Exosomes Derived from Colon Cancer Cells Promote Tumor Progression and Affect the Tumor Microenvironment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Exosome Isolation and Treatment
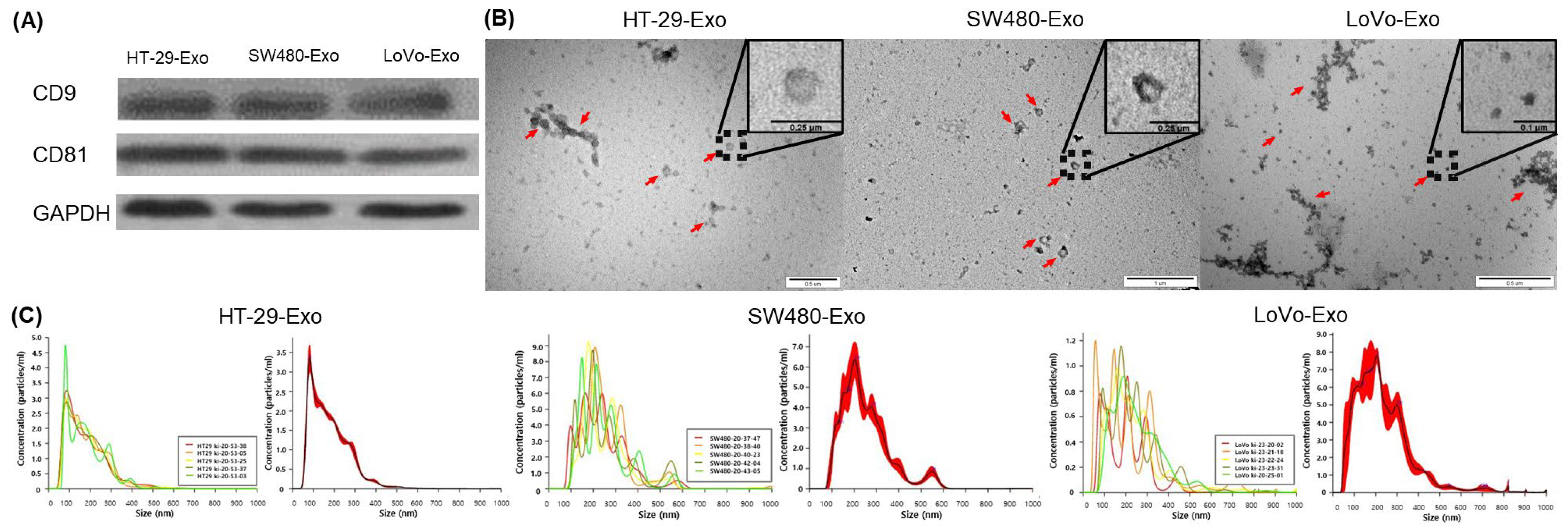
2.3. Immunoblotting
2.4. Transmission Electron Microscopy (TEM)
2.5. NanoSight Tracking Analysis (NTA)
2.6. MTT Assay
2.7. Wound-Healing Assay
2.8. Isolation and Culture of Cancer-Associated Fibroblasts (CAFs)
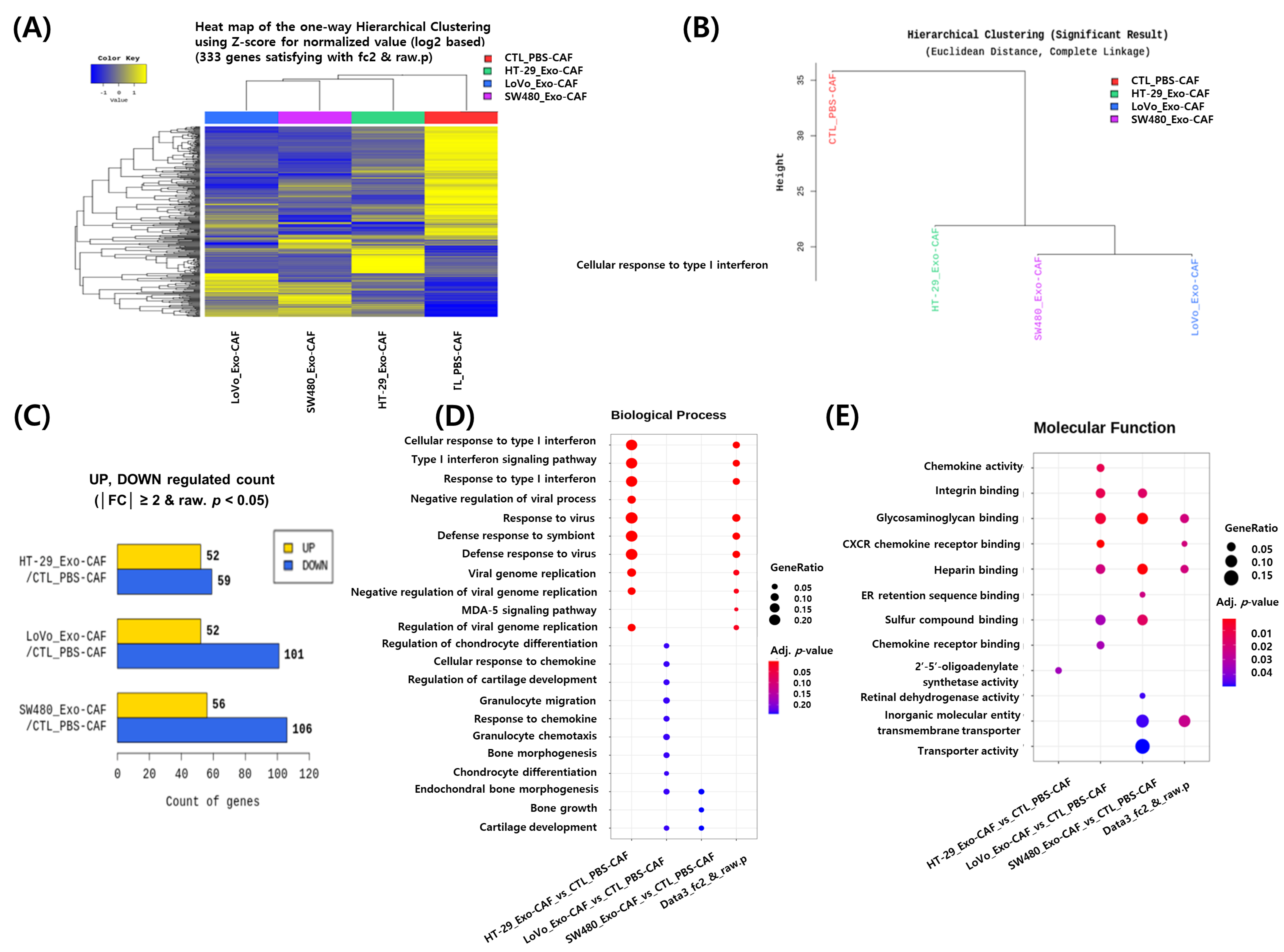
2.9. RNA Sequencing
2.10. Statistical Analysis
3. Results
3.1. Isolation of Colon Cancer-Cell-Derived Exosomes
3.2. Cellular Proliferation Induced by Colon Cancer-Cell-Derived Exosomes
3.3. EMT Induction by Colon Cancer-Cell-Derived Exosomes
3.4. RNA Sequencing Analysis of CAFs Treated with Colon Cancer Cell-Derived Exosomes
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, B.; Wang, Y.; Yan, Z.; Sun, Y.; Su, C. Colorectal cancer cell-derived exosomes promote proliferation and decrease apoptosis by activating the ERK pathway. Int. J. Clin. Exp. Pathol. 2019, 12, 2485–2495. [Google Scholar] [PubMed]
- Ruiz-López, L.; Blancas, I.; Garrido, J.M.; Mut-Salud, N.; Moya-Jódar, M.; Osuna, A.; Rodríguez-Serrano, F. The role of exosomes on colorectal cancer: A review. J. Gastroenterol. Hepatol. 2018, 33, 792–799. [Google Scholar] [CrossRef] [Green Version]
- Andaloussi, E.S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Ren, R.; Sun, H.; Ma, C.; Liu, J.; Wang, H. Colon cancer cells secrete exosomes to promote self-proliferation by shortening mitosis duration and activation of STAT3 in a hypoxic environment. Cell Biosci. 2019, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Barger, J.F.; Lovat, F.; Gao, M.; Otterson, G.A.; Nana-Sinkam, P. Lung cancer exosomes as drivers of epithelial mesenchymal transition. Oncotarget 2016, 7, 54852–54866. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. Tumor-derived exosomes and their role in cancer progression. Adv. Clin. Chem. 2016, 74, 103–141. [Google Scholar] [CrossRef] [Green Version]
