Abstract
(1) Background: Circulating tumor DNA (ctDNA) has emerged as a promising biomarker for many kinds of tumors. However, whether ctDNA could be an accurate diagnostic biomarker in colorectal cancer (CRC) remains to be clarified. The aim of this study was to evaluate the diagnostic accuracy of ctDNA in CRC. (2) Methods: PubMed, Web of Science, and Cochrane databases were searched to identify studies reporting the use of ctDNA to screen and diagnose CRC, and all relevant studies published until October 2022 were enrolled for our analysis. These studies were divided into three primer subgroups: the subgroup of quantitative or qualitative analysis of ctDNA and the subgroup of septin9 (SEPT9) methylation assay. (3) Results: A total of 79 qualified articles with 25,240 subjects were incorporated into our meta-analysis. For quantitative studies, the combined sensitivity (SEN), specificity (SPE), and diagnostic odds ratio (DOR) were 0.723 (95% CI: 0.623–0.803), 0.920 (95% CI: 0.827–0.966), and 23.305 (95% CI: 9.378–57.906), respectively, yielding an AUC of 0.860. The corresponding values for qualitative studies were 0.610 (95% CI: 0.566–0.651), 0.891 (95% CI: 0.878–0.909), 12.569 (95% CI: 9.969–15.848), and 0.823, respectively. Detection of SEPT9 methylation depicted an AUC of 0.879, with an SEN of 0.679 (95% CI: 0.622–0.732), an SPE of 0.903 (95% CI: 0.878–0.923), and a DOR of 20.121 (95% CI:14.404–28.106), respectively. (4) Conclusion: Blood-based ctDNA assay would be a potential novel biomarker for CRC screening and diagnosis. Specifically, quantitative analysis of ctDNA or qualitative analysis of SEPT9 methylation exhibited satisfying diagnostic efficiency. Larger sample studies are needed to further confirm our conclusions and to make the ctDNA approach more sensitive and specific.
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second most common cause of cancer death worldwide, which accounts for nearly 10% of cancer cases and 9% of cancer deaths [1]. Even though there have been dramatic advances in understanding the molecular mechanism and clinical treatment over decades, helping to double the overall survival of advanced CRC to three years, there was estimated over 0.9 million CRC-related death in 2020 [2]. Since there are significant differences in prognosis and therapy response between different clinical stages in CRC patients, it is extremely important to diagnose and treat CRC at an early stage [3]. However, most CRC patients are asymptomatic at an early stage. When common symptoms such as constipation, bleeding, anemia, or abdominal pain are noted, CRC may already develop to advanced stages [4]. Currently, several methods, including fecal occult blood test (FOBT), fecal immunochemical test (FIT), and colonoscopy, have been applied in CRC screening, which significantly reduced CRC mortality [5]. However, there are some defects in the current screening method. The sensitivity (SEN) and specificity (SPE) of FOBT and FIT for early CRC are unsatisfactory [6,7]. Additionally, the FIT demonstrated poorer compliance in CRC monitoring compared to other simple methods, such as blood-based tests [8,9]. Although colonoscopy has been regarded as the gold standard for CRC screening, it has low acceptance by general people due to the invasive procedure, troublesome diet restriction, and bowel preparation [10,11]. Moreover, a colonoscopy might not reach the entire population in developing countries as it requires special instruments and qualified endoscopists to which they have limited access. Therefore, a simple, noninvasive, and cost-efficienct approach is urgently needed to screen and diagnose CRC.
Since the detection of higher levels of circulating free DNA (cfDNA), which consists of fragments of extracellular, in the blood of patients with various types of cancer [12], accumulating evidence has demonstrated that cfDNA could serve as a promising biomarker in multiple kinds of disease [13]. While most cfDNA originates from the hematopoietic system in a healthy person [14], apoptotic or necrotic cancer cells release DNA called circulating tumor DNA (ctDNA) into the biological fluid in cancer as a small proportion of cfDNA in cancer patients [15,16]. Although ctDNA may only account for less than 1% of cfDNA [17], ctDNA has been extensively explored with improving assay techniques. The analysis of ctDNA has been regarded as a promising tool in oncology as it can be detected by noninvasive methods and provide cancer-specific information, including nucleotide mutation [18], copy number aberration (CNA) [19], DNA methylation [20], and microsatellite instability (MSI) [21].
Alteration of ctDNA has been detected in CRC patients by the quantitative method, which measures the quantity of ctDNA [22], and the qualitative method, by reporting cancer-specific changes, including DNA methylation, can also detect alteration of ctDNA in CRC patients [23]. Although accumulating evidence indicated that ctDNA is a promising biomarker in CRC screening and diagnosis, there are some concerns about the application of ctDNA in CRC diagnosis due to the tumor heterogeneous, unstandardized protocol and concentration threshold [24]. Therefore, it is necessary to conduct a comprehensive analysis and evaluation of the diagnostic values of ctDNA in CRC before large-scale clinical application. Thus, we implemented a meta-analysis to assess the diagnostic performance of ctDNA assays for CRC, which could potentially contribute to the technology development and modify the clinical decision.
2. Materials and Methods
2.1. Protocol and Registration
This systematic review and meta-analysis were performed according to the Preferred Items for Systematic Review and Meta-Analysis (PRISMA) [25] and Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group [26]. The study was also registered at the International Prospective Register of Systematic Reviews (PROSPERO) database (Number: CRD42022370118).
2.2. Search Strategy
PubMed, Web of Science and The Cochrane Library were searched with query term: “(circulating tumor DNA OR circulating DNA OR ctDNA OR Cell Free Tumor DNA OR Plasma DNA OR Serum DNA) AND (Colonic Neoplasm OR Colon Neoplasm OR Colorectal cancer OR Colorectal Carcinoma OR Colonic Carcinoma OR Colonic Cancer OR Colon Cancer OR Colonic tumor OR Colon tumor) AND (Diagnosis OR Detection OR Screen OR Sensitivity OR Specificity)”. The language was limited to English. All potentially relevant articles published until October 2022 were retrieved and independently queried by two authors. We also manually screened the reference from the included studies and relevant reviews for enlarged retrieval.
2.3. Inclusion and Exclusion Criteria
The articles that conformed to the following criteria were included: (a) ctDNA was used for first-diagnosed CRC patients rather than the recurrent patients; (b) the numerical value of SEN and SPE of each detection method could be calculated in each study; (c) specimens were extracted from peripheral blood. The exclusion criteria were as follows: (a) review, case report or case series, comments, letter, or conference abstract; (b) sample size of studies was less than 10; (c) duplicated or overlapping publications that utilized the same patient specimens and identical gene modification; (d) publications with no full-text available. Two authors independently evaluated the eligibility of all publications. A third author would assess studies when there was a discrepancy.
2.4. Data Extraction
Data from eligible publications were extracted and compiled by two independent authors. The information extracted from included studies was as follows: the first author’s name, publication year, region/country, study design, sample size, control type, source of specimens, sampling time, detection method, reference gene, cut-off values, diagnostic performance (including SEN and SPE), true positive (TP), true negative (TN), false positive (FP), false negative (FN), positive likelihood ratio (PLR) and negative likelihood ratio (NLR), and diagnostic concordance.
2.5. Quality Assessment
The quality of all included studies was evaluated by two independent authors according to Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2), which assesses the studies on four key domains, including patient selection, index test, reference standard, and flow timing [27]. Two authors independently labeled “low risk”, “some concerns”, and “high risk” for each domain in each study, and divergence was discussed until an agreement was reached. Publications would be excluded if they were regarded as poor quality by both authors.
2.6. Statistical Analysis
We utilized Revman Manager 5.4 and R program (R Foundation, Vienna, Austria, version 4.2.1) with “meta” [28], “mada” [29], and “metafor” [30] packages to conduct this diagnostic meta-analysis. The pooled sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), positive likelihood ratio (NLR), diagnostic odds ratio (DOR), and corresponding 95% confidence interval (95% CI) were calculated as an indicator of test performance [31]. The summary receiver operating characteristic (SROC) curve and corresponding area under the curve (AUC) were formulated to assess the overall test accuracy [32,33]. Generally, the AUC ranges of 0.7–0.8, 0.8–0.9, and 0.9–1.0 were interpreted as acceptable, excellent, and outstanding accuracy, respectively [34,35]. The Spearman correlation coefficient and corresponding p-value were used to identify the presence of the threshold effect [36]. Heterogeneity among the studies was evaluated by chi-square and I2 tests. p < 0.10 or I2 > 50% indicated a significant heterogeneity, and the random effect model should be used [37]. Subgroup analysis and meta-regression were conducted to further reveal the source of heterogeneity [38]. Potential publication bias was checked by Deek’s funnel plot asymmetry test [39].
