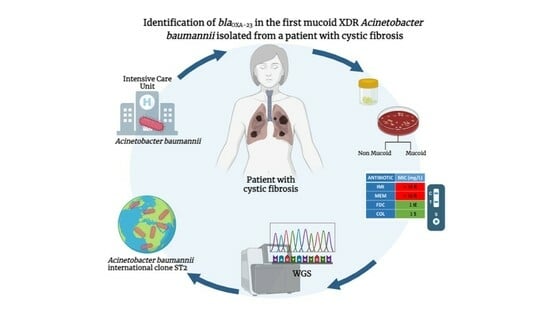
Identification of the blaOXA-23 Gene in the First Mucoid XDR Acinetobacter baumannii Isolated from a Patient with Cystic Fibrosis
Abstract
1. Introduction
2. Case Description
3. Microbiological Investigation
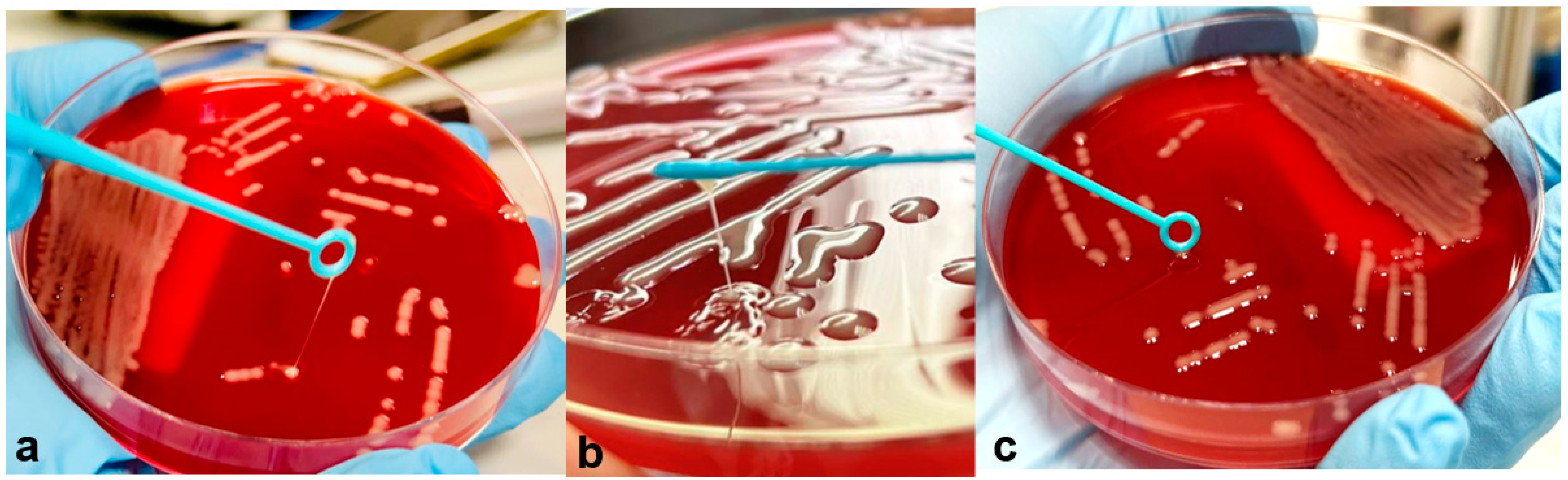
3.1. Isolates and Antimicrobial Susceptibility Testing
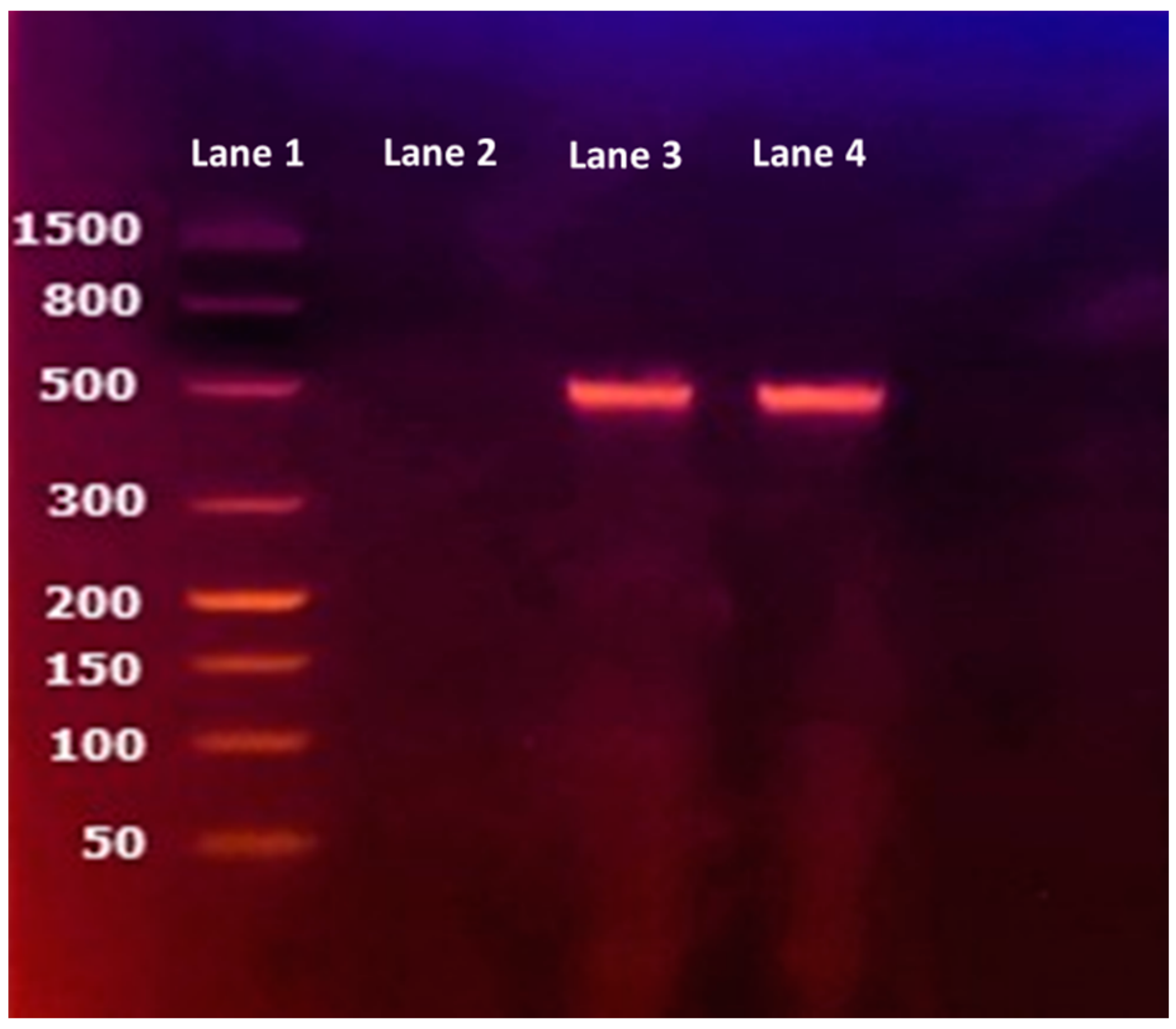
3.2. Carbapenemase Detection
3.3. Molecular Typing
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nowak, J.; Zander, E.; Stefanik, D.; Higgins, P.G.; Roca, I.; Vila, J.; McConnell, M.J.; Cisneros, J.M.; Seifert, H.; Garnacho-Montero, J.; et al. High Incidence of Pandrug-Resistant Acinetobacter baumannii Isolates Collected from Patients with Ventilator-Associated Pneumonia in Greece, Italy and Spain as Part of the MagicBullet Clinical Trial. J. Antimicrob. Chemother. 2017, 72, 3277–3282. [Google Scholar] [CrossRef] [PubMed]
- Mentasti, M.; Prime, K.; Sands, K.; Khan, S.; Wootton, M. Rapid Detection of OXA-23-like, OXA-24-like, and OXA-58-like Carbapenemases from Acinetobacter Species by Real-Time PCR. J. Hosp. Infect. 2020, 105, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Moubareck, C.A.; Halat, D.H. Insights into Acinetobacter baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Zhou, Z.; Yang, Q.; Shi, Q.; Xu, Q.; Wang, J.; Shi, K.; Zhao, F.; Sun, L.; Ruan, Z.; et al. Evolution of Acinetobacter baumannii in Vivo: International Clone II, More Resistance to Ceftazidime, Mutation In Ptk. Front. Microbiol. 2017, 8, 1256. [Google Scholar] [CrossRef] [PubMed]
