Role of α-Defensin and the Microbiome in Prosthetic Joint Infection: A Prospective Cohort Study in Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
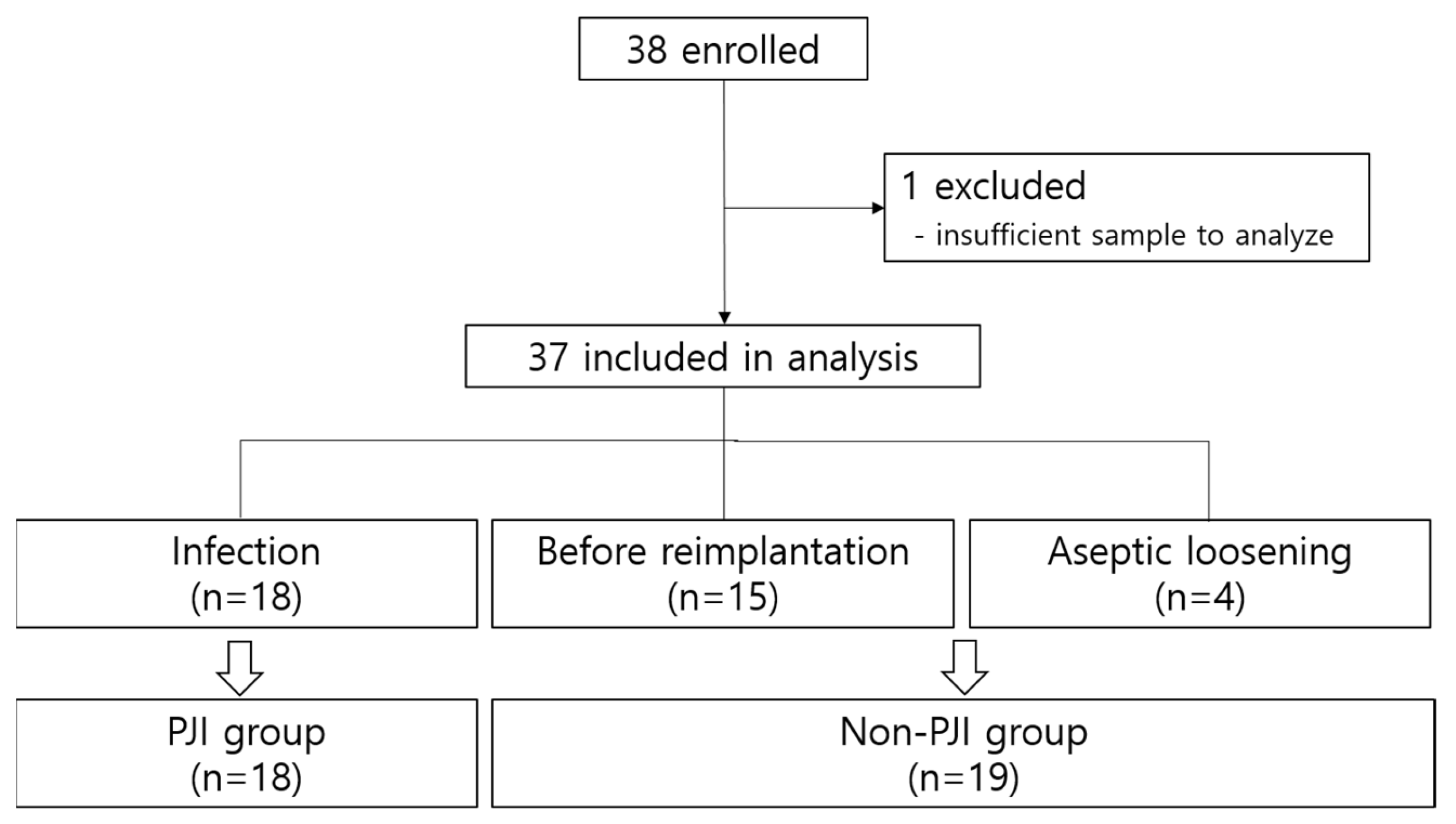
2.2. Data and Sample Collection
2.3. Definitions
2.4. Laboratory Methods
2.5. Statistical Analysis
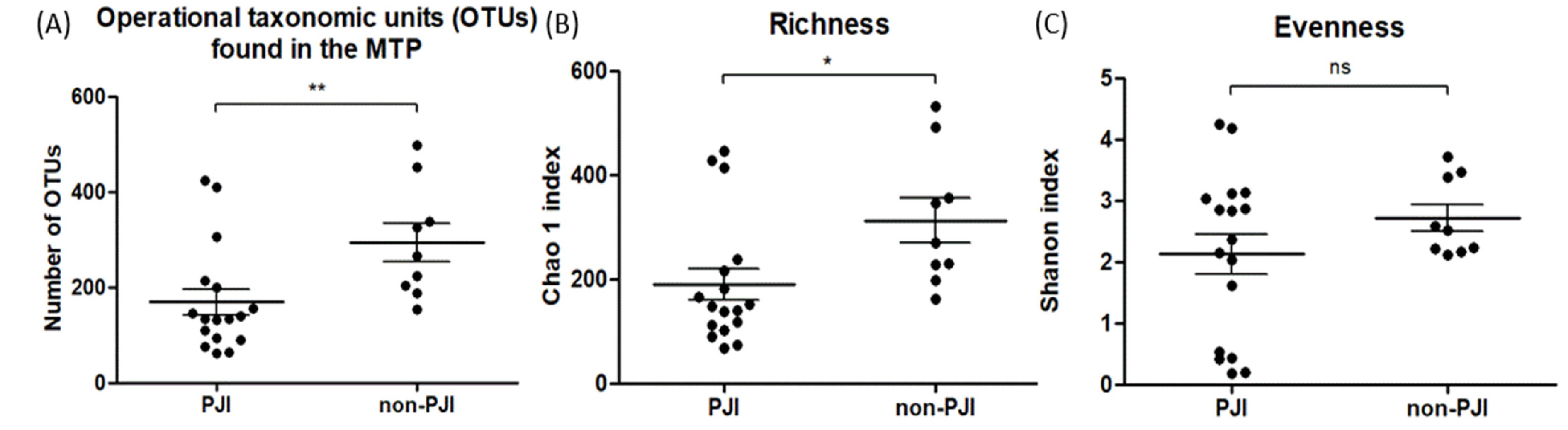
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sandiford, N.A.; Franceschini, M.; Kendoff, D. The burden of prosthetic joint infection (PJI). Ann. Jt. 2020, 6, 25. [Google Scholar] [CrossRef]
- Kim, H.S.; Park, J.W.; Moon, S.-Y.; Lee, Y.-K.; Ha, Y.-C.; Koo, K.-H. Current and Future Burden of Periprosthetic Joint Infection from National Claim Database. J. Korean Med. Sci. 2020, 35, e410. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.M.A.; Lau, E.C.; Son, M.S.; Chang, E.T.; Zimmerli, W.; Parvizi, J. Are We Winning or Losing the Battle with Periprosthetic Joint Infection: Trends in Periprosthetic Joint Infection and Mortality Risk for the Medicare Population. J. Arthroplast. 2018, 33, 3238–3245. [Google Scholar] [CrossRef]
- Parvizi, J.; Zmistowski, B.; Berbari, E.F.; Bauer, T.W.; Springer, B.D.; Della Valle, C.J.; Garvin, K.L.; Mont, M.A.; Wongworawat, M.D.; Zalavras, C.G. New definition for periprosthetic joint infection: From the Workgroup of the Musculoskeletal Infection Society. Clin. Orthop. Relat. Res. 2011, 469, 2992–2994. [Google Scholar] [CrossRef] [PubMed]
