Genetic Characteristics of Latvian Patients with Familial Hypercholesterolemia: The First Analysis from Genome-Wide Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. DNA Extraction and Whole-Genome Sequencing
2.3. Variant Calling and Annotation
2.4. Data Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. Characteristics Based on Genetic Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABCG5 | gene encoding ATP-binding cassette sub-family G member 5 |
| ABCG8 | gene encoding ATP-binding cassette sub-family G member 8 |
| ACMG | the American College of Medical Genetics and Genomics |
| AMP | the Association for Molecular Pathology |
| APOB | gene encoding Apolipoprotein B |
| APOE | gene encoding Apolipoprotein E |
| CAD | coronary artery disease |
| CNV | copy number variation |
| CYP27A1 | gene encoding cytochrome P450 family 27 subfamily A member 1 or sterol 27-hydroxylase |
| DLCN | Dutch Lipid Clinic Network |
| DNA | deoxyribonucleic acid |
| FH | familial hypercholesterolemia |
| HDL | high-density lipoprotein |
| IQR | interquartile range |
| LDL | low-density lipoprotein |
| LDLR | gene encoding low-density lipoprotein receptor |
| LDLRAP1 | gene encoding low-density lipoprotein receptor adaptor protein 1 |
| LGDB | the Genome Database of Latvian Population |
| LIPA | gene encoding lipase A, the lysosomal acid lipase |
| LBMC | the Latvian Biomedical Research and Study Centre |
| LLM | lipid lowering medication |
| LP | likely pathogenic |
| LPA | gene encoding lipoprotein(a) |
| LRFH | Latvian Registry of Familial Hypercholesterolemia |
| MLPA | Multiplex Ligation-dependent Probe Amplification |
| P | pathogenic |
| PCSK9 | gene encoding Proprotein convertase subtilisin/kexin type 9 |
| PRS | polygenic risk score |
| SD | standard deviation |
| TG | triglycerides |
| VCEP | Variant Curation Expert Panel |
| VEP | Variant Effect Predictor |
| VUS | Variant of Uncertain Significance |
References
- Nordestgaard, B.G.; Chapman, M.J.; Humphries, S.E.; Ginsberg, H.N.; Masana, L.; Descamps, O.S.; Wiklund, O.; Hegele, R.A.; Raal, F.J.; Defesche, J.C.; et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: Guidance for clinicians to prevent coronary heart disease: Consensus Statement of the European Atherosclerosis Society. Eur. Heart J. 2013, 34, 3478–3490. [Google Scholar] [CrossRef] [PubMed]
- Henderson, R.; O’kane, M.; McGilligan, V.; Watterson, S. The genetics and screening of familial hypercholesterolaemia. J. Biomed. Sci. 2016, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Tada, M.T.; Rocha, V.Z.; Lima, I.R.; Oliveira, T.G.M.; Chacra, A.P.; Miname, M.H.; Nunes, V.S.; Nakandakare, E.R.; Castelo, M.H.C.G.; Jannes, C.E.; et al. Screening of ABCG5 and ABCG8 Genes for Sitosterolemia in a Familial Hypercholesterolemia Cascade Screening Program. Circ. Genom. Precis. Med. 2022, 15, e003390. [Google Scholar] [CrossRef] [PubMed]