- Dabitao, D.; Margolick, J.B.; Lopez, J.; Bream, J.H. Multiplex measurement of proinflammatory cytokines in human serum: Comparison of the Meso Scale Discovery electrochemiluminescence assay and the Cytometric Bead Array. J. Immunol. Methods 2011, 372, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.Y.; Ahn, K.S.; Kim, Y.H.; Kim, T.S.; Baek, W.K.; Suh, S.I.; Kang, K.J. Circulating microRNAs as biomarkers in bile-derived exosomes of cholangiocarcinoma. Ann. Surg. Treat. Res. 2021, 101, 140–150. [Google Scholar] [CrossRef]
- Kahlert, C.; Kalluri, R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef] [Green Version]
- Rak, J.; Guha, A. Extracellular vesicles--Vehicles that spread cancer genes. BioEssays 2012, 34, 489–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noh, G.T.; Kwon, J.; Kim, J.; Park, M.; Choi, D.W.; Cho, K.A.; Woo, S.Y.; Oh, B.Y.; Lee, K.Y.; Lee, R.A. Verification of the role of exosomal microRNA in colorectal tumorigenesis using human colorectal cancer cell lines. PLoS ONE 2020, 15, e0242057. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Pucci, F.; Pittet, M.J. Molecular pathways: Tumor-derived microvesicles and their interactions with immune cells in vivo. Clin. Cancer Res. 2013, 19, 2598–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Nedawi, K.; Meehan, B.; Micallef, J.; Lhotak, V.; May, L.; Guha, A.; Rak, J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008, 10, 619–624. [Google Scholar] [CrossRef]
- Harada, T.; Yamamoto, H.; Kishida, S.; Kishida, M.; Awada, C.; Takao, T.; Kikuchi, A. Wnt5b-associated exosomes promote cancer cell migration and proliferation. Cancer Sci. 2017, 108, 42–52. [Google Scholar] [CrossRef]
- Hong, B.S.; Cho, J.H.; Kim, H.; Choi, E.J.; Rho, S.; Kim, J.; Kim, J.H.; Choi, D.S.; Kim, Y.K.; Hwang, D.; et al. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genom. 2009, 10, 556. [Google Scholar] [CrossRef] [Green Version]
