Pivotal Clinical Study to Evaluate the Efficacy and Safety of Assistive Artificial Intelligence-Based Software for Cervical Cancer Diagnosis
Abstract
1. Introduction
2. Materials and Methods
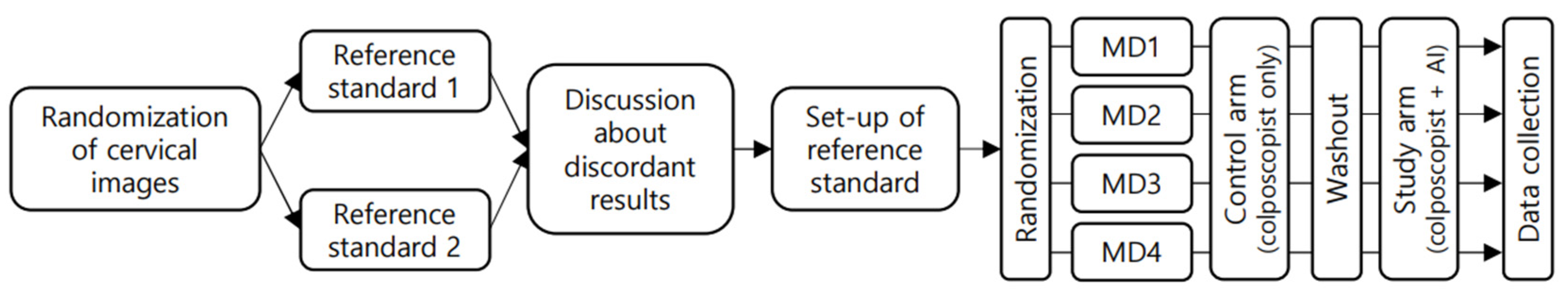
2.1. Study Design and Terminology
2.2. Preparation of Machine Learning System
2.3. Interpretation of Colposcopic Images
2.4. Statistical Analysis
3. Results
3.1. Patient and Disease Characteristics
3.2. Primary Endpoint
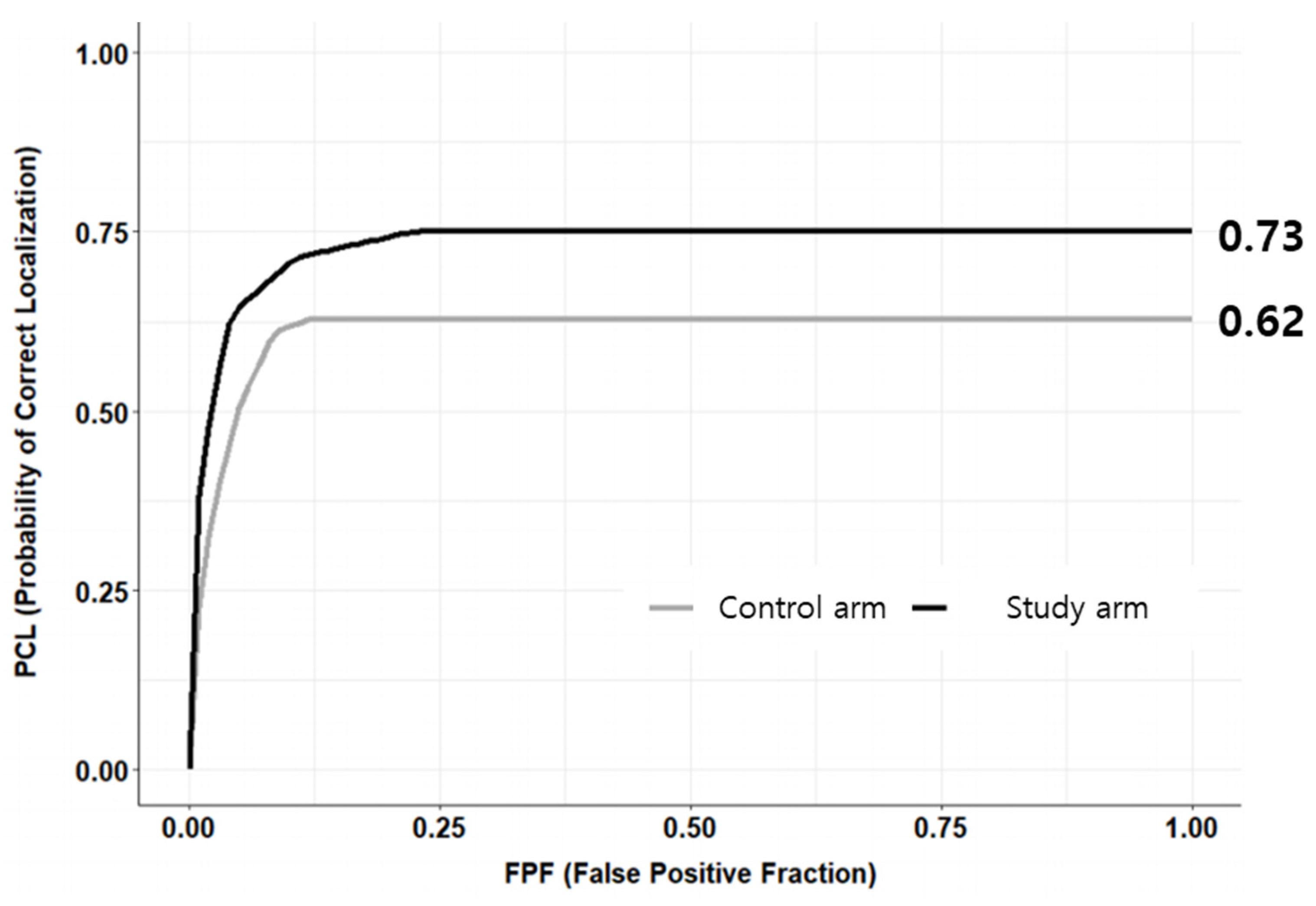
3.3. Secondary Endpoints
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- De Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.K.; Monk, B.J.; Brewer, C.; Keefe, A.K.; Osann, K.; McMeekin, S.; Rose, G.S.; Youssef, M.; Wilczynski, S.P.; Meyskens, F.L.; et al. HPV infection and number of lifetime sexual partners are strong predictors for ‘natural’ regression of CIN 2 and 3. Br. J. Cancer 2003, 89, 1062–1066. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO Guidelines for Treatment of Cervical Intraepithelial Neoplasia 2-3 and Adenocarcinoma In Situ: Cryotherapy, Large Loop Excision of the Transformation Zone, and Cold Knife Conization; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Bouvard, V.; Wentzensen, N.; Mackie, A.; Berkhof, J.; Brotherton, J.; Giorgi-Rossi, P.; Kupets, R.; Smith, R.; Arrossi, S.; Bendahhou, K.; et al. The IARC Perspective on Cervical Cancer Screening. N. Engl. J. Med. 2021, 385, 1908–1918. [Google Scholar] [CrossRef]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef]
- Hull, R.; Mbele, M.; Makhafola, T.; Hicks, C.; Wang, S.; Reis, R.M.; Mehrotra, R.; Mkhize-Kwitshana, Z.; Kibiki, G.; Bates, D.O.; et al. Cervical cancer in low and middle-income countries (Review). Oncol. Lett. 2020, 20, 2058–2074. [Google Scholar] [CrossRef]
- Hu, L.; Bell, D.; Antani, S.; Xue, Z.; Yu, K.; Horning, M.P.; Gachuhi, N.; Wilson, B.; Jaiswal, M.S.; Befano, B.; et al. An Observational Study of Deep Learning and Automated Evaluation of Cervical Images for Cancer Screening. J. Natl. Cancer Inst. 2019, 111, 923–932. [Google Scholar] [CrossRef]
- Gyawali, P.; Kc, S.; Ghimire, S. Role of Colposcopy in Detection of Dysplastic Cervical Lesion as a Screening Tool. J. Glob. Oncol. 2018, 4, 33s. [Google Scholar] [CrossRef]