- Chora, J.R.; Alves, A.C.; Medeiros, A.M.; Mariano, C.; Lobarinhas, G.; Guerra, A.; Mansilha, H.; Cortez-Pinto, H.; Bourbon, M. Lysosomal acid lipase deficiency: A hidden disease among cohorts of familial hypercholesterolemia? J. Clin. Lipidol. 2017, 11, 477–484.e2. [Google Scholar] [CrossRef] [PubMed]
- Khera, A.V.; Won, H.-H.; Peloso, G.M.; Lawson, K.S.; Bartz, T.M.; Deng, X.; van Leeuwen, E.M.; Natarajan, P.; Emdin, C.A.; Bick, A.G.; et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients with Severe Hypercholesterolemia. J. Am. Coll. Cardiol. 2016, 67, 2578–2589. [Google Scholar] [CrossRef]
- Vallejo-Vaz, A.J.; Seshasai, S.R.K.; Cole, D.; Hovingh, G.K.; Kastelein, J.J.; Mata, P.; Raal, F.J.; Santos, R.D.; Soran, H.; Watts, G.F.; et al. Familial hypercholesterolaemia: A global call to arms. Atherosclerosis 2015, 243, 257–259. [Google Scholar] [CrossRef]
- Sturm, A.C.; Knowles, J.W.; Gidding, S.S.; Ahmad, Z.S.; Ahmed, C.D.; Ballantyne, C.M.; Baum, S.J.; Bourbon, M.; Carrié, A.; Cuchel, M.; et al. Clinical Genetic Testing for Familial Hypercholesterolemia: JACC Scientific Expert Panel. J. Am. Coll. Cardiol. 2018, 72, 662–680. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Anesth. Analg. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Chora, J.R.; Medeiros, A.M.; Alves, A.C.; Bourbon, M. Analysis of publicly available LDLR, APOB, and PCSK9 variants associated with familial hypercholesterolemia: Application of ACMG guidelines and implications for familial hypercholesterolemia diagnosis. Genet. Med. 2018, 20, 591–598. [Google Scholar] [CrossRef]
- Chora, J.R.; Iacocca, M.A.; Tichý, L.; Wand, H.; Kurtz, C.L.; Zimmermann, H.; Leon, A.; Williams, M.; Humphries, S.E.; Hooper, A.J.; et al. The Clinical Genome Resource (ClinGen) Familial Hypercholesterolemia Variant Curation Expert Panel consensus guidelines for LDLR variant classification. Genet. Med. 2022, 24, 293–306. [Google Scholar] [CrossRef]
- Latkovskis, G.; Saripo, V.; Gilis, D.; Nesterovics, G.; Upena-Roze, A.; Erglis, A. Latvian registry of familial hypercholesterolemia: The first report of three-year results. Atherosclerosis 2018, 277, 347–354. [Google Scholar] [CrossRef]