- Parvizi, J.; Gehrke, T. International Consensus Group on Periprosthetic Joint Infection. Definition of periprosthetic joint infection. J. Arthroplast. 2014, 29, 1331. [Google Scholar] [CrossRef] [PubMed]
- Aalirezaie, A.; Bauer, T.W.; Fayaz, H.; Griffin, W.; Higuera, C.A.; Krenn, V.; Krenn, V.; Molano, M.; Moojen, D.-J.; Restrepo, C.; et al. Hip and Knee Section, Diagnosis, Reimplantation: Proceedings of International Consensus on Orthopedic Infections. J. Arthroplast. 2019, 34, S369–S379. [Google Scholar] [CrossRef] [PubMed]
- Lehrer, R.I.; Ganz, T. Defensins: Endogenous antibiotic peptides from human leukocytes. Ciba Found. Symp. 1992, 171, 276–290. [Google Scholar]
- Miyamae, Y.; George, J.; Klika, A.K.; Barsoum, W.K.; Higuera, C.A. Diagnostic Accuracy of the Alpha-Defensin Test for Periprosthetic Joint Infection in Patients With Inflammatory Diseases. J. Arthroplast. 2019, 34, 1767–1771. [Google Scholar] [CrossRef]
- Wetters, N.G.; Berend, K.R.; Lombardi, A.V.; Morris, M.J.; Tucker, T.L.; Della Valle, C.J. Leukocyte esterase reagent strips for the rapid diagnosis of periprosthetic joint infection. J. Arthroplast. 2012, 27, 8–11. [Google Scholar] [CrossRef]
