Deep Learning in Coeliac Disease: A Systematic Review on Novel Diagnostic Approaches to Disease Diagnosis
Abstract
:1. Introduction
2. Methods
2.1. Search Strategy
2.2. Study Selection
2.2.1. Inclusion and Exclusion Criteria
2.2.2. Data Extraction
2.2.3. Risk of Bias
2.3. Data Analysis
2.4. DLT Overview
3. Results
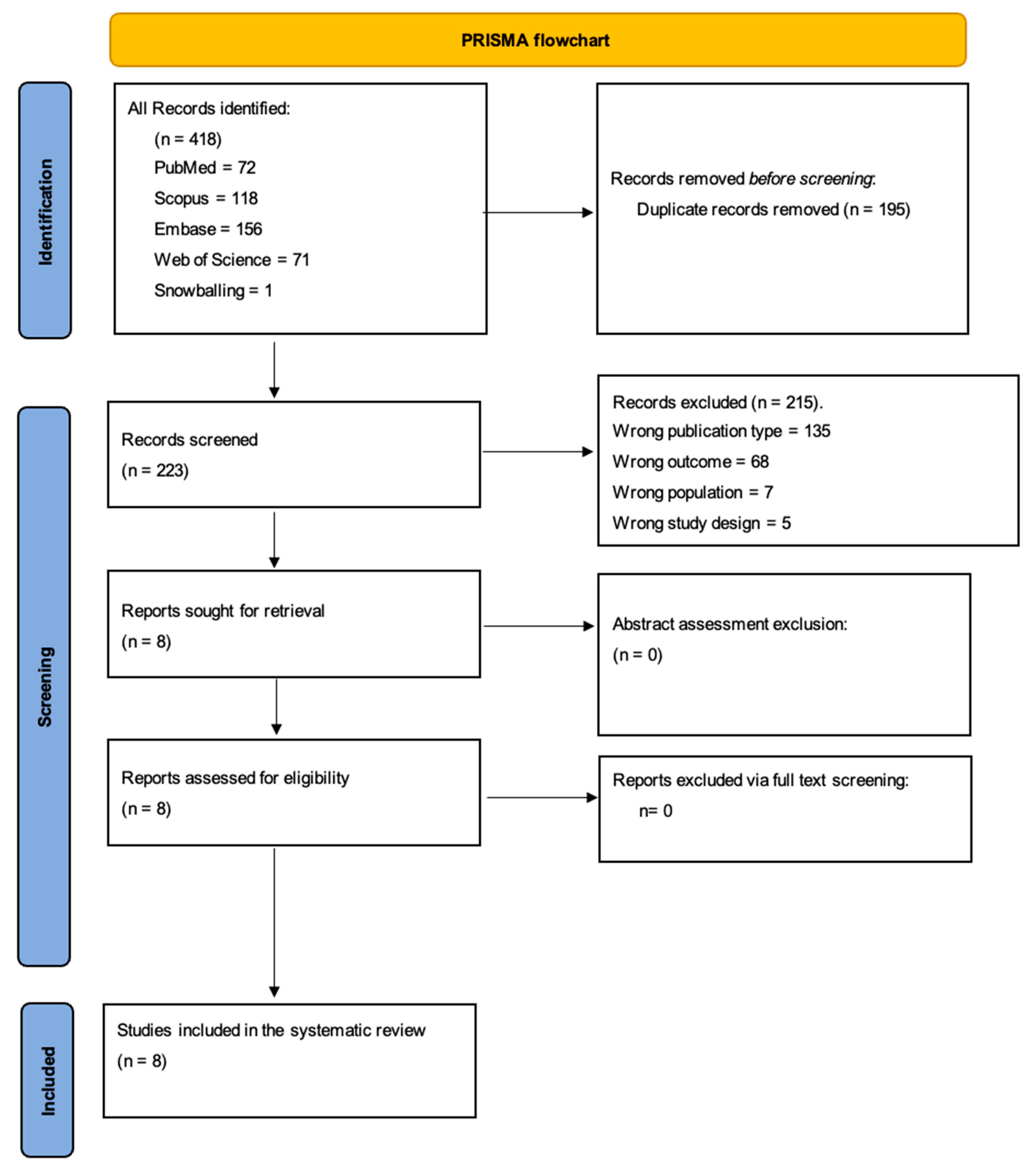
3.1. Search Results and Study Selection
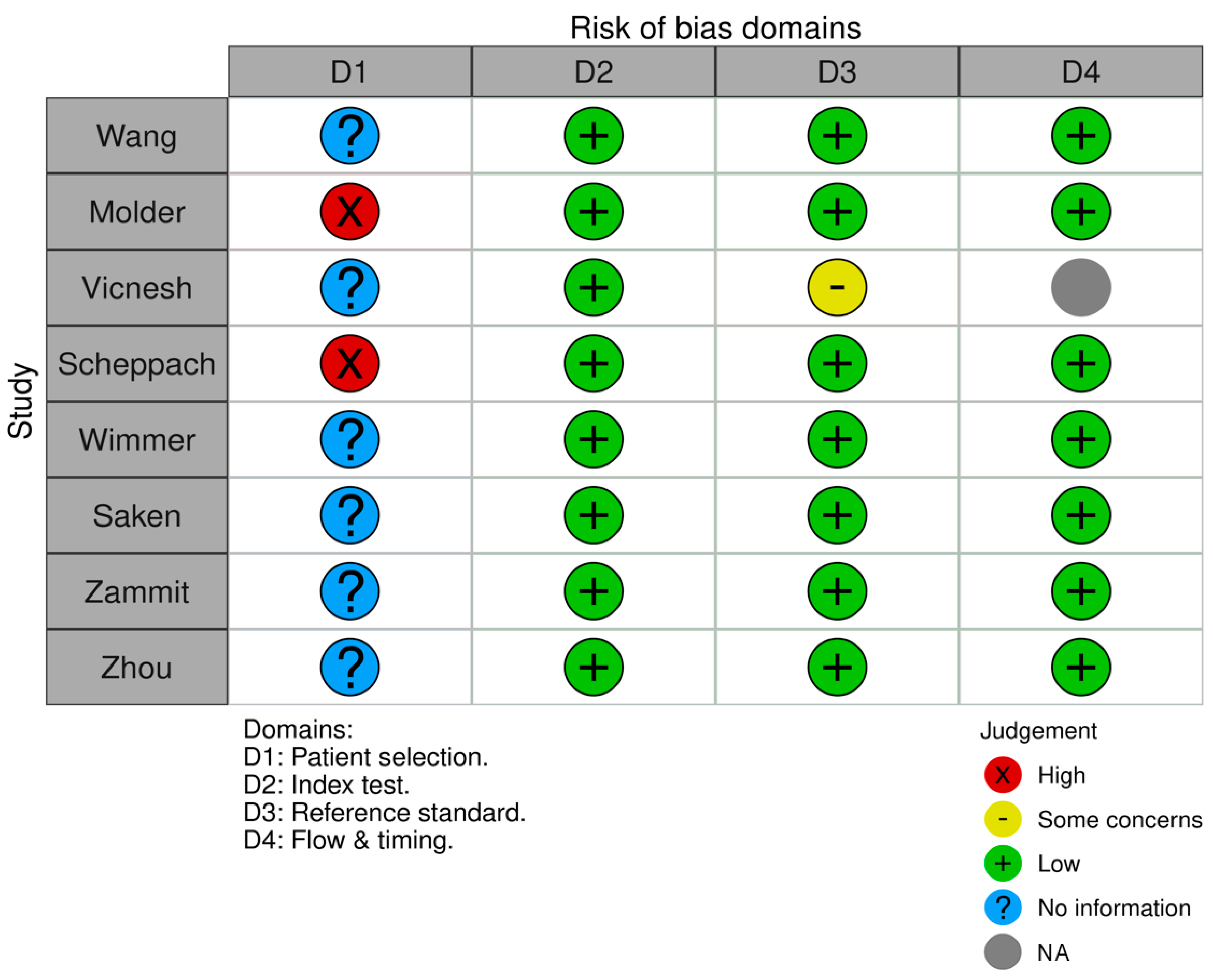
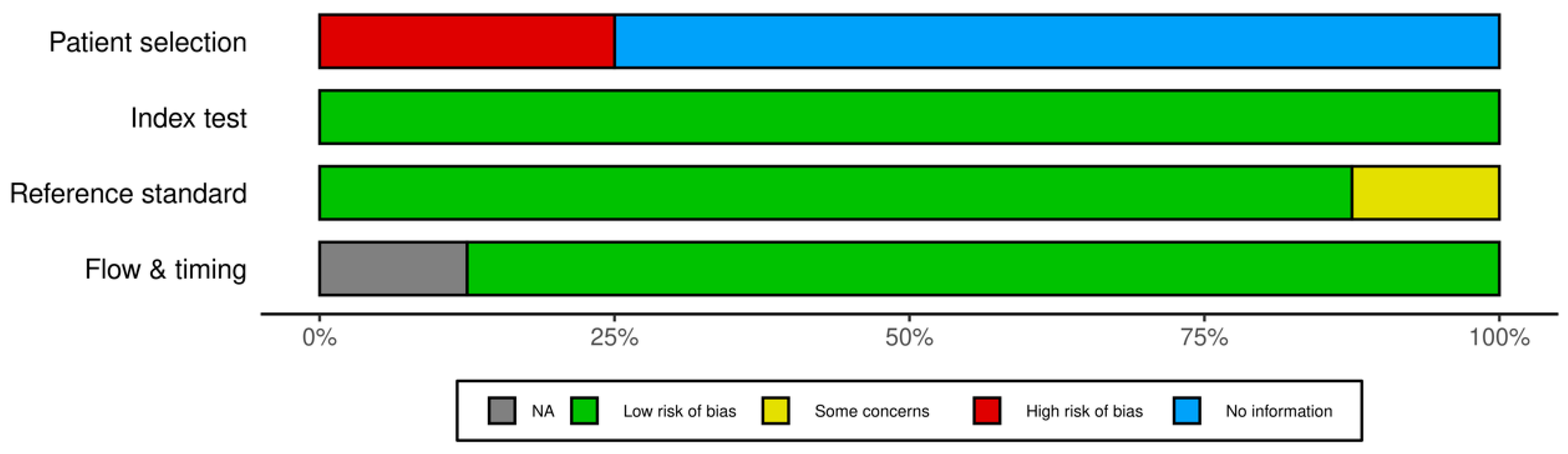
3.2. Risk of Bias
3.3. Overview of Included Studies
3.4. Narrative Synthesis of Deep-Learning Approaches in Coeliac Disease Research
- Harnessing video capsule endoscopy (VCE) images for diagnosis:
- 2.
- DLTTargeting duodenal endoscopy images:
- 3.
- Hybrid techniques and comparative analyses:
- 4.