3. Results
3.1. Studies Characteristics
Figure 1 shows a PRISMA flow diagram depicting the retrieval strategy of databases to incorporate qualified studies. A total of 7212 publications were queried based on our search strategy at first, and 79 eligible articles published from 2008 to 2022 were incorporated into our meta-analysis after examination of the abstract and comprehension of the full text (Table S1). All included studies consisted of quantitative analysis of ctDNA concentration (n = 11) [40,41,42,43,44,45,46,47,48,49,50] and qualitative analysis of tumor-specific gene methylation in ctDNA (n = 67) [51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118] as well as both quantitative and qualitative analysis (n = 1) [119]. Among these publications, 34 articles [51,59,60,62,63,64,65,66,67,68,70,71,72,74,75,78,79,81,82,83,90,91,96,100,101,102,104,105,107,108,110,112,115,116] evaluated the diagnostic efficiency of septin9 (SEPT9) methylation for CRC.
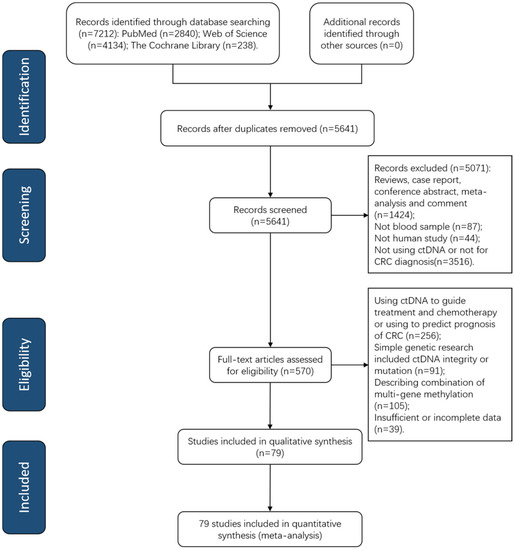
Figure 1.
A PRISMA flow diagram of paper search. CRC, colorectal cancer; ctDNA, circulating tumor DNA; PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analysis.
In our meta-analysis, a total population of 8076 CRC patients and 17164 control subjects were included. The primary characteristics of all studies are summarized in Table 1. The majority of participants were from the Asia area (n= 13,841). There were 18 prospective studies and 8 retrospective studies, while 53 studies did not clarify the study design. Among 50 studies with known sample collecting time, all samples were collected before surgery or treatment. While the majority of studies collected plasma for ctDNA analysis (n = 55), serum specimens were also analyzed in some research (n = 24). The methods applied to measure the concentration of ctDNA include real-time quantitative polymerase chain reaction (RT-qPCR) (n = 8), droplet digital polymerase chain reaction (ddPCR) (n = 2), branched DNA assay (bDNA) (n = 1) and florescent dye assay (n = 1). For qualitative analysis of ctDNA, methylation-specific polymerase chain reaction (MSP) was the most common method applied (n = 64).

Table 1.
Summary of diagnostic accuracy of ctDNA assay for CRC in multiple subgroups.
3.2. Quality Assessment
The quality assessment was based on QUADAS-2, and the outcome of eligible studies is depicted in Figure 2 and Figure S1. The majority of incorporated studies possessed a moderate–high quality, indicating that the overall quality of the included studies was highly acceptable. However, there were some concerns about the patient selection of 39 studies, such as whether the patient selection was consecutive or random in a specific time period. Moreover, predefined threshold information was absent in 22 studies, which potentially generated some concerns about the index test. All enrolled patients obtained definite pathological diagnoses.
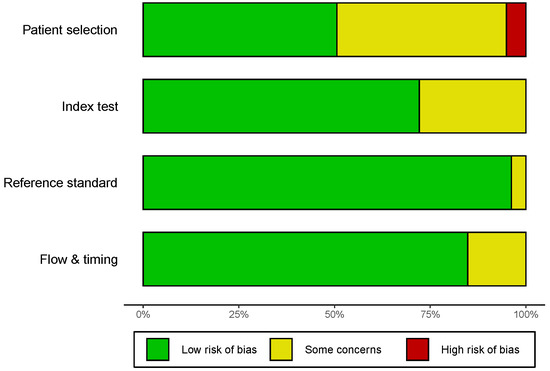
Figure 2.
Quality assessment of the included studies by the revised QUADAS-2. QUADAS-2, Quality Assessment of Diagnostic Accuracy Studies-2.
3.3. Diagnostic Value of Quantitative and Qualitative Analysis of ctDNA for CRC
The quantitative analysis group, including 12 studies, yields an SEN of 0.723 (95% CI: 0.623–0.803) and an SPE of 0.920 (95% CI: 0.827–0.966) (Figure 3a,b). The numeric value of PLR, NLR, and DOR were 6.820 (95% CI: 3.335–12.800), 0.326 (95% CI: 0.227–0.446), and 23.305 (95% CI: 9.380–57.906) respectively. The corresponding SROC yield an AUC of 0.860, indicating excellent accuracy in discriminating CRC patients from control subjects (Figure 4a). Notably, the significant heterogeneity among these quantitative studies was observed (SEN: I2 = 87.5%, p = 0.000; SPE: I2 = 77.0%, p = 0.000; DOR: I2 = 82.3%, p = 0.000). The Spearman correlation coefficient was 0.008 (p = 0.983), indicating that heterogeneity among quantitative studies was derived from other factors rather than the threshold effect.
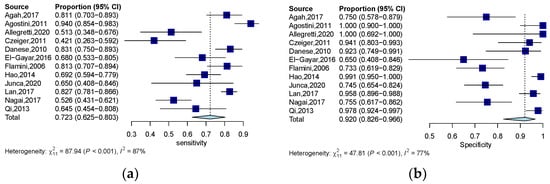
Figure 3.
Forest plots of SEN (a) and SPE (b) for the diagnostic value of ctDNA assay for CRC in the quantitative detection subgroup. 95% CI, 95% confidence interval; CRC, colorectal cancer; ctDNA, circulating tumor DNA; SEN, sensitivity; SPE, specificity; diamond shapes representing average effect; dark blue squares representing individual study results.
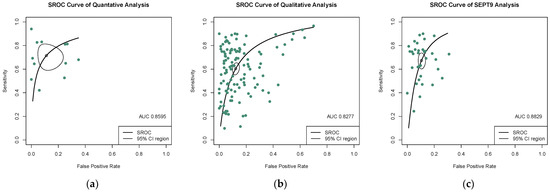
Figure 4.
SROC curves of diagnostic value for (a) the quantitative detection subgroup; (b) the qualitative detection subgroup; (c) the SEPT-9 methylation detection subgroup. 95% CI, 95% confidence interval; AUC, the area under the curve; SROC, summary receiver operating characteristic.
In the qualitative analysis group, an SEN of 0.609 (95% CI: 0.563–0.650), an SPE of 0.891 (95% CI: 0.864–0.910) (Figure 5a,b), and a DOR of 12.621 (95% CI: 10.004–15.922) were generated. The numeric values of PLR and NLR were 4.960 (95% CI: 4.190–5.860) and 0.453 (95% CI: 0.406–0.498). An AUC of 0.828 was calculated from the corresponding SROC, indicating excellent accuracy in discriminating CRC patients from control subjects (Figure 4b). Significant heterogeneity among these qualitative studies was also observed (SEN: I2 = 93.9%, p = 0.000; SPE: I2 = 93.5%, p = 0.000; DOR: I2 = 89.6%, p = 0.000). The Spearman correlation analysis was 0.149 (p = 0.118), depicting that heterogeneity among quantitative studies was derived from a non-threshold effect.
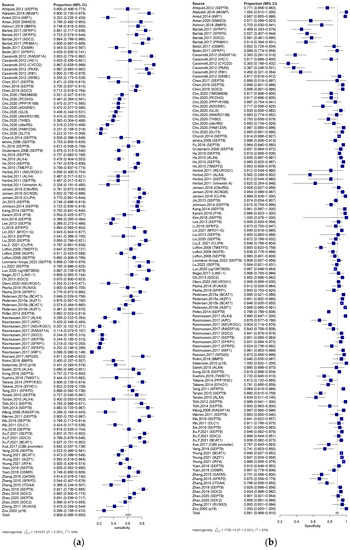
Figure 5.
Forest plots of SEN (a) and SPE (b) for diagnostic value of ctDNA assay for CRC in the qualitative detection subgroup. 95% CI, 95% confidence interval; CRC, colorectal cancer; ctDNA, circulating tumor DNA; SEN, sensitivity; SPE, specificity; diamond shapes representing average effect; darkblue squares representing individual study results.
Methylation of SEPT9 was the most frequently detected epigenetic change in the qualitative analysis of ctDNA in CRC. Therefore, we also analyzed the diagnostic accuracy of SEPT9 methylation in discriminating CRC subjects from control. In 34 studies that measured the methylation of SEPT9, the pooled SEN, SPE, and DOR were 0.679 (95% CI: 0.622–0.732), 0.904 (95% CI: 0.881–0.923) (Figure 6a,b), and 20.551 (95% CI: 14.684–28.760), respectively. The pooled PLR and NLR were 6.420 (95% CI: 5.110–7.970) and 0.367 (95% CI: 0.307–0.430). The corresponding SROC yield an AUC of 0.883, indicating excellent accuracy in discriminating CRC patients from control subjects (Figure 4c). Significant heterogeneity among these studies was also observed (SEN: I2 = 91.8%, p = 0.000; SPE: I2 = 89.8%, p = 0.000; DOR: I2 = 82.3%, p = 0.000). The result of Spearman correlation analysis was 0.02 (p = 0.993), indicating that heterogeneity among quantitative studies was derived from the non-threshold effect (Table 1).