- Barut, M.U.; Kale, A.; Kuyumcuoğlu, U.; Bozkurt, M.; Ağaçayak, E.; Özekinci, S.; Gül, T. Analysis of Sensitivity, Specificity, and Positive and Negative Predictive Values of Smear and Colposcopy in Diagnosis of Premalignant and Malignant Cervical Lesions. Med. Sci. Monit. 2015, 21, 3860–3867. [Google Scholar] [CrossRef]
- Stuebs, F.A.; Schulmeyer, C.E.; Mehlhorn, G.; Gass, P.; Kehl, S.; Renner, S.K.; Renner, S.P.; Geppert, C.; Adler, W.; Hartmann, A.; et al. Accuracy of colposcopy-directed biopsy in detecting early cervical neoplasia: A retrospective study. Arch. Gynecol. Obstet. 2019, 299, 525–532. [Google Scholar] [CrossRef]
- Giansanti, D. Artificial Intelligence in Public Health: Current Trends and Future Possibilities. Int. J. Environ. Res. Public Health 2022, 19, 11907. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Luo, G. Progressive sampling-based Bayesian optimization for efficient and automatic machine learning model selection. Health Inf. Sci. Syst. 2017, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Koul, A.; Singla, R.; Ijaz, M.F. Artificial intelligence in disease diagnosis: A systematic literature review, synthesizing framework and future research agenda. J. Ambient. Intell. Humaniz. Comput. 2022, 14, 8459–8486. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Bi, H.; Zhang, X.; Zhao, Y.; Dong, Y.; Luo, X.; Zhou, D.; You, Z.; Wu, Y.; Liu, Z.; et al. Artificial intelligence-assisted cytology for detection of cervical intraepithelial neoplasia or invasive cancer: A multicenter, clinical-based, observational study. Gynecol. Oncol. 2020, 159, 171–178. [Google Scholar] [CrossRef]
- Xue, P.; Tang, C.; Li, Q.; Li, Y.; Shen, Y.; Zhao, Y.; Chen, J.; Wu, J.; Li, L.; Wang, W.; et al. Development and validation of an artificial intelligence system for grading colposcopic impressions and guiding biopsies. BMC Med. 2020, 18, 406. [Google Scholar] [CrossRef]
- Kim, S.; Lee, H.; Lee, S.; Song, J.-Y.; Lee, J.-K.; Lee, N.-W. Role of Artificial Intelligence Interpretation of Colposcopic Images in Cervical Cancer Screening. Healthcare 2022, 10, 468. [Google Scholar] [CrossRef]
- Chakraborty, D.P.; Yoon, H.-J. Operating characteristics predicted by models for diagnostic tasks involving lesion localization. Med. Phys. 2008, 35, 435–445. [Google Scholar] [CrossRef]