- Visseren, F.L.; Mach, F.; Smulders, Y.M.; Carballo, D.; Koskinas, K.C.; Bäck, M.; Benetos, A.; Biffi, A.; Boavida, J.-M.; Capodanno, D.; et al. 2021 ESC Guidelines on cardiovascular disease prevention in clinical practice: Developed by the Task Force for cardiovascular disease prevention in clinical practice with representatives of the European Society of Cardiology and 12 medical societies with the special contribution of the European Association of Preventive Cardiology (EAPC). Eur. Heart J. 2021, 42, 3227–3337. [Google Scholar] [CrossRef]
- Rovite, V.; Wolff-Sagi, Y.; Zaharenko, L.; Nikitina-Zake, L.; Grens, E.; Klovins, J. Genome Database of the Latvian Population (LGDB): Design, Goals, and Primary Results. J. Epidemiol. 2018, 28, 353–360. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- García-Alcalde, F.; Okonechnikov, K.; Carbonell, J.; Cruz, L.M.; Götz, S.; Tarazona, S.; Dopazo, J.; Meyer, T.F.; Conesa, A. Qualimap: Evaluating next-generation sequencing alignment data. Bioinformatics 2012, 28, 2678–2679. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Tange, O. GNU Parallel—The Command-Line Power Tool. Usenix Mag. 2011, 36, 42–47. [Google Scholar]
- Kurtzer, G.M.; Sochat, V.; Bauer, M.W. Singularity: Scientific containers for mobility of compute. PLoS ONE 2017, 12, e0177459. [Google Scholar] [CrossRef] [PubMed]
- Futema, M.; Taylor-Beadling, A.; Williams, M.; Humphries, S.E. Genetic testing for familial hypercholesterolemia—Past, present, and future. J. Lipid Res. 2021, 62, 100139. [Google Scholar] [CrossRef]
- Haralambos, K.; Humphries, S.; Whitmore, J.; Datta, D.; Cather, M.; Miedzybrodzka, Z.; Breen, J.; Gritzmacher, L.; Hamlen, A.; Potter, A.; et al. Familial hypercholesterolaemia (fh) genetic testing in the uk. Atheroscler. Suppl. 2018, 34, e4. [Google Scholar] [CrossRef]
- Grenkowitz, T.; Kassner, U.; Wühle-Demuth, M.; Salewsky, B.; Rosada, A.; Zemojtel, T.; Hopfenmüller, W.; Isermann, B.; Borucki, K.; Heigl, F.; et al. Clinical characterization and mutation spectrum of German patients with familial hypercholesterolemia. Atherosclerosis 2016, 253, 88–93. [Google Scholar] [CrossRef] [PubMed]
- D’Erasmo, L.; Minicocci, I.; Di Costanzo, A.; Pigna, G.; Commodari, D.; Ceci, F.; Montali, A.; Brancato, F.; Stanca, I.; Nicolucci, A.; et al. Clinical Implications of Monogenic Versus Polygenic Hypercholesterolemia: Long-Term Response to Treatment, Coronary Atherosclerosis Burden, and Cardiovascular Events. J. Am. Heart Assoc. 2021, 10, e018932. [Google Scholar] [CrossRef] [PubMed]
- Ajufo, E.; Cuchel, M. Improving the yield of genetic testing in familial hypercholesterolaemia. Eur. Heart J. 2016, 38, 574–576. [Google Scholar] [CrossRef][Green Version]