- Comparative analysis for enhanced understanding:
3.5. Summary of Diagnostic Accuracies
4. Discussion
Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. The Systematic Literature Review Boolean Strings
References
- Makharia, G.K.; Singh, P.; Catassi, C.; Sanders, D.S.; Leffler, D.; Ali, R.A.R.; Bai, J.C. The global burden of coeliac disease: Opportunities and challenges. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Sanchez, M.I.; Silvester, J.A.; Lebwohl, B.; Leffler, D.A.; Anderson, R.P.; Therrien, A.; Kelly, C.P.; Verdu, E.F. Society for the Study of Celiac Disease position statement on gaps and opportunities in coeliac disease. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Fasano, A. Celiac disease diagnosis: Simple rules are better than complicated algorithms. Am. J. Med. 2010, 123, 691–693. [Google Scholar] [CrossRef] [PubMed]
- Raiteri, A.; Granito, A.; Giamperoli, A.; Catenaro, T.; Negrini, G.; Tovoli, F. Current guidelines for the management of celiac disease: A systematic review with comparative analysis. World J. Gastroenterol. 2022, 28, 154–176. [Google Scholar] [CrossRef] [PubMed]
- Ciacci, C.; Bai, J.C.; Holmes, G.; Al-Toma, A.; Biagi, F.; Carroccio, A.; Ciccocioppo, R.; Di Sabatino, A.; Gingold-Belfer, R.; Jinga, M.; et al. Serum anti-tissue transglutaminase IgA and prediction of duodenal villous atrophy in adults with suspected coeliac disease without IgA deficiency (Bi.A.CeD): A multicentre, prospective cohort study. Lancet Gastroenterol. Hepatol. 2023, 8, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsson, J.F.; Bai, J.C.; Biagi, F.; Card, T.R.; Ciacci, C.; Ciclitira, P.J.; Green, P.H.R.; Hadjivassiliou, M.; Holdoway, A.; van Heel, D.A.; et al. Diagnosis and management of adult coeliac disease: Guidelines from the British Society of Gastroenterology. Gut 2014, 63, 1210–1228. [Google Scholar] [CrossRef]
- Owais, M.; Arsalan, M.; Choi, J.; Mahmood, T.; Park, K.R. Artificial Intelligence-Based Classification of Multiple Gastrointestinal Diseases Using Endoscopy Videos for Clinical Diagnosis. J. Clin. Med. 2019, 8, 986. [Google Scholar] [CrossRef]
- Leśniewska, M.; Patryn, R.; Kopystecka, A.; Kozioł, I.; Budzyńska, J. Third Eye? The Assistance of Artificial Intelligence (AI) in the Endoscopy of Gastrointestinal Neoplasms. J. Clin. Med. 2023, 12, 6721. [Google Scholar] [CrossRef]
- Sadoughi, F.; Ghaderzadeh, M. A hybrid particle swarm and neural network approach for detection of prostate cancer from benign hyperplasia of prostate. Stud. Health Technol. Inform. 2014, 205, 481–485. [Google Scholar]
- Ghaderzadeh, M. Clinical decision support system for early detection of prostate cancer from benign hyperplasia of prostate. Stud. Health Technol. Inform. 2013, 192, 928. [Google Scholar]
- Madhu, G.; Mohamed, A.W.; Kautish, S.; Shah, M.A.; Ali, I. Intelligent diagnostic model for malaria parasite detection and classification using imperative inception-based capsule neural networks. Sci. Rep. 2023, 13, 13377. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Eshraghi, M.A.; Taami, T.; Sadeghsalehi, H.; Hoseinzadeh, Z.; Ghaderzadeh, M.; Rafiee, M. A mobile application based on efficient lightweight CNN model for classification of B-ALL cancer from non-cancerous cells: A design and implementation study. Inform. Med. Unlocked 2023, 39, 101244. [Google Scholar] [CrossRef]
- Wang, X.; Qian, H.; Ciaccio, E.J.; Lewis, S.K.; Bhagat, G.; Green, P.H.; Xu, S.; Huang, L.; Gao, R.; Liu, Y. Celiac disease diagnosis from videocapsule endoscopy images with residual learning and deep feature extraction. Comput. Methods Programs Biomed. 2020, 187, 105236. [Google Scholar] [CrossRef]