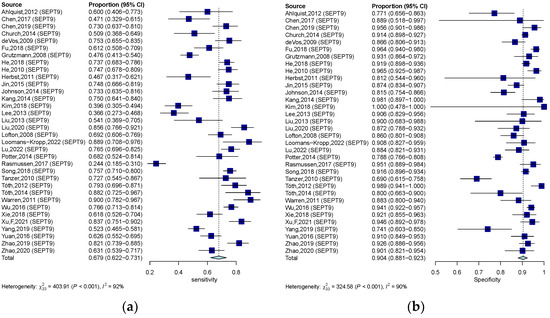
Figure 6.
Forest plots of SEN (a) and SPE (b) for diagnostic value of ctDNA assay for CRC in the SEPT-9 methylation detection subgroup. 95% CI, 95% confidence interval; CRC, colorectal cancer; ctDNA, circulating tumor DNA; SEN, sensitivity; SPE, specificity; diamond shapes representing average effect; dark blue squares representing individual study results.
3.4. Subgroup Analysis and Meta-Regression Analysis
Different covariates including region (Asia vs. non-Asia), sample size (≥100 vs. <100), control type (health control vs. non-CRC disease), sample source (plasma vs. serum), assay methods (RT-PCR vs. other methods in quantitative analysis; MSP vs. other methods in qualitative analysis), and methylation gene (SEPT9 vs. other genes in qualitative studies) were applied in the subgroup analysis (Table 2).

Table 2.
Subgroup analysis of diagnostic performance of ctDNA assay for CRC.
Subgroup analysis based on sample size revealed an improved diagnostic accuracy in quantitative studies with sample size ≥ 100 since the SEP, SEN, and AUC for quantitative studies with sample size ≥100 were 0.771 (95% CI: 0.681–0.842), 0.924 (95% CI: 0.815–0.971) and 0.878, whereas corresponding values were 0.545 (95% CI: 0.417–0.667), 0.904 (95% CI: 0.604–0.983), and 0.694 for quantitative studies with sample size < 100. Similarly, qualitative analysis of ctDNA with a large sample size also had a more robust diagnostic accuracy (SEN: 0.598 (95% CI: 0.552–0.643), SPE: 0.903 (95% CI: 0.884–0.919), AUC: 0.849) compared to qualitative studies with sample size < 100 (SEN: 0.662 (95% CI: 0.544–0.762), SPE: 0.819 (95% CI: 0.694–0.900), AUC: 0.759).
For the quantitative analysis of ctDNA, subgroup analysis associated with control type showed that the SEN, SPE, and AUC for quantitative analysis of ctDNA to discriminate CRC from healthy control were 0.723 (95% CI: 0.626–0.803), 0.927 (95% CI: 0.833–0.967) and 0.861, while the corresponding values for quantitative analysis of ctDNA to discriminate CRC form non-CRC disease were 0.647 (95% CI: 0.508–0.765), 0.878 (95% CI: 0.563–0.976), and 0.641, respectively. While subgroup analysis in qualitative studies depicted an SEN of 0.601 (95% CI: 0.554–0.647), an SPE of 0.915 (95% CI: 0.895–0.932), and an AUC of 0.844 in qualitative studies with healthy people as control, and an SEN of 0.672 (95% CI: 0.617–0.723), an SPE of 0.809 (95% CI: 0.737–0.865) and an AUC of 0.774 in qualitative studies used non-CRC disease as control. These results indicated that studies utilizing healthy people as control provided a more satisfactory diagnostic efficiency compared to studies that used non-CRC patients as a control in both quantitative and qualitative analysis of ctDNA, which highlighted a more robust capability of ctDNA assay to distinguish CRC from healthy individuals than from non-CRC disease patients. Multiple levels of meta-regression also revealed that the parameter “control type” could contribute to the heterogeneity (Table 3). Moreover, heterogeneity was also derived from the parameter “methylation gene” as the corresponding coefficient was 0.661 (p = 0.01), which was consistent with the primary outcome of a clinical trial of SEPT9 methylation for CRC detection, indicating SEPT9 as a promising CRC diagnostic biomarker.

Table 3.
Meta-regression of impacts of study features on diagnostic value of ctDNA for CRC.
3.5. Publication Bias
Deek’s funnel plot asymmetry test [39] was used to check the potential publication bias, and a p-value less than 0.05 indicated significance. No significant publication bias was observed in the quantitative analysis of ctDNA (Figure 7a, p = 0.307). Nevertheless, our results revealed a significant publication bias in the qualitative analysis of ctDNA (Figure 7b, p = 0.000). In addition, among these qualitative analysis studies, our analysis demonstrated that no significant publication bias existed in the SEPT9 methylation group (Figure 7c, p = 0.731), while a significant publication bias was found in the non-SEPT9 methylation group (Figure 7d, p = 0.000).
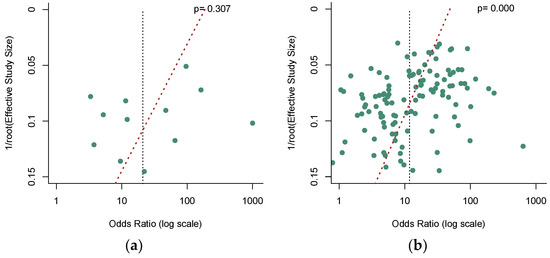
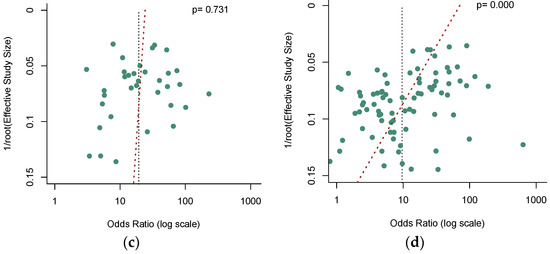
Figure 7.
Funnel plots to evaluate the publication bias for (a) the quantitative detection subgroup; (b) the qualitative detection subgroup; (c) the SEPT-9 methylation detection subgroup; (d) the non-SEPT-9 methylation detection subgroup.
4. Discussion
CRC is one of the most common malignancies with poor prognosis and high mortality, which is highly correlated with the low early diagnostic rate [1]. When detected at an early stage, the survival rate of CRC would be over 90%, compared to approximately 7% for late-stage disease [120]. Although colonoscopy has demonstrated perfect diagnostic efficiency, it has low acceptance by general people due to the invasive procedure, troublesome diet restriction, and bowel preparation [10,11]. Thus, novel noninvasive diagnostic methods with robust diagnostic efficiency are in urgent need. Advance in precise ctDNA detection has made it possible for “liquid biopsy” of ctDNA to become a potential diagnostic biomarker of CRC [121]. In this diagnostic meta-analysis, we aimed to incorporate the diagnostic accuracy of ctDNA for CRC.
In our meta-analysis, quantitative analysis of ctDNA exhibited a higher SEN (0.723 vs. 0.609) and a comparable SPE (0.920 vs. 0.891) compared to qualitative analysis, which is consistent with a higher AUC (0.860 vs. 0.828). This might be explained by the fact that some genetic loci selected for the test were not commonly expressed in CRC, which might generate a false negative in the test. Various septins have been associated with tumorigenesis, and SEPT9 hypermethylation has been detected in CRC [122]. Furthermore, the detection of SEPT9 methylation in cfDNA has been regarded as a potential biomarker for CRC diagnosis and has become the first blood-based CRC screening test approved by the FDA [123]. When we looked into the SEPT9 methylation analysis among the qualitative analysis, the corresponding AUC was 0.883, relatively higher than the AUC of the quantitative analysis, with the value of the SEN (0.679) and SPE (0.904) similar compared to the quantitative analysis. These results depicted the superiority of SEPT9 methylation detection over other gene locus analyses in CRC diagnosis.
DOR was also applied to evaluate the diagnostic accuracy, and the discriminatory test performance would be classified as satisfactory when the value of DOR was over 10 [31]. Both quantitative and qualitative analyses of ctDNA exhibited satisfactory discriminatory test performance as the value of DOR was 23.305 and 12.621, respectively. Specifically, SEPT9 methylation assay among qualitative analysis revealed a higher DOR of 20.551 than that of qualitative analysis of ctDNA, indicating a better discriminatory test performance of SEPT9 methylation assay than other single gene methylation assay in qualitative analysis.