- Damgaard, D.; Larsen, M.L.; Nissen, P.H.; Jensen, J.M.; Jensen, H.K.; Soerensen, V.R.; Jensen, L.G.; Faergeman, O. The relationship of molecular genetic to clinical diagnosis of familial hypercholesterolemia in a Danish population. Atherosclerosis 2005, 180, 155–160. [Google Scholar] [CrossRef]
- Iacocca, M.A.; Chora, J.R.; Carrié, A.; Freiberger, T.; Leigh, S.E.; Defesche, J.C.; Kurtz, C.L.; DiStefano, M.T.; Santos, R.D.; Humphries, S.E.; et al. ClinVar database of global familial hypercholesterolemia-associated DNA variants. Hum. Mutat. 2018, 39, 1631–1640. [Google Scholar] [CrossRef]
- Hegele, R.A.; Ban, M.R.; Cao, H.; McIntyre, A.D.; Robinson, J.F.; Wang, J. Targeted Next-Generation Sequencing in Monogenic Dyslipidemias. Curr. Opin. Lipidol. 2015, 26, 103–113. [Google Scholar] [CrossRef]
- Futema, M.; Plagnol, V.; Li, K.; Whittall, R.A.; Neil, H.A.W.; Seed, M.; Bertolini, S.; Calandra, S.; Descamps, O.S.; Graham, C.A.; et al. Whole exome sequencing of familial hypercholesterolaemia patients negative for LDLR/APOB/PCSK9 mutations. J. Med. Genet. 2014, 51, 537–544. [Google Scholar] [CrossRef] [PubMed]
- Marmontel, O.; Charrière, S.; Simonet, T.; Bonnet, V.; Dumont, S.; Mahl, M.; Jacobs, C.; Nony, S.; Chabane, K.; Bozon, D.; et al. Single, short in-del, and copy number variations detection in monogenic dyslipidemia using a next-generation sequencing strategy. Clin. Genet. 2018, 94, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Iacocca, M.A.; Dron, J.S.; Hegele, R.A. Progress in Finding Pathogenic DNA Copy Number Variations in Dyslipidemia. Curr. Opin. Lipidol. 2019, 30, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, P.; Peloso, G.M.; Zekavat, S.M.; Montasser, M.; Ganna, A.; Chaf, M.; Khera, A.V.; Zhou, W.; Bloom, J.M.; Engreitz, J.M.; et al. Deep-Coverage Whole Genome Sequences and Blood Lipids among 16,324 Individuals. Nat. Commun. 2018, 9, 3391. [Google Scholar] [CrossRef]
- Olmastroni, E.; Gazzotti, M.; Arca, M.; Averna, M.; Pirillo, A.; Catapano, A.L.; Casula, M.; Bertolini, S.; Calandra, S.; Tarugi, P.; et al. Twelve Variants Polygenic Score for Low-Density Lipoprotein Cholesterol Distribution in a Large Cohort of Patients with Clinically Diagnosed Familial Hypercholesterolemia With or Without Causative Mutations. J. Am. Heart Assoc. 2022, 11, e023668. [Google Scholar] [CrossRef]
- Talmud, P.J.; Shah, S.; Whittall, R.; Futema, M.; Howard, P.; Cooper, J.A.; Harrison, S.C.; Li, K.; Drenos, F.; Karpe, F.; et al. Use of low-density lipoprotein cholesterol gene score to distinguish patients with polygenic and monogenic familial hypercholesterolaemia: A case-control study. Lancet 2013, 381, 1293–1301. [Google Scholar] [CrossRef]