- Wang, C.; Li, R.; Wang, Q.; Wang, C. Synovial Fluid Leukocyte Esterase in the Diagnosis of Peri-Prosthetic Joint Infection: A Systematic Review and Meta-Analysis. Surg. Infect. 2018, 19, 245–253. [Google Scholar] [CrossRef]
- Parvizi, J.; Tan, T.L.; Goswami, K.; Higuera, C.; Della Valle, C.; Chen, A.F.; Shohat, N. The 2018 Definition of Periprosthetic Hip and Knee Infection: An Evidence-Based and Validated Criteria. J. Arthroplast. 2018, 33, 1309–1314.e2. [Google Scholar] [CrossRef] [PubMed]
- McNally, M.; Sousa, R.; Wouthuyzen-Bakker, M.; Chen, A.F.; Soriano, A.; Vogely, H.C.; Clauss, M.; Higuera, C.A.; Trebše, R. The EBJIS definition of periprosthetic joint infection. Bone Jt. J. 2021, 103, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Evidence-Based Comparison of the Synovasure®® Alpha Defensin ELISA Test and Alpha Defensin Lateral Flow Test. Available online: https://www.zimmerbiomet.com/content/dam/zb-corporate/en/products/specialties/diagnostics/synovasure-alpha-defensin-lateral-flow-test/23771GLBLenEvidenceBasedComparison.pdf (accessed on 10 February 2023).
- Synovasure Alpha Defensin Lateral Flow Test. Available online: https://www.zimmerbiomet.com/content/dam/zb-corporate/en/products/specialties/diagnostics/synovasure-alpha-defensin-lateral-flow-test/2468.2-GLBL-en%20Synovasure%20Lateral%20Flow%20Test%20Brochure-A4%20final.pdf.coredownload.pdf (accessed on 10 February 2023).