In this study, the value of PLR of quantitative analysis and SEPT9 methylation were 6.820 and 6.420, respectively, indicating that CRC subjects have an approximately six to seven folds higher chance of being positive in quantitative analysis and SEPT9 methylation analysis of ctDNA. However, qualitative analysis of ctDNA depicted a PLR of 4.920, which is lower than the value of quantitative analysis and SEPT9 methylation. Moreover, the qualitative analysis of ctDNA yielded an NLR of 0.496, implying that the probability for cases with negative qualitative assay results to have CRC is 49.6%, whereas the value of NLR of quantitative analysis and SEPT9 methylation detection of ctDNA were 0.326 and 0.367, respectively. These results might be explained that some gene locus methylation might not exist in all CRC cases, and using single gene methylation as a biomarker might lead to false negatives. However, SEPT9 methylation demonstrated a comparable PLR and NLR with quantitative analysis of ctDNA, depicting the robust diagnostic performance of SEPT9 methylation assay. Additionally, the SEPT9 assay demonstrated better compliance than FIT and colonoscopy in CRC screening [8,9].
Publication bias was evaluated by Deek’s funnel plot asymmetry test, and no publication bias was revealed in the quantitative analysis of ctDNA (p = 0.307). However, it was highly possible that publication bias was presented in the qualitative analysis group (p = 0.000). As previous analysis demonstrated the disparity between the SEPT9 methylation assay and other gene methylation assays within the qualitative analysis, we further evaluated the publication bias based on gene loci. The Deek’s funnel plot asymmetry test revealed that publication biases were likely to exist in the non-SEPT9 methylation assay (p = 0.000) but not in the SEPT9 methylation assay (p = 0.731). Furthermore, a meta-regression analysis was adapted to explore the potential source of heterogeneity. In the quantitative analysis of ctDNA, none of the parameters of region, control types, sample size, ample source, and assay methods represented a primary source of heterogeneity; thus, heterogeneity might be generated from subjects’ ages, TMN stages, tumor size, tumor metastasis, or different protocol, which failed to be evaluated in this meta-analysis due to the lack of complete information. In the qualitative analysis of ctDNA, among parameters of region, control types, sample size, ample source, assay methods, and gene loci methylation, “control type” (coefficient = 0.502, p = 0.034) and “gene loci methylation” (coefficient = 0.661, p = 0.010) were potential primary sources of heterogeneity. We further analyzed the heterogeneity in SEPT9 methylation studies and revealed that “region” (coefficient = 0.676, p = 0.045) and “control type” (coefficient = 0.733, p = 0.035) could be the primary sources of heterogeneity. Thus, further large clinical trials should reasonably select the control subjects and various ethnicities or be performed in multiple regions to boost the diagnostic performance of ctDNA of CRC.
Remarkably, there were several limitations that should be discussed in this meta-analysis. Firstly, even though we searched various databases, there were several publications we did not include in our meta-analysis as we failed to access the full texts. Moreover, there were only a small number of studies that described quantitative analysis of ctDNA, which might diminish the statistical significance. Thirdly, we failed to include some parameters, such as TMN stages, as incomplete information in included articles. Additionally, potential publication bias within the qualitative analysis that detects non-SEPT9 methylation, which might diminish the confidence of the analysis. Eventually, single gene loci were analyzed in qualitative analysis, while the combination of different gene alterations or other detection techniques might help to improve the diagnostic performance [124]. Therefore, more large-scale clinical trials with complete information should be performed to delineate the diagnostic value of ctDNA detection for CRC to further clarify the conclusions in this meta-analysis.
5. Conclusions
In summary, this is the first integrated meta-analysis on the overall diagnostic accuracy of circulating tumor DNA assays in CRC. Both quantitative and qualitative analyses of ctDNA exhibited excellent diagnostic efficiency. Specifically, quantitative analysis of ctDNA or qualitative analysis of SEPT9 methylation could potentially serve as effective diagnostic methods for CRC. Larger sample studies are needed to further confirm our conclusions and to make the ctDNA approach more sensitive and specific.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jcm12020408/s1, Table S1: Summary of most relevant features of the enrolled publications. Figure S1: Traffic-light plot of quality assessment by the revised QUADAS-2. QUADAS-2, Quality Assessment of Diagnostic Accuracy Studies-2.
Author Contributions
J.C. designed/planned the study; L.M. and J.C. performed publication search and data extraction; L.M. performed the data analysis and wrote the manuscript. M.Y. and D.L. review and revise the manuscript. L.M., J.C., M.Y. and D.L. participated in the discussion. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China [82070547 (Deliang Liu)].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors thank Liwen Wang from the third Xiangya Hospital, who kindly provided help during the data retrieval and analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global Colorectal Cancer Burden in 2020 and Projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Matsuda, T.; Di Carlo, V.; Harewood, R.; Matz, M.; Nikšić, M.; Bonaventure, A.; Valkov, M.; Johnson, C.J.; Estève, J.; et al. Global Surveillance of Trends in Cancer Survival 2000-14 (CONCORD-3): Analysis of Individual Records for 37,513,025 Patients Diagnosed with One of 18 Cancers from 322 Population-Based Registries in 71 Countries. Lancet 2018, 391, 1023–1075. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef] [PubMed]
- Ladabaum, U.; Dominitz, J.A.; Kahi, C.; Schoen, R.E. Strategies for Colorectal Cancer Screening. Gastroenterology 2020, 158, 418–432. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Sargent, D.J.; Loprinzi, C.L.; Levin, T.R.; Rex, D.K.; Ahnen, D.J.; Knigge, K.; Lance, M.P.; Burgart, L.J.; Hamilton, S.R.; et al. Stool DNA and Occult Blood Testing for Screen Detection of Colorectal Neoplasia. Ann. Intern. Med. 2008, 149, 441—450, W81. [Google Scholar] [CrossRef]
- Morikawa, T.; Kato, J.; Yamaji, Y.; Wada, R.; Mitsushima, T.; Shiratori, Y. A Comparison of the Immunochemical Fecal Occult Blood Test and Total Colonoscopy in the Asymptomatic Population. Gastroenterology 2005, 129, 422–428. [Google Scholar] [CrossRef]
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The Performance of the SEPT9 Gene Methylation Assay and a Comparison with Other CRC Screening Tests: A Meta-Analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef]
- Adler, A.; Geiger, S.; Keil, A.; Bias, H.; Schatz, P.; deVos, T.; Dhein, J.; Zimmermann, M.; Tauber, R.; Wiedenmann, B. Improving Compliance to Colorectal Cancer Screening Using Blood and Stool Based Tests in Patients Refusing Screening Colonoscopy in Germany. BMC Gastroenterol. 2014, 14, 183. [Google Scholar] [CrossRef]
- Niu, F.; Wen, J.; Fu, X.; Li, C.; Zhao, R.; Wu, S.; Yu, H.; Liu, X.; Zhao, X.; Liu, S.; et al. Stool DNA Test of Methylated Syndecan-2 for the Early Detection of Colorectal Neoplasia. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1411–1419. [Google Scholar] [CrossRef]
- Meissner, H.I.; Breen, N.; Klabunde, C.N.; Vernon, S.W. Patterns of Colorectal Cancer Screening Uptake among Men and Women in the United States. Cancer Epidemiol. Biomark. Prev. 2006, 15, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.; Shapiro, B.; Sklaroff, D.; Yaros, M. Free DNA in the Serum of Cancer Patients and the Effect of Therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Han, D.S.C.; Lo, Y.M.D. The Nexus of CfDNA and Nuclease Biology. Trends Genet. 2021, 37, 758–770. [Google Scholar] [CrossRef] [PubMed]
- Lui, Y.Y.; Chik, K.-W.; Chiu, R.W.; Ho, C.-Y.; Lam, C.W.; Lo, Y.D. Predominant Hematopoietic Origin of Cell-Free DNA in Plasma and Serum after Sex-Mismatched Bone Marrow Transplantation. Clin. Chem. 2002, 48, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.W.; Chan, K.A.; Sun, H.; Jiang, P.; Su, X.; Chen, E.Z.; Lun, F.M.; Hung, E.C.; Lee, V.; Wong, J. Nonhematopoietically Derived DNA Is Shorter than Hematopoietically Derived DNA in Plasma: A Transplantation Model. Clin. Chem. 2012, 58, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Hasenleithner, S.O.; Speicher, M.R. A Clinician’s Handbook for Using CtDNA throughout the Patient Journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Yi, X.; Ma, J.; Guan, Y.; Chen, R.; Yang, L.; Xia, X. The Feasibility of Using Mutation Detection in CtDNA to Assess Tumor Dynamics. Int. J. Cancer 2017, 140, 2642–2647. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating Tumor DNA as a Liquid Biopsy for Cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef]
- Roy, D.; Tiirikainen, M. Diagnostic Power of DNA Methylation Classifiers for Early Detection of Cancer. Trends Cancer 2020, 6, 78–81. [Google Scholar] [CrossRef]
- Gilson, P.; Merlin, J.-L.; Harlé, A. Detection of Microsatellite Instability: State of the Art and Future Applications in Circulating Tumour DNA (CtDNA). Cancers 2021, 13, 1491. [Google Scholar] [CrossRef] [PubMed]
- Symonds, E.L.; Pedersen, S.K.; Yeo, B.; Al Naji, H.; Byrne, S.E.; Roy, A.; Young, G.P. Assessment of Tumor Burden and Response to Therapy in Patients with Colorectal Cancer Using a Quantitative CtDNA Test for Methylated BCAT1/IKZF1. Mol. Oncol. 2022, 16, 2031–2041. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Lefort, F.; Vasioukhin, V.; Lyautey, J.; Lederrey, C.; Chen, X.Q.; Stroun, M.; Mulcahy, H.E.; Farthing, M. K-Ras Mutations Are Found in DNA Extracted from the Plasma of Patients with Colorectal Cancer. Gastroenterology 1997, 112, 1114–1120. [Google Scholar] [CrossRef] [PubMed]
- Vymetalkova, V.; Cervena, K.; Bartu, L.; Vodicka, P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int. J. Mol. Sci. 2018, 19, 3356. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-Analysis of Observational Studies in Epidemiology: A Proposal for Reporting. Meta-Analysis Of Observational Studies in Epidemiology (MOOSE) Group. JAMA 2000, 283, 2008–2012. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M.; QUADAS-2 Group. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Schwarzer, G. Meta: An R Package for Meta-Analysis. R News 2007, 7, 40–45. [Google Scholar]
- Doebler, P.; Holling, H. Meta-Analysis of Diagnostic Accuracy with Mada. R Packag. 2015, 1, 15. [Google Scholar]
- Viechtbauer, W.; Viechtbauer, M.W. Package ‘Metafor’. The Comprehensive R Archive Network. Package ‘Metafor’. 2015. Available online: http://cran.r-project.org/web/packages/metafor/metafor.pdf (accessed on 20 November 2022).