- Trinder, M.; Li, X.; DeCastro, M.L.; Cermakova, L.; Sadananda, S.; Jackson, L.M.; Azizi, H.; Mancini, G.J.; Francis, G.A.; Frohlich, J.; et al. Risk of Premature Atherosclerotic Disease in Patients with Monogenic Versus Polygenic Familial Hypercholesterolemia. J. Am. Coll. Cardiol. 2019, 74, 512–522. [Google Scholar] [CrossRef]
- Todorovova, V.; Altschmiedova, T.; Vrablik, M.; Ceska, R. Familial Hypercholesterolemia: Real-World Data of 1236 Patients Attending a Czech Lipid Clinic. A Retrospective Analysis of Experience in More than 50 years. Part I: Genetics and Biochemical Parameters. Front. Genet. 2022, 13, 849008. [Google Scholar] [CrossRef]
- Benn, M.; Watts, G.F.; Tybjærg-Hansen, A.; Nordestgaard, B.G. Mutations causative of familial hypercholesterolaemia: Screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur. Heart J. 2016, 37, 1384–1394. [Google Scholar] [CrossRef]
- NCD Risk Factor Collaboration. Available online: https://ncdrisc.org/country-profile.html (accessed on 26 June 2023).
- De Cario, R.; Kura, A.; Suraci, S.; Magi, A.; Volta, A.; Marcucci, R.; Gori, A.M.; Pepe, G.; Giusti, B.; Sticchi, E. Sanger Validation of High-Throughput Sequencing in Genetic Diagnosis: Still the Best Practice? Front. Genet. 2020, 11, 592588. [Google Scholar] [CrossRef]
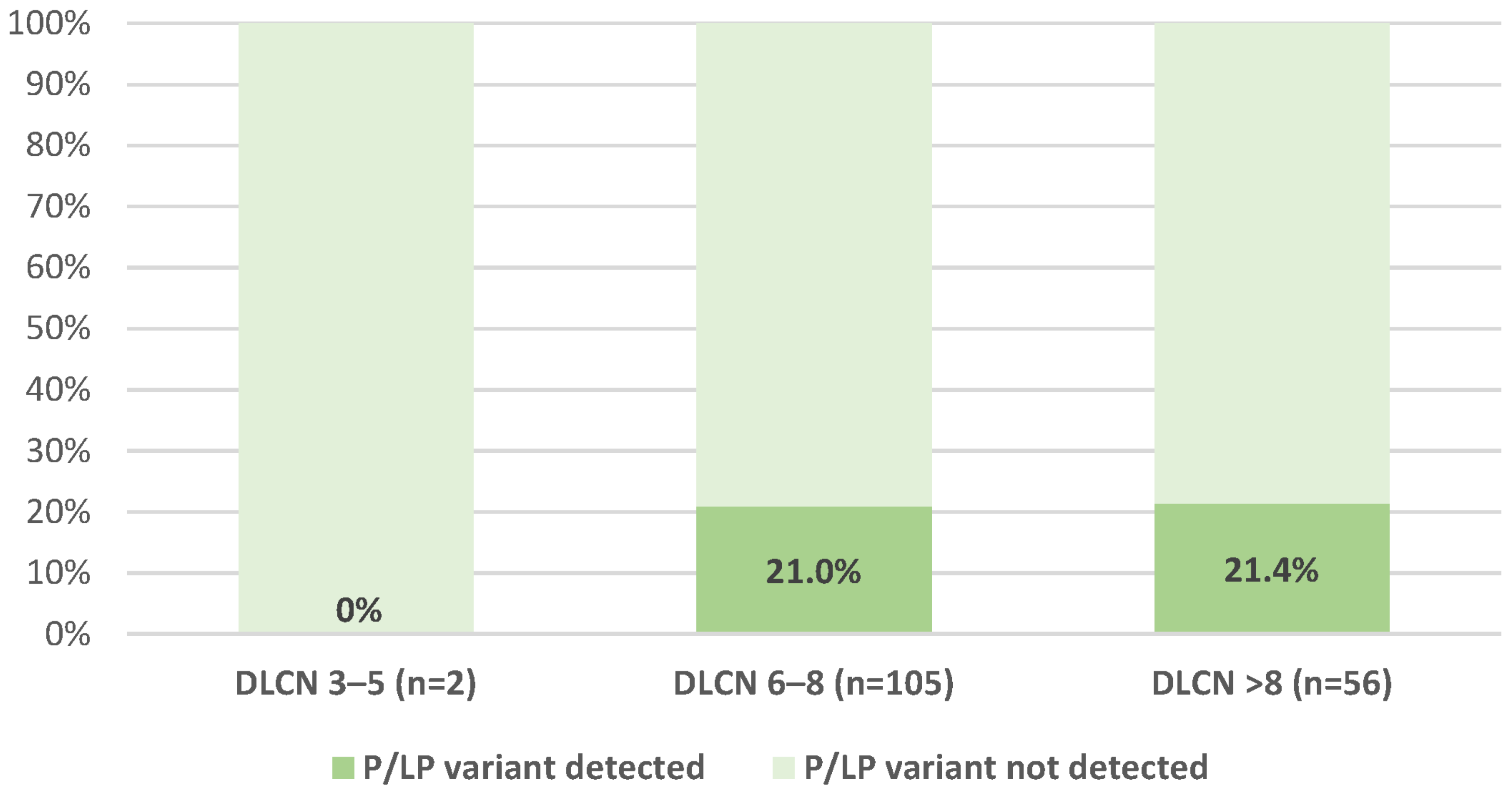
| All Subjects (n = 163) | |
|---|---|
| Men a | 53 (32.5%) |
| Age, years b | 52.91 ± 11.40 |
| High risk c | 24 (14.7%) |
| Very high risk d | 139 (85.3%) |
| Total cholesterol (highest documented, mmol/L) a | 9.82 ± 1.92 |
| LDL-cholesterol (mmol/L) b | |
| Highest documented | 7.47 ± 1.60 |
| At inclusion in the Registry | 5.43 ± 2.09 |
| Lowest on-treatment e | 3.30 ± 1.55 |
| At the latest follow-up | 4.30 ± 2.19 |
| Triglycerides (highest documented, mmol/) f | 1.78 (1.28–2.29) |
| HDL-cholesterol (lowest documented, mmol/L) b | 1.48 ± 0.38 |
| LLM at the latest follow-up | |
| Statin a | 116 (71.2%) |
| High-intensity statin a | 83 (50.9%) |
| Ezetimibe a | 49 (30.1%) |
| PCSK9 inhibitor a | 6 (3.7%) |
| FH diagnosis (based on DLCN criteria) g | |
| Definite FH a | 56 (34.4%) |
| Probable FH a | 105 (64.4%) |
| Possible FH a | 2 (1.2%) |
| CAD a | 102 (62.6%) |
| Premature CAD a | 74 (45.4%) |
| Arterial hypertension a | 59 (36.2%) |
| Diabetes mellitus a | 8 (4.9%) |
| Type 1 a | 1 (0.6%) |
| Type 2 a | 7 (4.3%) |
| BMI (kg/m2) b | 26.99 ± 4.11 |
| Obesity a,h | 32 (19.6%) |
| Smoking status | |
| Current or ex-smokers a | 59 (36.2%) |
| Non-smokers a | 104 (63.8%) |
| Tendon xanthomas a | 35 (21.5%) |
| Corneal arcus before age 45 a | 7 (4.3%) |
| Xanthelasms a | 9 (5.5%) |
| Family history of premature ASCVD a | 62 (38.0%) |