- Sato, M.; Horie, K.; Hara, A.; Miyamoto, Y.; Kurihara, K.; Tomio, K.; Yokota, H. Application of deep learning to the classification of images from colposcopy. Oncol. Lett. 2018, 15, 3518–3523. [Google Scholar] [CrossRef]
- Simões, P.W.; Izumi, N.B.; Casagrande, R.S.; Venson, R.; Veronezi, C.D.; Moretti, G.P.; da Rocha, E.L.; Cechinel, C.; Ceretta, L.B.; Comunello, E.; et al. Classification of Images Acquired with Colposcopy Using Artificial Neural Networks. Cancer Inform. 2014, 13, 119–124. [Google Scholar] [CrossRef]
- Cho, B.-J.; Choi, Y.J.; Lee, M.-J.; Kim, J.H.; Son, G.-H.; Park, S.-H.; Kim, H.-B.; Joo, Y.-J.; Cho, H.-Y.; Kyung, M.S.; et al. Classification of cervical neoplasms on colposcopic photography using deep learning. Sci. Rep. 2020, 10, 13652. [Google Scholar] [CrossRef] [PubMed]
- Fan, A.; Wang, C.; Zhang, L.; Yan, Y.; Han, C.; Xue, F. Diagnostic value of the 2011 International Federation for Cervical Pathology and Colposcopy Terminology in predicting cervical lesions. Oncotarget 2018, 9, 9166–9176. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, M.F.; Schottenfeld, D.; Tortolero-Luna, G.; Cantor, S.B.; Richards-Kortum, R. Colposcopy for the diagnosis of squamous intraepithelial lesions: A meta-analysis. Obstet. Gynecol. 1998, 91, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, R.A.; Santesso, N.; Khatib, R.; Mustafa, A.A.; Wiercioch, W.; Kehar, R.; Gandhi, S.; Chen, Y.; Cheung, A.; Hopkins, J.; et al. Systematic reviews and meta-analyses of the accuracy of HPV tests, visual inspection with acetic acid, cytology, and colposcopy. Int. J. Gynecol. Obstet. 2016, 132, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Khongthip, Y.; Manchana, T.; Oranratanaphan, S.; Lertkhachonsuk, R. Learning curve in colposcopic training among gynecologic oncology fellows. Eur. J. Gynaecol. Oncol. 2019, 40, 647–651. [Google Scholar] [CrossRef]
| Reference Standard | |||||
|---|---|---|---|---|---|
| Negative | Positive | ||||
| Normal (n = 90) | CIN1 (n = 90) | CIN2/3 (n = 360) | CIN3+ (n = 346) | ||
| Histologic diagnosis | Benign (n = 89) | 86 | 2 | 1 | 0 |
| CIN1 (n = 89) | 0 | 76 | 12 | 1 | |
| CIN2/3 (n = 354) | 0 | 9 | 331 | 14 | |
| CIN3+ (n = 354) | 4 | 3 | 16 | 331 | |
| Negative Mean (SD) | Positive Mean (SD) | p-Value | ||
|---|---|---|---|---|
| Age (years) | 41.54 (12.98) | 41.65 (11.49) | 0.917 | |