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M.M. The Diagnostic Odds Ratio: A Single Indicator of Test Performance. J. Clin. Epidemiol. 2003, 56, 1129–1135. [Google Scholar] [CrossRef]
- Arends, L.R.; Hamza, T.H.; van Houwelingen, J.C.; Heijenbrok-Kal, M.H.; Hunink, M.G.M.; Stijnen, T. Bivariate Random Effects Meta-Analysis of ROC Curves. Med. Decis. Mak. 2008, 28, 621–638. [Google Scholar] [CrossRef] [PubMed]
- Reitsma, J.B.; Glas, A.S.; Rutjes, A.W.S.; Scholten, R.J.P.M.; Bossuyt, P.M.; Zwinderman, A.H. Bivariate Analysis of Sensitivity and Specificity Produces Informative Summary Measures in Diagnostic Reviews. J. Clin. Epidemiol. 2005, 58, 982–990. [Google Scholar] [CrossRef] [PubMed]
- Mandrekar, J.N. Receiver Operating Characteristic Curve in Diagnostic Test Assessment. J. Thorac. Oncol. 2010, 5, 1315–1316. [Google Scholar] [CrossRef] [PubMed]
- Walter, S. Properties of the Summary Receiver Operating Characteristic (SROC) Curve for Diagnostic Test Data. Stat. Med. 2002, 21, 1237–1256. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H. Overview of the Process of Conducting Meta-Analyses of the Diagnostic Test Accuracy. J. Rheum. Dis. 2018, 25, 3–10. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring Inconsistency in Meta-Analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Thompson, S.G.; Higgins, J.P.T. How Should Meta-Regression Analyses Be Undertaken and Interpreted? Stat. Med. 2002, 21, 1559–1573. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for Examining and Interpreting Funnel Plot Asymmetry in Meta-Analyses of Randomised Controlled Trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef]
- Agah, S.; Akbari, A.; Talebi, A.; Masoudi, M.; Sarveazad, A.; Mirzaei, A.; Nazmi, F. Quantification of Plasma Cell-Free Circulating DNA at Different Stages of Colorectal Cancer. Cancer Investig. 2017, 35, 625–632. [Google Scholar] [CrossRef]
- Agostini, M.; Pucciarelli, S.; Enzo, M.V.; Del Bianco, P.; Briarava, M.; Bedin, C.; Maretto, I.; Friso, M.L.; Lonardi, S.; Mescoli, C.; et al. Circulating Cell-Free DNA: A Promising Marker of Pathologic Tumor Response in Rectal Cancer Patients Receiving Preoperative Chemoradiotherapy. Ann. Surg. Oncol. 2011, 18, 2461–2468. [Google Scholar] [CrossRef]
- Allegretti, M.; Cottone, G.; Carboni, F.; Cotroneo, E.; Casini, B.; Giordani, E.; Amoreo, C.A.; Buglioni, S.; Diodoro, M.; Pescarmona, E.; et al. Cross-Sectional Analysis of Circulating Tumor DNA in Primary Colorectal Cancer at Surgery and during Post-Surgery Follow-up by Liquid Biopsy. J. Exp. Clin. Cancer Res. 2020, 39, 12. [Google Scholar] [CrossRef] [PubMed]
- Czeiger, D.; Shaked, G.; Eini, H.; Vered, I.; Belochitski, O.; Avriel, A.; Ariad, S.; Douvdevani, A. Measurement of Circulating Cell-Free DNA Levels by a New Simple Fluorescent Test in Patients With Primary Colorectal Cancer. Am. J. Clin. Pathol. 2011, 135, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Danese, E.; Montagnana, M.; Minicozzi, A.M.; De Matteis, G.; Scudo, G.; Salvagno, G.L.; Cordiano, C.; Lippi, G.; Guidi, G.C. Real-Time Polymerase Chain Reaction Quantification of Free DNA in Serum of Patients with Polyps and Colorectal Cancers. Clin. Chem. Lab. Med. 2010, 48, 1665–1668. [Google Scholar] [CrossRef] [PubMed]
- El-Gayar, D.; El-Abd, N.; Hassan, N.; Ali, R. Increased Free Circulating DNA Integrity Index as a Serum Biomarker in Patients with Colorectal Carcinoma. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 939–944. [Google Scholar] [CrossRef]
- Flamini, E.; Mercatali, L.; Nanni, O.; Calistri, D.; Nunziatini, R.; Zoli, W.; Rosetti, P.; Gardini, N.; Lattuneddu, A.; Verdecchia, G.M.; et al. Free DNA and Carcinoembryonic Antigen Serum Levels: An Important Combination for Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2006, 12, 6985–6988. [Google Scholar] [CrossRef]
- Hao, T.B.; Shi, W.; Shen, X.J.; Qi, J.; Wu, X.H.; Wu, Y.; Tang, Y.Y.; Ju, S.Q. Circulating Cell-Free DNA in Serum as a Biomarker for Diagnosis and Prognostic Prediction of Colorectal Cancer. Br. J. Cancer 2014, 111, 1482–1489. [Google Scholar] [CrossRef]
- Junca, A.; Tachon, G.; Evrard, C.; Villalva, C.; Frouin, E.; Karayan-Tapon, L.; Tougeron, D. Detection of Colorectal Cancer and Advanced Adenoma by Liquid Biopsy (Decalib Study): The DdPCR Challenge. Cancers 2020, 12, 1482. [Google Scholar] [CrossRef]
- Lan, Y.T.; Chen, M.H.; Fang, W.L.; Hsieh, C.C.; Lin, C.H.; Jhang, F.Y.; Yang, S.H.; Lin, J.K.; Chen, W.S.; Jiang, J.K.; et al. Clinical Relevance of Cell-Free DNA in Gastrointestinal Tract Malignancy. Oncotarget 2017, 8, 3009–3017. [Google Scholar] [CrossRef]
- Qi, J.; Qian, C.; Shi, W.; Wu, X.H.; Jing, R.R.; Zhang, L.R.; Wang, Z.W.; Ju, S.Q. Alu-Based Cell-Free DNA: A Potential Complementary Biomarker for Diagnosis of Colorectal Cancer. Clin. Biochem. 2013, 46, 64–69. [Google Scholar] [CrossRef]
- Ahlquist, D.A.; Taylor, W.R.; Mahoney, D.W.; Zou, H.; Domanico, M.; Thibodeau, S.N.; Boardman, L.A.; Berger, B.M.; Lidgard, G.P. The Stool DNA Test Is More Accurate than the Plasma Septin 9 Test in Detecting Colorectal Neoplasia. Clin. Gastroenterol. Hepatol. 2012, 10, 272–277.e1. [Google Scholar] [CrossRef]
- Alizadeh Naini, M.; Kavousipour, S.; Hasanzarini, M.; Nasrollah, A.; Monabati, A.; Mokarram, P. O6-Methyguanine-DNA Methyl Transferase (MGMT) Promoter Methylation in Serum DNA of Iranian Patients with Colorectal Cancer. Asian Pac. J. Cancer Prev. APJCP 2018, 19, 1223–1227. [Google Scholar] [CrossRef]
- Amiot, A.; Mansour, H.; Baumgaertner, I.; Delchier, J.C.; Tournigand, C.; Furet, J.P.; Carrau, J.P.; Canoui-Poitrine, F.; Sobhani, I.; the CRC Group of Val De Marne. The Detection of the Methylated Wif-1 Gene Is More Accurate than a Fecal Occult Blood Test for Colorectal Cancer Screening. PLoS ONE 2014, 9, e99233. [Google Scholar] [CrossRef] [PubMed]
- Ansar, M.; Wang, C.-J.; Wang, Y.-H.; Shen, T.-H.; Hung, C.-S.; Chang, S.-C.; Lin, R.-K. SMAD3 Hypomethylation as a Biomarker for Early Prediction of Colorectal Cancer. Int. J. Mol. Sci. 2020, 21, 7395. [Google Scholar] [CrossRef]
- Ashoori, H.; Ghamarchehreh, M.E.; Tavallaei, M.; Ganji, S.M.; Hosseini, M.; Zolfaghari, M.; Ghamarchehreh, Z.; Vahidian, F. Evaluation of the Epigenetic Biomarker Bone Morphogenic Protein 3 for Colorectal Cancer Diagnosis. J. Clin. Diagn. Res. 2018, 12, GC07–GC09. [Google Scholar] [CrossRef]