- Chiba, M.; Kimura, M.; Asari, S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol. Rep. 2012, 28, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Jutzy, J.M.; Aspe, J.R.; McGregor, D.W.; Neidigh, J.W.; Wall, N.R. Survivin is released from cancer cells via exosomes. Apoptosis. 2011, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Thiery, J.P. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Gopal, S.K.; Mathias, R.A.; Liu, L.; Sheng, J.; Zhu, H.J.; Simpson, R.J. Emerging roles of exosomes during epithelial-mesenchymal transition and cancer progression. Semin. Cell Dev. Biol. 2015, 40, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Bigagli, E.; Luceri, C.; Guasti, D.; Cinci, L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol. Ther. 2016, 17, 1062–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-cadherin and N-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef] [Green Version]
- Osman, N.H.; Jailani, R.F.; Rahman, H.A.; Hamid, N.A. Gene and protein expression of epithelial to mesenchymal transition for intestinal and anal fistula: A systematic review. Ann. Coloproctol. 2021, 39, 106–114. [Google Scholar] [CrossRef]
- Kim, W.K.; Kwon, Y.; Jang, M.; Park, M.; Kim, J.; Cho, S.; Jang, D.G.; Lee, W.B.; Jung, S.H.; Choi, H.J.; et al. β-catenin activation down-regulates cell-cell junction-related genes and induces epithelial-to-mesenchymal transition in colorectal cancers. Sci. Rep. 2019, 9, 18440. [Google Scholar] [CrossRef] [Green Version]
- Lucchetti, D.; Ricciardi Tenore, C.; Colella, F.; Sgambato, A. Extracellular vesicles and cancer: A focus on metabolism, cytokines, and immunity. Cancers 2020, 12, 171. [Google Scholar] [CrossRef] [Green Version]
- Lucchetti, D.; Zurlo, I.V.; Colella, F.; Ricciardi-Tenore, C.; Di Salvatore, M.; Tortora, G.; De Maria, R.; Giuliante, F.; Cassano, A.; Basso, M.; et al. Mutational status of plasma exosomal KRAS predicts outcome in patients with metastatic colorectal cancer. Sci. Rep. 2021, 11, 22686. [Google Scholar] [CrossRef]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schäfer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J.; et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef] [Green Version]
