/276) |
| [103] (2020) | 120 | First- and second-generation EGFR-TKIs | Easy EGFR, Therascreen EGFR RGQ PCR and Cobas EGFR Mutation Test v2 | T790M:25.8% (31/120) |
| [104] (2020) | 104 | First- and second-generation EGFR-TKIs | Cobas EGFR Mutation Test v2 | T790M: 49% (34/104) |
| [94] (2018) | 25 | Afatinib | dPCR and NGS | T790M dPCR: 56.5% (13/23) NGS: 43.5% (10/23) |
| [98] (2019) | 67 | Afatinib | ddPCR | T790M: 73.1% (49/67) |
| [47] (2019) | 22 | First- and second-generation EGFR-TKIs | dPCR and NGS | T790M: 86% (19/22) Progression to osimertinib (16), EGFR C797S (3), A750P (1), S464L (1), amplification (1), PIK3CA E545A (3) and E545K (1) |
| [108] (2021) | 122 | First- and second-generation EGFR-TKIs | NGS for 9-genes panel | T790M: 32% (39/122) EGFR amplification: 6.6% (8/122) PIK3CA: 3.3% (4/122) MET amplification: 3.3% (4/122) HER2 amplification: 4.1% (5/122) |
| [102] (2020) | 11 | Third -generation EGFR-TKIs | Nanowire-based colorimetric cfDNA assay (EGFR mutation and MET amplification) | Plasma cfDNA profiles Drug-sensitive EGFR founder mutations: 36.3% (4/11) De novo EGFR C797S: 18.2% (2/11) MET amplification: 18.2% (2/11) EGFR T790M: 18.2% (2/11) CSF-cfDNA: Drug-sensitive EGFR founder mutations: 45.5% (5/11) De novo EGFR C797S: 36.3% (4/11) MET amplification: 18.2% (2/11) EGFR T790M: 18.2% (2/11) |
| [109] (2022) | 49 | Third-generation EGFR-TKIs | ddPCR (MET copy number gain) | MET CNG: 26.5% (13/49) MET amplification:16.3% (8/49) |
| [100] (2020) | 26 | Osimertinib | ddPCR | EGFR C797S: 20% (3/15) Loss of T790M: 33.3% (4/15) |
| [107] (2021) | 56 | Osimertinib | NGS for 39-genes panel | Second-line osimertinib (n = 41) EGFR C797S: 39% (16/41) Non-C797S EGFR mutations: 12% (5/41) V843I, L718Q, C724S, L792H and one patient with L718V, L718Q, L792H and G796S RB1 and TP53 inactivating mutations: 7% (3/41) EGFR amplification: 10% (4/41) MET amplification: 7% (3/41) CTNNB1 point mutations: 7% (3/41) KRAS mutations: 5% (2/41) PIK3CA activating mutations: 5% (2/41) ERBB2, PTEN, mTOR and RET mutations: 2% (1/41 each) AGK-BRAF, RET-RUFY1, TACC-FGFR3 and DLG1-BRAF fusion: 2% (1/41 each) Loss of T790M: 34% (14/41) First-line osimertinib (n = 7) EGFR alterations (EGFR C797S and EGFR T854A): 28.5% (2/7) MET amplification: 14.2% (1/7) EML4-ALK fusions: 14.2% (1/7) SCC/SCLC switch [RB1(R787*)]: 14.2% (1/7) |
| [93] (2017) | 19 | Osimertinib | NGS for 73-genes panel | MET amplification: 5.3% (n = 1) EGFR and KRAS amplification: 5.3% (n = 1) MEK1, KRAS or PIK3CA mutations: 5.3% (n = 1 each) EGFR C797S: 10.6% (n = 2) JAK2 mutation: 5.3% (n = 1) HER2 exon 20 insertion: 5.3% (n = 1) |
| [92] (2022) | - | EGFR-TKIs | NGS for 74-genes panel | First- and second-generation EGFR-TKIs (n = 490) EGFR T790M: 48.0% (235/490) MET amplification: 7.1% (35/490) BRAF V600E: 1.0% (5/490) KRAS mutations: 3.6% (20/490) Third-generation EGFR-TKIs (n = 205) MET amplification: 8.9% (16/205) EGFR C797S: 5.6% (10/205) BRAF V600E: 4.5% (8/205) KRAS mutation: 3.4% (7/205) |
| References (Year) | Sample | Stage | Detection Methods/Study Design | Median Follow-Up Time (Month) | Detection Time | Clinical Relevance | ctDNA Positivity Precedes Radiological Recurrence by a Median Lead Time (Month) |
|---|---|---|---|---|---|---|---|
| [131] (2019) | 26 | I–III | NGS for a 9-gene panel/Pro | 532 days for all patients and 629 days for patients who were free from progression |
B: 5 min; C: 30 min; D: 2 h
|
| NR |
| [132] (2020) | 20 | IIA–IIIA | NGS for a 197-gene panel/Pro | 12 |
|
| NR |
| [133] (2020) | 38 | IB–III | NGS for a 425-gene panel/Pro | 15.8 |
|
| NR |
| [134] (2021) | 174 | I–III | ARMs for EGFR/Pro | NA |
|
| NR |
| [135] (2021) | 116 | I–IV | NGS for a 139-gene panel/Pro | NA |
|
Proportion of patients who were ctDNA-positive after surgery: 21.2% (18/85); after the completion of ACT: 12.5% (8/64)
| 2.93 |
| [136] (2021) | 77 | I–IV | cSMART for a 127-gene panel/Pro | 46 |
|
| 12.6 |
| [137] (2021) | 119 | I–IIIA | NGS for a 425-gene panel/Pro | 30.7 |
|
| 8.71 |
| [138] (2022) | 21 | IA–IIIB | NGS for an18-gene panel/Pro | 26.2 |
|
| 10.31 |
| [139] (2022) | 330 | I–III | NGS for a 769-gene panel/Pro | 35.6 |
|
| NR |
| [140] (2022) | 88 | IA–IIIB | RaDaRTMNGS | 36 |
surgery + adjuvant chemotherapy/radiotherapy (n = 8); chemoradiotherapy (n = 19) |
| 7.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-Z.; Kong, S.-N.; Liu, Y.-P.; Yang, Y.; Zhang, H.-M. Can Liquid Biopsy Based on ctDNA/cfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC? J. Clin. Med. 2023, 12, 1438. https://doi.org/10.3390/jcm12041438
Li Y-Z, Kong S-N, Liu Y-P, Yang Y, Zhang H-M. Can Liquid Biopsy Based on ctDNA/cfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC? Journal of Clinical Medicine. 2023; 12(4):1438. https://doi.org/10.3390/jcm12041438
Chicago/Turabian StyleLi, Yi-Ze, Sheng-Nan Kong, Yun-Peng Liu, Yue Yang, and Hong-Mei Zhang. 2023. "Can Liquid Biopsy Based on ctDNA/cfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC?" Journal of Clinical Medicine 12, no. 4: 1438. https://doi.org/10.3390/jcm12041438
APA StyleLi, Y.-Z., Kong, S.-N., Liu, Y.-P., Yang, Y., & Zhang, H.-M. (2023). Can Liquid Biopsy Based on ctDNA/cfDNA Replace Tissue Biopsy for the Precision Treatment of EGFR-Mutated NSCLC? Journal of Clinical Medicine, 12(4), 1438. https://doi.org/10.3390/jcm12041438