- Zhou, T.; Han, G.; Li, B.N.; Lin, Z.; Ciaccio, E.J.; Green, P.H.; Qin, J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput. Biol. Med. 2017, 85, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Scheppach, M.W.; Rauber, D.; Stallhofer, J.; Muzalyova, A.; Otten, V.; Manzeneder, C.; Schwamberger, T.; Wanzl, J.; Schlottmann, J.; Tadic, V.; et al. Detection of duodenal villous atrophy on endoscopic images using a deep learning algorithm. Gastrointest. Endosc. 2023, 97, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, G.; Vécsei, A.; Häfner, M.; Uhl, A. Fisher encoding of convolutional neural network features for endoscopic image classification. J. Med. Imaging 2018, 5, 034504. [Google Scholar] [CrossRef] [PubMed]
- Vicnesh, J.; Wei, J.K.E.; Ciaccio, E.J.; Oh, S.L.; Bhagat, G.; Lewis, S.K.; Green, P.H.; Acharya, U.R. Automated diagnosis of celiac disease by video capsule endoscopy using DAISY Descriptors. J. Med. Syst. 2019, 43, 157. [Google Scholar] [CrossRef]
- Zammit, S.C.; McAlindon, M.E.; Greenblatt, E.; Maker, M.; Siegelman, J.; Leffler, D.A.; Yardibi, O.; Raunig, D.; Brown, T.; Sidhu, R. Quantification of Celiac Disease Severity Using Video Capsule Endoscopy: A Comparison of Human Experts and Machine Learning Algorithms. Curr. Med. Imaging 2023, 19, 1455–1662. [Google Scholar]
- Molder, A.; Balaban, D.V.; Molder, C.-C.; Jinga, M.; Robin, A. Computer-Based Diagnosis of Celiac Disease by Quantitative Processing of Duodenal Endoscopy Images. Diagnostics 2023, 13, 2780. [Google Scholar] [CrossRef]
- Saken, M.; Banzragch Yagci, M.; Yumusak, N. Impact of image segmentation techniques on celiac disease classification using scale invariant texture descriptors for standard flexible endoscopic systems. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 598–615. [Google Scholar] [CrossRef]
- Rubio-Tapia, A.; Ludvigsson, J.F.; Brantner, T.L.; Murray, J.A.; Everhart, J.E. The Prevalence of Celiac Disease in the United States. Am. J. Gastroenterol. 2012, 107, 1538–1544. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Mulak, A.; Jasińska, M.; Paradowski, L. Diagnostic challenges in celiac disease. Adv. Clin. Exp. Med. 2017, 26, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Biagi, F.; Rondonotti, E.; Campanella, J.; Villa, F.; Bianchi, P.I.; Klersy, C.; De Franchis, R.; Corazza, G.R. Video capsule endoscopy and histology for small-bowel mucosa evaluation: A comparison performed by blinded observers. Clin. Gastroenterol. Hepatol. 2006, 4, 998–1003. [Google Scholar] [CrossRef] [PubMed]
- Petroniene, R.; Dubcenco, E.; Baker, J.P.; Ottaway, C.A.; Tang, S.-J.; Zanati, S.; Streutker, C.J.; Gardiner, G.W.; Et Walrren, R.; Jeejeebhoy, K.N. Given capsule endoscopy in celiac disease: Evaluation of diagnostic accuracy and interobserver agreement. Am. J. Gastroenterol. 2005, 100, 685–694. [Google Scholar] [CrossRef]
| No. | Author, Year, Ref | Main Objective | Data Type | Methodology | Validation Type | Sample Size | Main Results |
|---|---|---|---|---|---|---|---|
| 1 | Vicnesh et al., 2019 [17] | Develop CAD for coeliac from capsule images | Endoscopy Images | DAISY descriptors, Shannon entropy, PSO | 10-fold cross-validation | Total number of video clips: 52 with coeliac disease (CD), 55 healthy. Total number of images used for analysis: 2140 (1100 healthy mucosa, 1040 damaged mucosa). | 89.82% accuracy, 89.17% PPV, 94.35% sensitivity, 83.20% specificity. |
| 2 | ChetcutiZammit et al., 2023 [18] | Compare CD severity assessment with VCE by expert human readers and an MLA. | VCE Images | Scoring by humans and MLA, 4-point scale | Primary validation with 36 VCE test set, ensembled prediction across orientations, and curve fit post-processing. | Total patients: 34 (18 with coeliac disease, 16 controls) Total images: 66 from coeliac disease patients, 16 from controls | Inter-reader agreement on coeliac villous damage, alpha = 0.924. Excellent MLA agreement with expert readers. |