- Cherubini, S.; Perilli, M.; Segatore, B.; Fazii, P.; Parruti, G.; Frattari, A.; Amicosante, G.; Piccirilli, A. Whole-Genome Sequencing of ST2 A. baumannii Causing Bloodstream Infections in COVID-19 Patients. Antibiotics 2022, 11, 955. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Zhao, Q.; Wu, Y.; Zhou, H.; Ding, S.; Zhu, K. Mucoid Acinetobacter baumannii Enhances Anti-Phagocytosis through Reducing C3b Deposition. Front. Med. 2022, 9, 879361. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Zhang, H.; Kan, J.; Yin, M.; Zhang, J.; Wan, L.; Chang, R.; Li, M. Acquired Mucoid Phenotype of Acinetobacter baumannii: Impact for the Molecular Characteristics and Virulence. Microbiol. Res. 2021, 246, 126702. [Google Scholar] [CrossRef] [PubMed]
- Nawfal Dagher, T.; Al-Bayssari, C.; Chabou, S.; Antar, N.; Diene, S.M.; Azar, E.; Rolain, J.M. Investigation of Multidrug-Resistant ST2 Acinetobacter baumannii Isolated from Saint George Hospital in Lebanon. BMC Microbiol. 2019, 19, 29. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, L.L.; Botelho, L.A.B.; Barbosa, L.C.; Mattos, C.S.; Carballido, J.M.; de Castro, C.L.T.; Mondino, P.J.J.; de Paula, G.R.; de Mondino, S.S.B.; de Mendonça-Souza, C.R.V. Detection of BlaOXA-23 in Acinetobacter spp. Isolated from Patients of a University Hospital. Braz. J. Infect. Dis. 2012, 16, 521–526. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rocha, G.A.; Lima, D.F.; Rodrigues, E.R.; Leão, R.S.; Folescu, T.W.; Firmida, M.C.; Cohen, R.W.F.; Albano, R.M.; Marques, E.A. Species Distribution, Sequence Types and Antimicrobial Resistance of Acinetobacter spp. from Cystic Fibrosis Patients. Epidemiol. Infect. 2018, 146, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Nuñez, M.; Garcia-Gonzalez, M.; Pomares, X.; Montón, C.; Millares, L.; Quero, S.; Prina, E.; Asensio, O.; Bosque, M.; Capilla, S.; et al. The Respiratory Microbiome in Cystic Fibrosis: Compartment Patterns and Clinical Relationships in Early Stage Disease. Front. Microbiol. 2020, 11, 1463. [Google Scholar] [CrossRef] [PubMed]
- Liebchen, U.; Paal, M.; Jung, J.; Schroeder, I.; Frey, L.; Zoller, M.; Scharf, C. Therapeutic Drug Monitoring-Guided High Dose Meropenem Therapy of a Multidrug Resistant Acinetobacter baumannii—A Case Report. Respir. Med. Case Rep. 2020, 29, 100966. [Google Scholar] [CrossRef] [PubMed]
- SIFC-Società Italiana per lo Studio della Fibrosi Cistica. Available online: https://www.sifc.it/documento/aggiornamento-raccomandazioni-per-lesecuzione-delle-indagini-microbiologiche-di-campioni-delle-vie-respiratorie-di-pazienti-con-fibrosi-cistica/ (accessed on 30 May 2023).
- EUCAST-European Committee on Antimicrobial Susceptibility Testing. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 30 May 2023).