- Bartak, B.K.; Kalmar, A.; Peterfia, B.; Patai, A.V.; Galamb, O.; Valcz, G.; Spisak, S.; Wichmann, B.; Nagy, Z.B.; Toth, K.; et al. Colorectal Adenoma and Cancer Detection Based on Altered Methylation Pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in Plasma Samples. Epigenetics 2017, 12, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Bedin, C.; Enzo, M.V.; Del Bianco, P.; Pucciarelli, S.; Nitti, D.; Agostini, M. Diagnostic and Prognostic Role of Cell-Free DNA Testing for Colorectal Cancer Patients. Int. J. Cancer 2017, 140, 1888–1898. [Google Scholar] [CrossRef] [PubMed]
- Cassinotti, E.; Melson, J.; Liggett, T.; Melnikov, A.; Yi, Q.L.; Replogle, C.; Mobarhan, S.; Boni, L.; Segato, S.; Levenson, V. DNA Methylation Patterns in Blood of Patients with Colorectal Cancer and Adenomatous Colorectal Polyps. Int. J. Cancer 2012, 131, 1153–1157. [Google Scholar] [CrossRef]
- Chen, C.H.; Yan, S.L.; Yang, T.H.; Chen, S.F.; Yeh, Y.H.; Ou, J.J.; Lin, C.H.; Lee, Y.T.; Chen, C.H. The Relationship between the Methylated Septin-9 DNA Blood Test and Stool Occult Blood Test for Diagnosing Colorectal Cancer in Taiwanese People. J. Clin. Lab. Anal. 2017, 31, e22013. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Zhao, G.; Sun, C.; Ma, Y.; Zhang, L.; Zheng, M.; Li, H. Performance of a Novel Blood-Based Early Colorectal Cancer Screening Assay in Remaining Serum after the Blood Biochemical Test. Dis. Mrk. 2019, 2019, 5232780. [Google Scholar] [CrossRef]
- Cho, N.-Y.; Park, J.-W.; Wen, X.; Shin, Y.-J.; Kang, J.-K.; Song, S.-H.; Kim, H.-P.; Kim, T.-Y.; Bae, J.M.; Kang, G.H. Blood-Based Detection of Colorectal Cancer Using Cancer-Specific DNA Methylation Markers. Diagnostics 2021, 11, 51. [Google Scholar] [CrossRef]
- Church, T.R.; Wandell, M.; Lofton-Day, C.; Mongin, S.J.; Burger, M.; Payne, S.R.; Castanos-Velez, E.; Blumenstein, B.A.; Rosch, T.; Osborn, N.; et al. Prospective Evaluation of Methylated SEPT9 in Plasma for Detection of Asymptomatic Colorectal Cancer. Gut 2014, 63, 317–325. [Google Scholar] [CrossRef] [PubMed]
- deVos, T.; Tetzner, R.; Model, F.; Weiss, G.; Schuster, M.; Distler, J.; Steiger, K.V.; Grützmann, R.; Pilarsky, C.; Habermann, J.K.; et al. Circulating Methylated SEPT9 DNA in Plasma Is a Biomarker for Colorectal Cancer. Clin. Chem. 2009, 55, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Yan, P.; Zhang, S.; Lu, Y.; Pan, L.; Tang, W.Q.; Chen, S.; Chen, S.F.; Zhang, A.Q.; Liu, W. Cell-Free Circulating Methylated SEPT9 for Noninvasive Diagnosis and Monitoring of Colorectal Cancer. Dis. Markers 2018, 2018, 6437104. [Google Scholar] [CrossRef] [PubMed]
- Grutzmann, R.; Molnar, B.; Pilarsky, C.; Habermann, J.K.; Schlag, P.M.; Saeger, H.D.; Miehlke, S.; Stolz, T.; Model, F.; Roblick, U.J.; et al. Sensitive Detection of Colorectal Cancer in Peripheral Blood by Septin 9 DNA Methylation Assay. PLoS ONE 2008, 3, e3759. [Google Scholar] [CrossRef]
- He, N.; Song, L.L.; Kang, Q.; Jin, P.; Cai, G.X.; Zhou, J.F.; Zhou, G.P.; Sheng, J.Q.; Cai, S.J.; Wang, J.M.; et al. The Pathological Features of Colorectal Cancer Determine the Detection Performance on Blood CtDNA. Technol. Cancer Res. Treat. 2018, 17, 1533033818791794. [Google Scholar] [CrossRef]
- He, Q.O.; Chen, H.Y.; Bai, E.Q.; Luo, Y.X.; Fu, R.J.; He, Y.S.; Jiang, J.; Wang, H.Q. Development of a Multiplex MethyLight Assay for the Detection of Multigene Methylation in Human Colorectal Cancer. Cancer Genet. Cytogenet. 2010, 202, 1–10. [Google Scholar] [CrossRef]
- Herbst, A.; Rahmig, K.; Stieber, P.; Philipp, A.; Jung, A.; Ofner, A.; Crispin, A.; Neumann, J.; Lamerz, R.; Kolligs, F.T. Methylation of NEUROG1 in Serum Is a Sensitive Marker for the Detection of Early Colorectal Cancer. Am. J. Gastroenterol. 2011, 106, 1110–1118. [Google Scholar] [CrossRef]
- Jensen, S.; Øgaard, N.; Ørntoft, M.W.; Rasmussen, M.H.; Bramsen, J.B.; Kristensen, H.; Mouritzen, P.; Madsen, M.R.; Madsen, A.H.; Sunesen, K.G.; et al. Novel DNA Methylation Biomarkers Show High Sensitivity and Specificity for Blood-Based Detection of Colorectal Cancer-a Clinical Biomarker Discovery and Validation Study. Clin. Epigenetics 2019, 11, 158. [Google Scholar] [CrossRef]
- Jin, P.; Kang, Q.; Wang, X.; Yang, L.; Yu, Y.; Li, N.; He, Y.Q.; Han, X.L.; Hang, J.; Zhang, J.; et al. Performance of a Second-Generation Methylated SEPT9 Test in Detecting Colorectal Neoplasm. J. Gastroenterol. Hepatol. 2015, 30, 830–833. [Google Scholar] [CrossRef]
- Johnson, D.A.; Barclay, R.L.; Mergener, K.; Weiss, G.; König, T.; Beck, J.; Potter, N.T. Plasma Septin9 versus Fecal Immunochemical Testing for Colorectal Cancer Screening: A Prospective Multicenter Study. PLoS ONE 2014, 9, e98238. [Google Scholar] [CrossRef]
- Kang, Q.; Jin, P.; Yang, L.; Wang, X.; An, H.; Liu, L.; Li, N.; Sheng, J. Significance of Septin9 gene methylation detection of plasma circulation DNA in colorectal cancer screening. Zhonghua Yi Xue Za Zhi 2014, 94, 3839–3841. [Google Scholar] [PubMed]
- Karam, R.A.; Zidan, H.E.; Abd Elrahman, T.M.; Badr, S.A.; Amer, S.A. Study of P16 Promoter Methylation in Egyptian Colorectal Cancer Patients. J. Cell. Biochem. 2019, 120, 8581–8587. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Lee, J.K.; Hong, Y.J.; Moon, S.M.; Shin, U.S.; Kwon, H.; Shin, K.; Chang, Y.H. Detection of Methylated SEPT9 in Korean Colorectal Cancer Patients: Comparison with Previous Studies. Clin. Lab. 2018, 64, 1573–1579. [Google Scholar] [CrossRef]
- Lee, H.S.; Hwang, S.M.; Kim, T.S.; Kim, D.W.; Park, D.J.; Kang, S.B.; Kim, H.H.; Park, K.U. Circulating Methylated Septin 9 Nucleic Acid in the Plasma of Patients with Gastrointestinal Cancer in the Stomach and Colon. Transl. Oncol. 2013, 6, 290-U245. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, Z.Z.; Zhao, G.D.; Ma, Y.; Chen, Y.; Xue, Q.; Zheng, M.X.; Fei, S.J. Performance of a MethyLight Assay for Methylated SFRP2 DNA Detection in Colorectal Cancer Tissue and Serum. Int. J. Biol. Mrk. 2019, 34, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-H.; Xiao, J.; Ye, Z.-Y.; Wei, D.-L.; Zhai, X.-H.; Xu, R.-H.; Zeng, Z.-L.; Luo, H.-Y. Circulating Tumor DNA Methylation Marker MYO1-G for Diagnosis and Monitoring of Colorectal Cancer. Clin. Epigenetics 2021, 13, 232. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tham, C.K.; Ong, S.Y.; Ho, K.S.; Lim, J.F.; Chew, M.H.; Lim, C.K.; Zhao, Y.; Tang, C.L.; Eu, K.W. Serum Methylation Levels of TAC1. SEPT9 and EYA4 as Diagnostic Markers for Early Colorectal Cancers: A Pilot Study. Biomarkers 2013, 18, 399–405. [Google Scholar] [CrossRef]