| 3 | Wang et al., 2020 [13] | Develop a deep-learning module to diagnose coeliac disease from VCE images. | VCE Images | Recalibration in ResNet50, Inception-v3 | 10-fold cross-validation | Total participants: 37 (21 with coeliac disease, 16 healthy individuals) | 95.94% accuracy, 97.20% sensitivity, 95.63% specificity. |
| 4 | Molder et al., 2023 [19] | Automated detection of endoscopic markers during routine endoscopy examinations for COELIAC DISEASE diagnosis. | Endoscopy Images | ML, DL on images, histology reference | train–validation split | Total patients: 505 (182 with villous atrophy, 323 controls) Total images: 1704 (858 from patients with villous atrophy, 846 from controls) | Layered CNN best performance: 99.67% sensitivity and 98.07% PPV. |
| 5 | Scheppach et al., 2023 [15] | Apply AI for the macroscopic detection of VA during EGD to improve diagnostic performance. | Endoscopic Images | Trained ResNet18 on 858 images | 5-fold cross-validation | Total patients: 87 (11 with coeliac disease, 76 controls) Total images: 330 | AI algorithm: 90% sensitivity, 76% specificity, and 84% accuracy. Outperformed endoscopy fellows and experts. |
| 6 | Saken et al., 2021 [20] | Enhance the diagnostic accuracy of COELIAC DISEASE using CAD systems in endoscopy. | Endoscopy Images | Hybrid ML, multilevel thresholding, DWT | 10-fold cross-validation | Total patients: 353 Total images: 1661 (986 healthy mucosa, 675 affected by coeliac disease) | 94.79% accuracy, 94.29% sensitivity, 95.08% specificity. |
| 7 | Wimmer et al., 2018 [16] | Automate the diagnosis of CD and CP using Fisher encoding of [14,16] activations. | Endoscopy Images | Fisher on CNN activations, SVMs | 5-fold cross-validation | Total patients: 63 with biopsy-confirmed coeliac disease | Approach outperformed other CNN- and non-CNN-based approaches and required no training on the target dataset. |
| 8 | Zhou et al., 2017 [14] | Establish a DCNN for quantitative assessment of pathology in the small intestine from videocapsule endoscopy. | VCE Clips | GoogLeNet on preprocessed clips | 7-fold cross-validation | Total participants: 21 (11 with coeliac disease, 10 controls) | 100% sensitivity and specificity for the testing set. Evaluation confidence may relate to the severity level of small bowel mucosal lesions. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharif, K.; David, P.; Omar, M.; Sharif, Y.; Patt, Y.S.; Klang, E.; Lahat, A. Deep Learning in Coeliac Disease: A Systematic Review on Novel Diagnostic Approaches to Disease Diagnosis. J. Clin. Med. 2023, 12, 7386. https://doi.org/10.3390/jcm12237386
Sharif K, David P, Omar M, Sharif Y, Patt YS, Klang E, Lahat A. Deep Learning in Coeliac Disease: A Systematic Review on Novel Diagnostic Approaches to Disease Diagnosis. Journal of Clinical Medicine. 2023; 12(23):7386. https://doi.org/10.3390/jcm12237386
Chicago/Turabian StyleSharif, Kassem, Paula David, Mahmud Omar, Yousra Sharif, Yonatan Shneor Patt, Eyal Klang, and Adi Lahat. 2023. "Deep Learning in Coeliac Disease: A Systematic Review on Novel Diagnostic Approaches to Disease Diagnosis" Journal of Clinical Medicine 12, no. 23: 7386. https://doi.org/10.3390/jcm12237386
APA StyleSharif, K., David, P., Omar, M., Sharif, Y., Patt, Y. S., Klang, E., & Lahat, A. (2023). Deep Learning in Coeliac Disease: A Systematic Review on Novel Diagnostic Approaches to Disease Diagnosis. Journal of Clinical Medicine, 12(23), 7386. https://doi.org/10.3390/jcm12237386