| Number | Gene | Variant | rsID | Clinvar ID | Classification (Chora et al., 2022) [10] | Heterozygous or Homozygous, VEP Consequence | Number of Cases |
|---|---|---|---|---|---|---|---|
| 1 | LDLR | g.11089559G>A | rs201016593 | 250973 | Pathogenic | Heterozygous, stop gained | 1 |
| c.11G>A | |||||||
| (p.Trp4*) | |||||||
| 2 | LDLR | g.11105333T>A | rs875989901, | 920596 | Likely pathogenic | Heterozygous, missense variant | 1 |
| c.427T>A | |||||||
| (p.Cys143Ser) | |||||||
| 3 | LDLR | g.11105436C>T | rs121908026 | 3686 | Pathogenic | Heterozygous, missense variant | 2 |
| c.530C>T | |||||||
| (p.Ser177Leu) | |||||||
| 4 | LDLR | g.11105572C>A | rs756613387 | 251364 | Pathogenic | Heterozygous, stop gained | 1 |
| c.666C>A | |||||||
| (p.Cys222*) | |||||||
| 5 | LDLR | g.11106668T>A | rs139043155 | 161287 | Pathogenic | Heterozygous, missense variant | 1 |
| c.798T>A | |||||||
| (p.Asp266Glu) | |||||||
| 6 | LDLR | g.11107484G>A | rs121908030 | 3692 | Pathogenic | Heterozygous, missense variant | 2 |
| c.910G>A (p.Asp304Asn) | |||||||
| 7 | LDLR | g.11110697G>A | rs761954844 | 226344 | Pathogenic | Heterozygous, missense variant | 6 |
| c.986G>A | |||||||
| (p.Cys329Tyr) | |||||||
| 8 | LDLR | g.11113313G>A | rs137943601 | 36453 | Likely pathogenic | Heterozygous, missense variant | 1 |
| c.1222G>A (p.Glu408Lys) | |||||||
| 9 | LDLR | g.11113376G>A | rs28942078 | 3694 | Pathogenic | Heterozygous, missense variant | 2 |
| c.1285G>A | |||||||
| (p.Val429Met) | |||||||
| 10 | LDLR | g.11116928G>A | rs137929307 | 161271 | Pathogenic | Heterozygous, missense variant | 1 |
| c.1775G>A (p.Gly592Glu) | |||||||
| 11 | LDLR | g.11120224C>T | rs193922569 | 36458 | Likely pathogenic | Heterozygous, stop gained | 1 |
| c.1978C>T | |||||||
| (p.Gln660*) | |||||||
| 12 | LDLR | g.11120380G>A | rs752935814 | 252161 | Pathogenic | Heterozygous, stop gained | 3 |
| c.1998G>A | |||||||
| (p.Trp666*) | |||||||
| 13 | LDLR | g.11105531T>G | rs1600711065 | 684864 | Likely pathogenic | Heterozygous, missense variant | 1 |
| c.625T>G | |||||||
| (p.Cys209Gly) | |||||||
| 14 | LDLR | g.11113383C>T | NA | CA404084995 | Likely pathogenic | Heterozygous, missense variant | 2 |
| c.1292C>T | |||||||
| (p.Ala431Val) | |||||||
| 15 | APOB | g.21006288C>T | rs5742904 | 17890 | Pathogenic a | Heterozygous, missense variant | 9 |
| c.10580G>A | |||||||
| (p.Arg3527Gln) |
| Gene with P/LP Variant | DLCN Group | ||
|---|---|---|---|
| Possible (n = 2) | Probable (n = 105) | Definite (n = 56) | |
| LDLR | 0 (0%) | 17 (16.2%) | 8 (14.3%) |
| APOB | 0 (0%) | 5 (4.8%) | 4 (7.1%) |