- Rana, S.; Malinowska, K.; Zöller, M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia 2013, 15, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Hu, T.; Hu, J. Melanoma-derived exosomes induce reprogramming fibroblasts into cancer-associated fibroblasts via Gm26809 delivery. Cell Cycle 2019, 18, 3085–3094. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.L.; Wang, W.; Lan, X.L.; Zeng, Z.C.; Liang, Y.S.; Yan, Y.R.; Song, F.Y.; Wang, F.F.; Zhu, X.H.; Liao, W.J.; et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol. Cancer 2019, 18, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Li, Y.; Zou, L.; Zhu, Z. Role of exosomes in crosstalk between cancer-associated fibroblasts and cancer cells. Front. Oncol. 2019, 9, 356. [Google Scholar] [CrossRef] [PubMed]
- Sha, M.; Jeong, S.; Qiu, B.J.; Tong, Y.; Xia, L.; Xu, N.; Zhang, J.J.; Xia, Q. Isolation of cancer-associated fibroblasts and its promotion to the progression of intrahepatic cholangiocarcinoma. Cancer Med. 2018, 7, 4665–4677. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Son, I.T.; Noh, G.T.; Woo, S.-Y.; Lee, R.-A.; Oh, B.Y. Exosomes Derived from Colon Cancer Cells Promote Tumor Progression and Affect the Tumor Microenvironment. J. Clin. Med. 2023, 12, 3905. https://doi.org/10.3390/jcm12123905
Kim M, Son IT, Noh GT, Woo S-Y, Lee R-A, Oh BY. Exosomes Derived from Colon Cancer Cells Promote Tumor Progression and Affect the Tumor Microenvironment. Journal of Clinical Medicine. 2023; 12(12):3905. https://doi.org/10.3390/jcm12123905
Chicago/Turabian StyleKim, Minsung, Il Tae Son, Gyoung Tae Noh, So-Youn Woo, Ryung-Ah Lee, and Bo Young Oh. 2023. "Exosomes Derived from Colon Cancer Cells Promote Tumor Progression and Affect the Tumor Microenvironment" Journal of Clinical Medicine 12, no. 12: 3905. https://doi.org/10.3390/jcm12123905
APA StyleKim, M., Son, I. T., Noh, G. T., Woo, S.-Y., Lee, R.-A., & Oh, B. Y. (2023). Exosomes Derived from Colon Cancer Cells Promote Tumor Progression and Affect the Tumor Microenvironment. Journal of Clinical Medicine, 12(12), 3905. https://doi.org/10.3390/jcm12123905