- Liu, Y.; Zhao, G.D.; Miao, J.; Li, H.; Ma, Y.; Liu, X.Y.; Li, S.M.; Zhu, Y.; Xiong, S.M.; Zheng, M.X.; et al. Performance Comparison Between Plasma and Stool Methylated SEPT9 Tests for Detecting Colorectal Cancer. Front. Genet. 2020, 11, 324. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, H.; Zhang, W.; Wang, J.; Wan, L.; Li, X.; Ji, Y.; Kong, N.; Zhang, Y.; Wang, J.; et al. Coupling of Serum CK20 and Hyper-Methylated CLIP4 as Promising Biomarker for Colorectal Cancer Diagnosis: From Bioinformatics Screening to Clinical Validation. Aging 2021, 13, 26161–26179. [Google Scholar] [CrossRef]
- Lofton-Day, C.; Model, F.; Devos, T.; Tetzner, R.; Distler, J.; Schuster, M.; Song, X.; Lesche, R.; Liebenberg, V.; Ebert, M.; et al. DNA Methylation Biomarkers for Blood-Based Colorectal Cancer Screening. Clin. Chem. 2008, 54, 414–423. [Google Scholar] [CrossRef]
- Loomans-Kropp, H.A.; Song, Y.; Gala, M.; Parikh, A.R.; Van Seventer, E.E.; Alvarez, R.; Hitchins, M.P.; Shoemaker, R.H.; Umar, A. Methylated Septin9 (m SEPT9): A Promising Blood-Based Biomarker for the Detection and Screening of Early-Onset Colorectal Cancer. Cancer Res. Commun. 2022, 2, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Zhang, Q.; Li, L.; Luo, X.; Liang, B.; Lu, Y.; Hu, B.; Jiang, H. Methylated Septin9 Has Moderate Diagnostic Value in Colorectal Cancer Detection in Chinese Population: A Multicenter Study. BMC Gastroenterol. 2022, 22, 232. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.Y.; Zhao, Q.; Wei, W.; Zheng, L.H.; Yi, S.H.; Li, G.; Wang, W.Q.; Sheng, H.; Pu, H.Y.; Mo, H.Y.; et al. Circulating Tumor DNA Methylation Profiles Enable Early Diagnosis, Prognosis Prediction, and Screening for Colorectal Cancer. Sci. Transl. Med. 2020, 12, eaax7533. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.; Kim, N.; Moon, Y.; Kim, M.S.; Hoehn, B.D.; Park, C.H.; Kim, T.S.; Kim, N.K.; Chung, H.C.; An, S. Genome-Wide Identification and Validation of a Novel Methylation Biomarker, SDC2, for Blood-Based Detection of Colorectal Cancer. J. Mol. Diagn. 2013, 15, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Otero-Estevez, O.; Gallardo-Gomez, M.; de la Cadena, M.P.; Rodriguez-Berrocal, F.J.; Cubiella, J.; Ramirez, V.H.; Garcia-Nimo, L.; De Chiara, L. Value of Serum NEUROG1 Methylation for the Detection of Advanced Adenomas and Colorectal Cancer. Diagnostics 2020, 10, 437. [Google Scholar] [CrossRef]
- Pasha, H.F.; Radwan, M.I.; Yehia, A.M.; Toam, M.M. Circulating Methylated RUNX3 and SFRP1 Genes as a Noninvasive Panel for Early Detection of Colorectal Cancer. Eur. J. Gastroenterol. Hepatol. 2019, 31, 1342–1349. [Google Scholar] [CrossRef]
- Pedersen, S.K.; Baker, R.T.; McEvoy, A.; Murray, D.H.; Thomas, M.; Molloy, P.L.; Mitchell, S.; Lockett, T.; Young, G.P.; LaPointe, L.C. A Two-Gene Blood Test for Methylated DNA Sensitive for Colorectal Cancer. PLoS ONE 2015, 10, e125041. [Google Scholar] [CrossRef]
- Pedersen, S.K.; Symonds, E.L.; Baker, R.T.; Murray, D.H.; McEvoy, A.; Van Doorn, S.C.; Mundt, M.W.; Cole, S.R.; Gopalsamy, G.; Mangira, D.; et al. Evaluation of an Assay for Methylated BCAT1 and IKZF1 in Plasma for Detection of Colorectal Neoplasia. BMC Cancer 2015, 15, 654. [Google Scholar] [CrossRef]
- Potter, N.T.; Hurban, P.; White, M.N.; Whitlock, K.D.; Lofton-Day, C.E.; Tetzner, R.; Koenig, T.; Quigley, N.B.; Weiss, G. Validation of a Real-Time PCR-Based Qualitative Assay for the Detection of Methylated SEPT9 DNA in Human Plasma. Clin. Chem. 2014, 60, 1183–1191. [Google Scholar] [CrossRef]
- Rasmussen, S.L.; Krarup, H.B.; Sunesen, K.G.; Johansen, M.B.; Stender, M.T.; Pedersen, I.S.; Madsen, P.H.; Thorlacius-Ussing, O. Hypermethylated DNA, a Circulating Biomarker for Colorectal Cancer Detection. PLoS One 2017, 12, e180809. [Google Scholar] [CrossRef]
- Rezvani, N.; Alibakhshi, R.; Vaisi-Raygani, A.; Bashiri, F.; Saidijam, M. Detection of SPG20 Gene Promoter-Methylated DNA, as a Novel Epigenetic Biomarker, in Plasma for Colorectal Cancer Diagnosis Using the MethyLight Method. Oncol. Lett. 2017, 13, 3277–3284. [Google Scholar] [CrossRef] [PubMed]
- Rokni, P.; Shariatpanahi, A.M.; Sakhinia, E.; Kerachian, M.A. BMP3 Promoter Hypermethylation in Plasma-Derived Cell-Free DNA in Colorectal Cancer Patients. Genes Genom. 2018, 40, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, J.; Fujiya, M.; Okamoto, K.; Nata, T.; Inaba, Y.; Moriichi, K.; Tanabe, H.; Mizukami, Y.; Watari, J.; Ashida, T.; et al. Immunoprecipitation of Nucleosomal DNA Is a Novel Procedure to Improve the Sensitivity of Serum Screening for the P16 Hypermethylation Associated with Colon Cancer. Cancer Epidemiol. 2010, 34, 194–199. [Google Scholar] [CrossRef]
- Salehi, R.; Atapour, N.; Vatandoust, N.; Farahani, N.; Ahangari, F.; Salehi, A.R. Methylation Pattern of ALX4 Gene Promoter as a Potential Biomarker for Blood-Based Early Detection of Colorectal Cancer. Adv. Biomed. Res. 2015, 4, 252. [Google Scholar] [CrossRef]
- Song, L.; Wang, J.; Wang, H.; Chen, Y.; Jia, J.; Guo, S.; Liu, H.; Peng, X.; Xiao, W.; Gong, Y.; et al. The Quantitative Profiling of Blood MSEPT9 Determines the Detection Performance on Colorectal Tumors. Epigenomics 2018, 10, 1569–1583. [Google Scholar] [CrossRef] [PubMed]
- Suehiro, Y.; Hashimoto, S.; Higaki, S.; Fujii, I.; Suzuki, C.; Hoshida, T.; Matsumoto, T.; Yamaoka, Y.; Takami, T.; Sakaida, I.; et al. Blood Free-Circulating DNA Testing by Highly Sensitive Methylation Assay to Diagnose Colorectal Neoplasias. Oncotarget 2018, 9, 16974–16987. [Google Scholar] [CrossRef] [PubMed]
- Takane, K.; Midorikawa, Y.; Yagi, K.; Sakai, A.; Aburatani, H.; Takayama, T.; Kaneda, A. Aberrant Promoter Methylation of PPP1R3C and EFHD1 in Plasma of Colorectal Cancer Patients. Cancer Med. 2014, 3, 1235–1245. [Google Scholar] [CrossRef]
- Tang, D.; Liu, J.; Wang, D.R.; Yu, H.F.; Li, Y.K.; Zhang, J.Q. Diagnostic and Prognostic Value of the Methylation Status of Secreted Frizzled-Related Protein 2 in Colorectal Cancer. Clin. Investig. Med. 2011, 34, E88–E95. [Google Scholar] [CrossRef]
- Tanzer, M.; Balluff, B.; Distler, J.; Hale, K.; Leodolter, A.; Rocken, C.; Molnar, B.; Schmid, R.; Lofton-Day, C.; Schuster, T.; et al. Performance of Epigenetic Markers SEPT9 and ALX4 in Plasma for Detection of Colorectal Precancerous Lesions. PLoS ONE 2010, 5, e9061. [Google Scholar] [CrossRef]