- King, C.S.; Brown, A.W.; Aryal, S.; Ahmad, K.; Donaldson, S. Critical Care of the Adult Patient with Cystic Fibrosis. Chest 2019, 155, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Sood, N.; Paradowski, L.J.; Yankaskas, J.R. Outcomes of Intensive Care Unit Care in Adults with Cystic Fibrosis. Am. J. Respir. Crit. Care Med. 2001, 163, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Worlitzsch, D.; Tarran, R.; Ulrich, M.; Schwab, U.; Cekici, A.; Meyer, K.C.; Birrer, P.; Bellon, G.; Berger, J.; Weiss, T.; et al. Effects of Reduced Mucus Oxygen Concentration in Airway Pseudomonas Infections of Cystic Fibrosis Patients. J. Clin. Investig. 2002, 109, 317–325. [Google Scholar] [CrossRef]
- Karakonstantis, S.; Saridakis, I. Colistin Heteroresistance in Acinetobacter spp.: Systematic Review and Meta-Analysis of the Prevalence and Discussion of the Mechanisms and Potential Therapeutic Implications. Int. J. Antimicrob. Agents 2020, 56, 106065. [Google Scholar] [CrossRef] [PubMed]
- Saiman, L.; Siegel, J. Infection Control Recommendations for Patients with Cystic Fibrosis: Microbiology, Important Pathogens, and Infection Control Practices to Prevent Patient-to-Patient Transmission. Am. J. Infect. Control 2003, 31, S1–S62. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Rousaki, M.; Kritsotakis, E.I. Cefiderocol: Systematic Review of Mechanisms of Resistance, Heteroresistance and In Vivo Emergence of Resistance. Antibiotics 2022, 11, 723. [Google Scholar] [CrossRef] [PubMed]
- Karami-Zarandi, M.; Douraghi, M.; Vaziri, B.; Adibhesami, H.; Rahbar, M.; Yaseri, M. Variable Spontaneous Mutation Rate in Clinical Strains of Multidrug-Resistant Acinetobacter baumannii and Differentially Expressed Proteins in a Hypermutator Strain. Mutat. Res. 2017, 800–802, 37–45. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Antimicrobial Resistance Surveillance in Europe 2022–2020 Data; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
| Antimicrobial MIC (µg/mL) and Interpretation for: | ||||
|---|---|---|---|---|
| Antimicrobial Agent | A. baumannii HM | A. baumannii LM | ||
| MIC | INT | MIC | INT | |
| Ceftazidime/avibactam | >16 | - | >16 | - |
| Ceftolozane/tazobactam | 32 | - | 32 | - |
| Ciprofloxacin | ≥4 | R | ≥4 | R |
| Colistin | 1 | S | 0.5 | S |
| Imipenem | ≥16 | R | ≥16 | R |
| Meropenem | ≥16 | R | ≥16 | R |
| Piperacillin/tazobactam | ≥32 | IE | ≥32 | IE |
| Cefotaxime | ≥8 | - | ≥8 | - |
| Ceftazidime | ≥16 | - | ≥16 | - |
| Trimethopim/sulfametoxazole | ≥320 | R | ≥320 | R |
| Cefiderocol | 1 | IE | 0.5 | IE |
| Tigecycline | 1 | IE | 1 | IE |
| Amikacin a | ≥32 | NA | ≥32 | NA |
| Gentamicin a | ≥8 | NA | ≥8 | NA |
| Tobramycin a | ≥8 | NA | ≥8 | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossitto, M.; Vrenna, G.; Tuccio Guarna Assanti, V.; Essa, N.; De Santis, M.L.; Granaglia, A.; Fini, V.; Costabile, V.; Onori, M.; Cristiani, L.; et al. Identification of the blaOXA-23 Gene in the First Mucoid XDR Acinetobacter baumannii Isolated from a Patient with Cystic Fibrosis. J. Clin. Med. 2023, 12, 6582. https://doi.org/10.3390/jcm12206582
Rossitto M, Vrenna G, Tuccio Guarna Assanti V, Essa N, De Santis ML, Granaglia A, Fini V, Costabile V, Onori M, Cristiani L, et al. Identification of the blaOXA-23 Gene in the First Mucoid XDR Acinetobacter baumannii Isolated from a Patient with Cystic Fibrosis. Journal of Clinical Medicine. 2023; 12(20):6582. https://doi.org/10.3390/jcm12206582
Chicago/Turabian StyleRossitto, Martina, Gianluca Vrenna, Vanessa Tuccio Guarna Assanti, Nour Essa, Maria Luisa De Santis, Annarita Granaglia, Vanessa Fini, Valentino Costabile, Manuela Onori, Luca Cristiani, and et al. 2023. "Identification of the blaOXA-23 Gene in the First Mucoid XDR Acinetobacter baumannii Isolated from a Patient with Cystic Fibrosis" Journal of Clinical Medicine 12, no. 20: 6582. https://doi.org/10.3390/jcm12206582
APA StyleRossitto, M., Vrenna, G., Tuccio Guarna Assanti, V., Essa, N., De Santis, M. L., Granaglia, A., Fini, V., Costabile, V., Onori, M., Cristiani, L., Boni, A., Cutrera, R., Perno, C. F., & Bernaschi, P. (2023). Identification of the blaOXA-23 Gene in the First Mucoid XDR Acinetobacter baumannii Isolated from a Patient with Cystic Fibrosis. Journal of Clinical Medicine, 12(20), 6582. https://doi.org/10.3390/jcm12206582