| Parity | 1.18 (1.07) | 1.22 (1.05) | 0.646 | |
| HPV | Negative | 5 (2.78%) | 11 (1.56%) | 0.080 |
| Positive | 28 (15.56%) | 74 (10.48%) | ||
| Unknown | 147 (81.67%) | 621 (87.96%) | ||
| MD1 | MD2 | MD3 | MD4 | Total | ||
|---|---|---|---|---|---|---|
| Sensitivity | Control armed | 0.79 | 0.54 | 0.90 | 0.96 | 89.18 |
| (0.76, 0.82) | (0.51, 0.58) | (0.88, 0.92) | (0.95, 0.98) | (88.12, 90.24) | ||
| Study armed | 0.81 | 0.61 | 0.75 | 0.62 | 71.33 | |
| (0.78, 0.84) | (0.57, 0.65) | (0.72, 0.79) | (0.58, 0.65) | (69.69, 72.97) | ||
| p-value | 0.424 | 0.013 | <0.001 | <0.001 | <0.001 | |
| Specificity | Control armed | 0.96 | 0.99 | 0.89 | 0.78 | 96.68 |
| (0.93, 0.99) | (0.97, 1.00) | (0.85, 0.94) | (0.72, 0.84) | (95.42, 97.94) | ||
| Study armed | 0.91 | 0.95 | 0.92 | 0.88 | 92.16 | |
| (0.87, 0.95) | (0.92, 0.98) | (0.88, 0.96) | (0.83, 0.93) | (90.20, 94.11) | ||
| p-value | 0.052 | 0.032 | 0.471 | 0.012 | <0.001 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; An, H.; Cho, H.-W.; Min, K.-J.; Hong, J.-H.; Lee, S.; Song, J.-Y.; Lee, J.-K.; Lee, N.-W. Pivotal Clinical Study to Evaluate the Efficacy and Safety of Assistive Artificial Intelligence-Based Software for Cervical Cancer Diagnosis. J. Clin. Med. 2023, 12, 4024. https://doi.org/10.3390/jcm12124024
Kim S, An H, Cho H-W, Min K-J, Hong J-H, Lee S, Song J-Y, Lee J-K, Lee N-W. Pivotal Clinical Study to Evaluate the Efficacy and Safety of Assistive Artificial Intelligence-Based Software for Cervical Cancer Diagnosis. Journal of Clinical Medicine. 2023; 12(12):4024. https://doi.org/10.3390/jcm12124024
Chicago/Turabian StyleKim, Seongmin, Hyonggin An, Hyun-Woong Cho, Kyung-Jin Min, Jin-Hwa Hong, Sanghoon Lee, Jae-Yun Song, Jae-Kwan Lee, and Nak-Woo Lee. 2023. "Pivotal Clinical Study to Evaluate the Efficacy and Safety of Assistive Artificial Intelligence-Based Software for Cervical Cancer Diagnosis" Journal of Clinical Medicine 12, no. 12: 4024. https://doi.org/10.3390/jcm12124024
APA StyleKim, S., An, H., Cho, H.-W., Min, K.-J., Hong, J.-H., Lee, S., Song, J.-Y., Lee, J.-K., & Lee, N.-W. (2023). Pivotal Clinical Study to Evaluate the Efficacy and Safety of Assistive Artificial Intelligence-Based Software for Cervical Cancer Diagnosis. Journal of Clinical Medicine, 12(12), 4024. https://doi.org/10.3390/jcm12124024