- Deirmengian, C.; Kardos, K.; Kilmartin, P.; Cameron, A.; Schiller, K.; Booth, R.E.; Parvizi, J. The alpha-defensin test for periprosthetic joint infection outperforms the leukocyte esterase test strip. Clin. Orthop. Relat. Res. 2015, 473, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Kuiper, J.W.P.; Verberne, S.J.; Vos, S.J.; van Egmond, P.W. Does the Alpha Defensin ELISA Test Perform Better Than the Alpha Defensin Lateral Flow Test for PJI Diagnosis? A Systematic Review and Meta-analysis of Prospective Studies. Clin. Orthop. Relat. Res. 2020, 478, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, M.C.; Beswick, A.D.; Kunutsor, S.K.; Wilson, M.J.; Whitehouse, M.R.; Blom, A.W. The Alpha-Defensin Immunoassay and Leukocyte Esterase Colorimetric Strip Test for the Diagnosis of Periprosthetic Infection: A Systematic Review and Meta-Analysis. J. Bone Jt. Surg. Am. 2016, 98, 992–1000. [Google Scholar] [CrossRef]
- Ding, B.T.; Tan, K.G.; Kau, C.Y.; Chan, H.Y.H.; Fadil, M.F.B.M. Accuracy of the α-defensin lateral flow assay for diagnosing periprosthetic joint infection in Asians. J. Orthop. Surg. 2019, 27, 2309499019828459. [Google Scholar] [CrossRef]
- Zheng, Q.-Y.; Zhang, G.-Q. Application of leukocyte esterase strip test in the screening of periprosthetic joint infections and prospects of high-precision strips. Arthroplasty 2020, 2, 34. [Google Scholar] [CrossRef]
- Tan, J.; Liu, Y.; Ehnert, S.; Nüssler, A.K.; Yu, Y.; Xu, J.; Chen, T. The Effectiveness of Metagenomic Next-Generation Sequencing in the Diagnosis of Prosthetic Joint Infection: A Systematic Review and Meta-Analysis. Front. Cell Infect. Microbiol. 2022, 12, 875822. [Google Scholar] [CrossRef]
- Thoendel, M.J.; Jeraldo, P.R.; E Greenwood-Quaintance, K.; Yao, J.Z.; Chia, N.; Hanssen, A.D.; Abdel, M.P.; Patel, R. Identification of Prosthetic Joint Infection Pathogens Using a Shotgun Metagenomics Approach. Clin. Infect. Dis. 2018, 67, 1333–1338. [Google Scholar] [CrossRef]
- Hao, L.; Wen, P.; Song, W.; Zhang, B.; Wu, Y.; Zhang, Y.; Ma, T.; Qiu, Y. Direct detection and identification of periprosthetic joint infection pathogens by metagenomic next-generation sequencing. Sci. Rep. 2023, 13, 7897. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.-L.; Flurin, L.; Thoendel, M.J.; Wolf, M.J.; Abdel, M.P.; E Greenwood-Quaintance, K.; Patel, R. Targeted Versus Shotgun Metagenomic Sequencing-based Detection of Microorganisms in Sonicate Fluid for Periprosthetic Joint Infection Diagnosis. Clin. Infect. Dis. 2023, 76, e1456–e1462. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, B.; Li, S.; Yang, L.; Zhu, D.; Wang, Y.; Wang, H.; Wang, T.; Shi, B.; Gai, Z.; et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci. Rep. 2018, 8, 14305. [Google Scholar] [CrossRef] [PubMed]
- Gbejuade, H.O.; Lovering, A.M.; Webb, J.C. The role of microbial biofilms in prosthetic joint infections. Acta. Orthop. 2015, 86, 147–158. [Google Scholar] [CrossRef]

| Variables | PJI (n = 18) | Non-PJI (n = 19) | p-Value |
|---|---|---|---|
| Male | 5 (10.5) | 2 (27.8) | 0.23 |
| Age, years, median [IQR] | 73 [70–79] | 78 [71–81] | 0.63 |