- Tóth, K.; Sipos, F.; Kalmár, A.; Patai, A.V.; Wichmann, B.; Stoehr, R.; Golcher, H.; Schellerer, V.; Tulassay, Z.; Molnár, B. Detection of Methylated SEPT9 in Plasma Is a Reliable Screening Method for Both Left- and Right-Sided Colon Cancers. PLoS One 2012, 7, e46000. [Google Scholar] [CrossRef]
- Tóth, K.; Wasserkort, R.; Sipos, F.; Kalmár, A.; Wichmann, B.; Leiszter, K.; Valcz, G.; Juhász, M.; Miheller, P.; Patai, Á.V.; et al. Detection of Methylated Septin 9 in Tissue and Plasma of Colorectal Patients with Neoplasia and the Relationship to the Amount of Circulating Cell-Free DNA. PLoS One 2014, 9, e115415. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Yu, Z.H.; Liu, C.; Xu, L.Z.; Yu, W.; Lu, J.; Zhu, R.M.; Li, G.L.; Xia, X.Y.; Wei, X.W.; et al. Detection of RASSF1A Promoter Hypermethylation in Serum from Gastric and Colorectal Adenocarcinoma Patients. World J. Gastroenterol. 2008, 14, 3074–3080. [Google Scholar] [CrossRef] [PubMed]
- Warren, J.D.; Xiong, W.; Bunker, A.M.; Vaughn, C.P.; Furtado, L.V.; Roberts, W.L.; Fang, J.C.; Samowitz, W.S.; Heichman, K.A. Septin 9 Methylated DNA Is a Sensitive and Specific Blood Test for Colorectal Cancer. BMC Med. 2011, 9, 133. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Zhou, G.P.; Jin, P.; Zhu, J.Q.; Li, S.J.; Wu, Q.; Wang, G.Q.; Sheng, J.Q.; Wang, J.M.; Song, L.L.; et al. Detection of Colorectal Cancer Using a Simplified SEPT9 Gene Methylation Assay Is a Reliable Method for Opportunistic Screening. J. Mol. Diagn. 2016, 18, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.P.; Zou, J.H.; Tang, R.N.; Yao, Y.; You, C.Z. Detection and Clinical Significance of DLC1 Gene Methylation in Serum DNA from Colorectal Cancer Patients. Chin. J. Cancer Res. 2011, 23, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Jiang, X.Y.; Li, Q.; Sun, Z.J.; Quan, W.Q.; Duan, Y.P.; Li, D.; Chen, T.H. Diagnostic Value of Methylated Septin9 for Colorectal Cancer Detection. Front. Oncol. 2018, 8, 247. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Yu, S.; Han, J.; Zong, M.; Tan, Q.; Zeng, X.; Fan, L. Detection of Circulating Tumor DNA Methylation in Diagnosis of Colorectal Cancer. Clin. Transl. Gastroenterol. 2021, 12, e00386. [Google Scholar] [CrossRef]
- Xue, G.; Lu, C.J.; Pan, S.J.; Zhang, Y.L.; Miao, H.; Shan, S.; Zhu, X.T.; Zhang, Y. DNA Hypomethylation of CBS Promoter Induced by Folate Deficiency Is a Potential Noninvasive Circulating Biomarker for Colorectal Adenocarcinomas. Oncotarget 2017, 8, 51387–51401. [Google Scholar] [CrossRef]
- Yang, X.; Xu, Z.J.; Chen, X.; Zeng, S.S.; Qian, L.; Wei, J.; Peng, M.; Wang, X.; Liu, W.L.; Ma, H.Y.; et al. Clinical Value of Preoperative Methylated Septin 9 in Chinese Colorectal Cancer Patients. World J. Gastroenterol. 2019, 25, 2099–2109. [Google Scholar] [CrossRef]
- Young, G.P.; Symonds, E.L.; Nielsen, H.J.; Ferm, L.; Christensen, I.J.; Dekker, E.; van der Vlugt, M.; Mallant-Hent, R.C.; Boulter, N.; Yu, B.; et al. Evaluation of a Panel of Tumor-Specific Differentially-Methylated DNA Regions in IRF4, IKZF1 and BCAT1 for Blood-Based Detection of Colorectal Cancer. Clin. Epigenetics 2021, 13, 14. [Google Scholar] [CrossRef]
- Yuan, P.; Cheng, X.J.; Wu, X.J.; Li, L.; Zhang, L.H.; Li, Z.Y.; Xing, X.F.; Du, H.; Wang, X.H.; Hu, Y.; et al. OSMR and SEPT9: Promising Biomarkers for Detection of Colorectal Cancer Based on Blood-Based Tests. Transl. Cancer Res. 2016, 5, 131–139. [Google Scholar] [CrossRef]
- Zhan, J.; Li, X.; Yu, Z.; Yuan, Y.H.; Hou, J. Value of fecal and blood adenomatous polyposis coli gene and K-ras gene mutation detection in colorectal neoplasm screening. Nan Fang Yi Ke Da Xue Xue Bao J. South. Med. Univ. 2007, 27, 1018–1021. [Google Scholar]
- Zhang, X.; Song, Y.F.; Lu, H.N.; Wang, D.P.; Zhang, X.S.; Huang, S.L.; Sun, B.L.; Huang, Z.G. Combined Detection of Plasma GATA5 and SFRP2 Methylation Is a Valid Noninvasive Biomarker for Colorectal Cancer and Adenomas. World J. Gastroenterol. 2015, 21, 2629–2637. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.D.; Li, H.; Yang, Z.X.; Wang, Z.Z.; Xu, M.Q.; Xiong, S.M.; Li, S.M.; Wu, X.T.; Liu, X.Y.; Wang, Z.W.; et al. Multiplex Methylated DNA Testing in Plasma with High Sensitivity and Specificity for Colorectal Cancer Screening. Cancer Med. 2019, 8, 5619–5628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.D.; Ma, Y.; Li, H.; Li, S.M.; Zhu, Y.; Liu, X.Y.; Xiong, S.M.; Liu, Y.; Miao, J.; Fei, S.J.; et al. A Novel Plasma Based Early Colorectal Cancer Screening Assay Base on Methylated SDC2 and SFRP2. Clin. Chim. Acta 2020, 503, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhang, Y.W.; Huang, X.; Chen, L.B. Analysis of the RUNX3 Gene Methylation in Serum DNA from Esophagus Squamous Cell Carcinoma, Gastric and Colorectal Adenocarcinoma Patients. Hepato-Gastroenterology 2011, 58, 2007–2011. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.Z.; Yu, B.M.; Wang, Z.W.; Sun, J.Y.; Cang, H.; Gao, F.; Li, D.H.; Zhao, R.; Feng, G.G.; Yi, J. Detection of Aberrant P16 Methylation in the Serum of Colorectal Cancer Patients. Clin. Cancer Res. 2002, 8, 188–191. [Google Scholar]
- Nagai, Y.; Sunami, E.; Yamamoto, Y.; Hata, K.; Okada, S.; Murono, K.; Yasuda, K.; Otani, K.; Nishikawa, T.; Tanaka, T.; et al. LINE-1 Hypomethylation Status of Circulating Cell-Free DNA in Plasma as a Biomarker for Colorectal Cancer. Oncotarget 2017, 8, 11906–11916. [Google Scholar] [CrossRef]
- Bresalier, R.S.; Kopetz, S.; Brenner, D.E. Blood-Based Tests for Colorectal Cancer Screening: Do They Threaten the Survival of the FIT Test? Dig. Dis. Sci. 2015, 60, 664–671. [Google Scholar] [CrossRef]
- Ma, M.; Zhu, H.; Zhang, C.; Sun, X.; Gao, X.; Chen, G. “Liquid Biopsy”—CtDNA Detection with Great Potential and Challenges. Ann. Transl. Med. 2015, 3, 235. [Google Scholar] [CrossRef]
- Connolly, D.; Abdesselam, I.; Verdier-Pinard, P.; Montagna, C. Septin Roles in Tumorigenesis. Biol. Chem. 2011, 392, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Østevold, K.; Meléndez, A.V.; Lehmann, F.; Schmidt, G.; Aktories, K.; Schwan, C. Septin Remodeling Is Essential for the Formation of Cell Membrane Protrusions (Microtentacles) in Detached Tumor Cells. Oncotarget 2017, 8, 76686–76698. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, P.-M.; Liu, R.-B. Advance in Plasma SEPT9 Gene Methylation Assay for Colorectal Cancer Early Detection. World J. Gastrointest. Oncol. 2018, 10, 15–22. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).