| LDL-Cholesterol | With P/LP Variants | Without P/LP Variants | p Value | |
|---|---|---|---|---|
| Highest documented, mmol/L (n = 163) | Mean ± SD Median (IQR) | 7.80 ± 1.81 7.42 (6.60–8.52) | 7.38 ± 1.54 7.00 (6.52–7.98) | 0.176 0.234 |
| Baseline at enrollment in the Registry, mmol/L (n = 163) | Mean ± SD | 5.65 ± 2.00 | 5.38 ± 2.11 | 0.490 |
| Median (IQR) | 5.46 (4.07–7.37) | 5.30 (3.74–6.72) | 0.472 | |
| Lowest/best on treatment, mmol/L (n = 127) a | (n = 25) | (n = 102) | ||
| Mean ± SD | 3.64 ± 1.54 | 3.21 ± 1.55 | 0.219 | |
| Median (IQR) | 3.39 (2.59–4.20) | 2.91 (1.97–4.21) | 0.158 | |
| Latest documented at follow-up, mmol/L (n = 163) | Mean ± SD | 4.77 ± 2.10 | 4.18 ± 2.20 | 0.162 |
| Median (IQR) | 4.40 (2.99–5.92) | 3.91 (2.37–5.73) | 0.102 | |
| Percent reduction from highest documented LDL-cholesterol to lowest/best on treatment in patients with at least one follow-up visit (n = 84) b | (n = 17) | (n = 67) | ||
| Mean ± SD | 54.91 ± 20.13 | 63.37 ± 14.48 | 0.051 | |
| Median (IQR) | 56.37 (48.45–67.80) | 66.47 (53.60–75.13) | 0.080 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Latkovskis, G.; Rescenko-Krums, R.; Nesterovics, G.; Briviba, M.; Saripo, V.; Gilis, D.; Terauda, E.; Meiere, R.; Skudrina, G.; Erglis, A.; et al. Genetic Characteristics of Latvian Patients with Familial Hypercholesterolemia: The First Analysis from Genome-Wide Sequencing. J. Clin. Med. 2023, 12, 5160. https://doi.org/10.3390/jcm12155160
Latkovskis G, Rescenko-Krums R, Nesterovics G, Briviba M, Saripo V, Gilis D, Terauda E, Meiere R, Skudrina G, Erglis A, et al. Genetic Characteristics of Latvian Patients with Familial Hypercholesterolemia: The First Analysis from Genome-Wide Sequencing. Journal of Clinical Medicine. 2023; 12(15):5160. https://doi.org/10.3390/jcm12155160
Chicago/Turabian StyleLatkovskis, Gustavs, Raimonds Rescenko-Krums, Georgijs Nesterovics, Monta Briviba, Vita Saripo, Dainus Gilis, Elizabete Terauda, Ruta Meiere, Gunda Skudrina, Andrejs Erglis, and et al. 2023. "Genetic Characteristics of Latvian Patients with Familial Hypercholesterolemia: The First Analysis from Genome-Wide Sequencing" Journal of Clinical Medicine 12, no. 15: 5160. https://doi.org/10.3390/jcm12155160
APA StyleLatkovskis, G., Rescenko-Krums, R., Nesterovics, G., Briviba, M., Saripo, V., Gilis, D., Terauda, E., Meiere, R., Skudrina, G., Erglis, A., Chora, J. R., Bourbon, M., & Klovins, J. (2023). Genetic Characteristics of Latvian Patients with Familial Hypercholesterolemia: The First Analysis from Genome-Wide Sequencing. Journal of Clinical Medicine, 12(15), 5160. https://doi.org/10.3390/jcm12155160