| BMI, kg/m2, mean ± SD | 26.5 ± 3.7 | 26.9 ± 4.5 | 0.77 |
| Underlying disease | |||
| Hypertension | 14 (77.8) | 11 (57.9) | 0.30 |
| Diabetes mellitus | 6 (33.3) | 8 (42.1) | 0.74 |
| CAOD | 3 (16.7) | 0 (0) | 0.11 |
| CVA | 2 (11.1) | 5 (26.3) | 0.40 |
| Time after TKA, months, median [IQR] | 32 [21–49] | 97 [41.0–184] | 0.04 |
| Previous antibiotics, yes | 8 (50) | 5 (27.8) | 0.29 |
| Variables | PJI | Non-PJI | p-Value |
|---|---|---|---|
| Blood | |||
| ESR, mm/h, median [IQR] | 62 [29.5–114.0] | 18.5 [9.3–27.8] | <0.001 |
| CRP, mg/dL, median [IQR] | 97 [10.4–120] | 1.6 [1.0–4.9] | 0.003 |
| Culture 1 | 12 (66.7) | 0 | <0.001 |
| Synovial fluid | |||
| Alpha defensin, median [IQR] | 4698 [2986–8466] | 296 [241–429] | <0.001 |
| WBC count, /μL, median [IQR] | 30250 [20818–60300] | 550 [240–810] | <0.001 |
| PMN (%), median [IQR] | 91.5 [85.3–94.0] | 15.5 [3.0–54.8] | <0.001 |
| LE, positive (%) | 10 (62.5) | 4 (21.1) | 0.01 |
| Culture 1 (%) | 3 (23.1) | 0 | 0.07 |
| Tests | Sensitivity | Specificity | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|
| Blood | ||||
| ESR | 70.6 (48.99–92.2) | 77.8 (58.6–97.0) | 75 (53.8–96.2) | 73.7 (53.9–93.5) |
| CRP | 76.5 (56.3–96.6) | 77.8 (58.6–97.0) | 76.5 (56.3–96.6) | 77.8 (58.6–97.0) |
| Culture 1 | 15.4 (0–35.0) | 100 (81.6–100) | 100 (34.2–100) | 63.0 (44.7–81.2) |
| Synovial fluid | ||||
| Alpha defensin | 94.4 (83.9–100) | 89.5 (75.7–100) | 89.4 (75.7–100) | 94.4 (83.9–100) |
| WBC | 94.4 (83.9–100) | 100 (79.6–100) | 100 (90.2–100) | 93.8 (81.9–100) |
| PMN (%) | 88.9 (74.4–100) | 92.9 (79.4–100) | 94.1 (83.0–100) | 86.7 (69.5–100) |
| LE, positive | 37.5 (13.8–61.2) | 100 (83.2–100) | 100 (61.0–100) | 65.5 (48.2–82.8) |
| Culture 1 | 66.7 (44.9–88.4) | 100 (79.6–100) | 100 (75.8–84.1) | 71.4 (52.1–90.8) |
| MTP 2 | 100 (78.5–100) | 55.6 (23.1–88.0) | 77.8 (58.6–97.0) | 100 (56.6–100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baek, Y.J.; Lee, Y.-J.; Lee, J.A.; Kim, J.H.; Kwon, H.M.; Yeom, J.-S.; Park, K.K.; Jeong, S.J. Role of α-Defensin and the Microbiome in Prosthetic Joint Infection: A Prospective Cohort Study in Korea. J. Clin. Med. 2023, 12, 5964. https://doi.org/10.3390/jcm12185964
Baek YJ, Lee Y-J, Lee JA, Kim JH, Kwon HM, Yeom J-S, Park KK, Jeong SJ. Role of α-Defensin and the Microbiome in Prosthetic Joint Infection: A Prospective Cohort Study in Korea. Journal of Clinical Medicine. 2023; 12(18):5964. https://doi.org/10.3390/jcm12185964
Chicago/Turabian StyleBaek, Yae Jee, Youn-Jung Lee, Jung Ah Lee, Jung Ho Kim, Hyuck Min Kwon, Joon-Sup Yeom, Kwan Kyu Park, and Su Jin Jeong. 2023. "Role of α-Defensin and the Microbiome in Prosthetic Joint Infection: A Prospective Cohort Study in Korea" Journal of Clinical Medicine 12, no. 18: 5964. https://doi.org/10.3390/jcm12185964
APA StyleBaek, Y. J., Lee, Y.-J., Lee, J. A., Kim, J. H., Kwon, H. M., Yeom, J.-S., Park, K. K., & Jeong, S. J. (2023). Role of α-Defensin and the Microbiome in Prosthetic Joint Infection: A Prospective Cohort Study in Korea. Journal of Clinical Medicine, 12(18), 5964. https://doi.org/10.3390/jcm12185964







