Krüppel-like Factor 6 Suppresses the Progression of Pancreatic Cancer by Upregulating Activating Transcription Factor 3
Abstract
1. Introduction
2. Materials and Methods
2.1. PAAD Clinical Samples
2.2. Immunohistochemistry (IHC)
2.3. Cell Culture
2.4. RNA Extraction, Reverse Transcription, and Quantitative Polymerase Chain Reaction (qRT-PCR)
2.5. Western Blotting
2.6. Plasmid Construction, Lentiviral Transduction, and Generation of Stable Cell Lines
2.7. CCK8 Assay and Clone Formation Assay
2.8. Migration and Invasion Assays
2.9. Databases
2.10. Statistical Analysis
3. Results
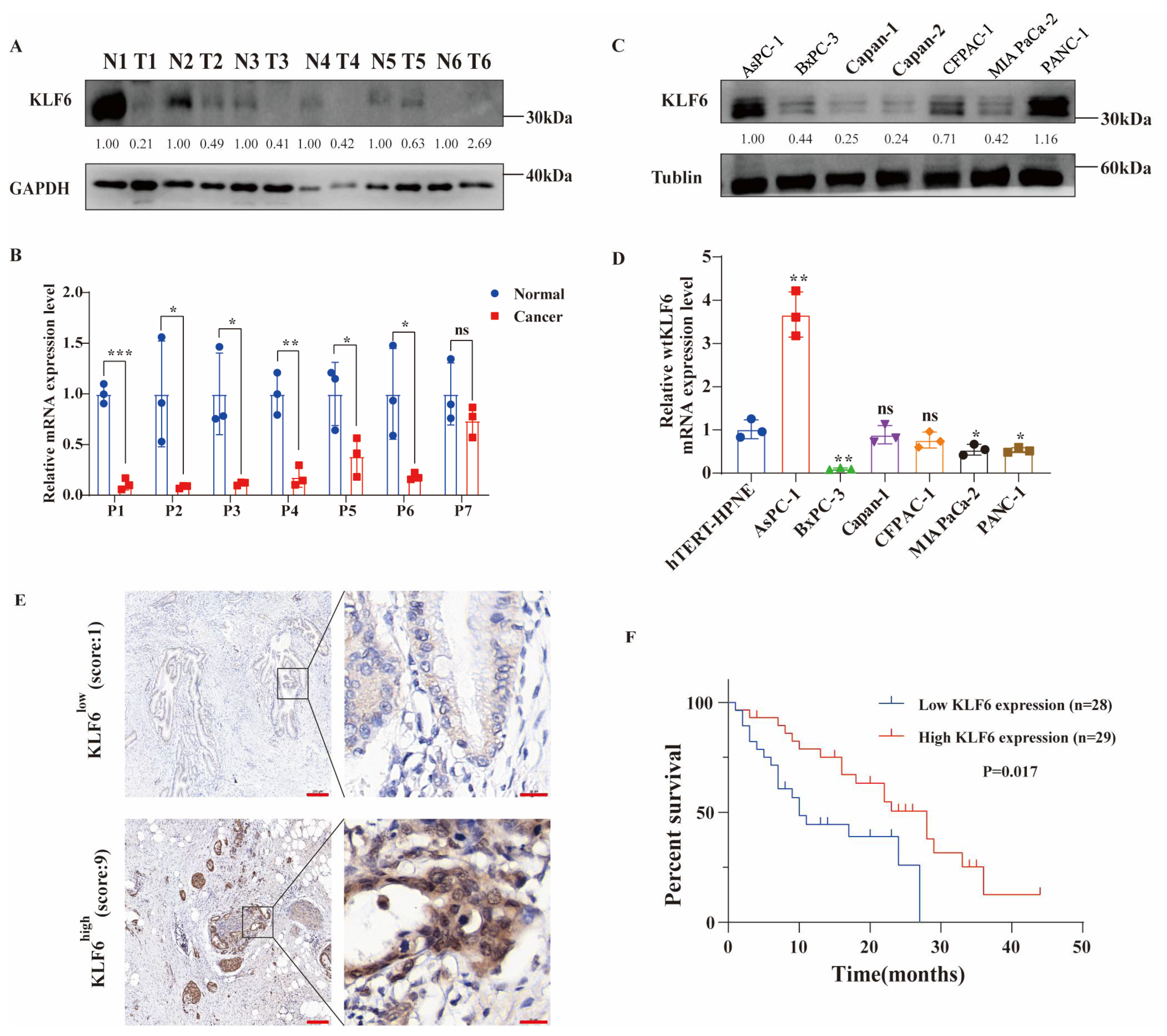
3.1. Evaluating the Expression Patterns and Prognostic Value of KLF6 in PAAD Patients and Pancreatic Cancer Cell Lines
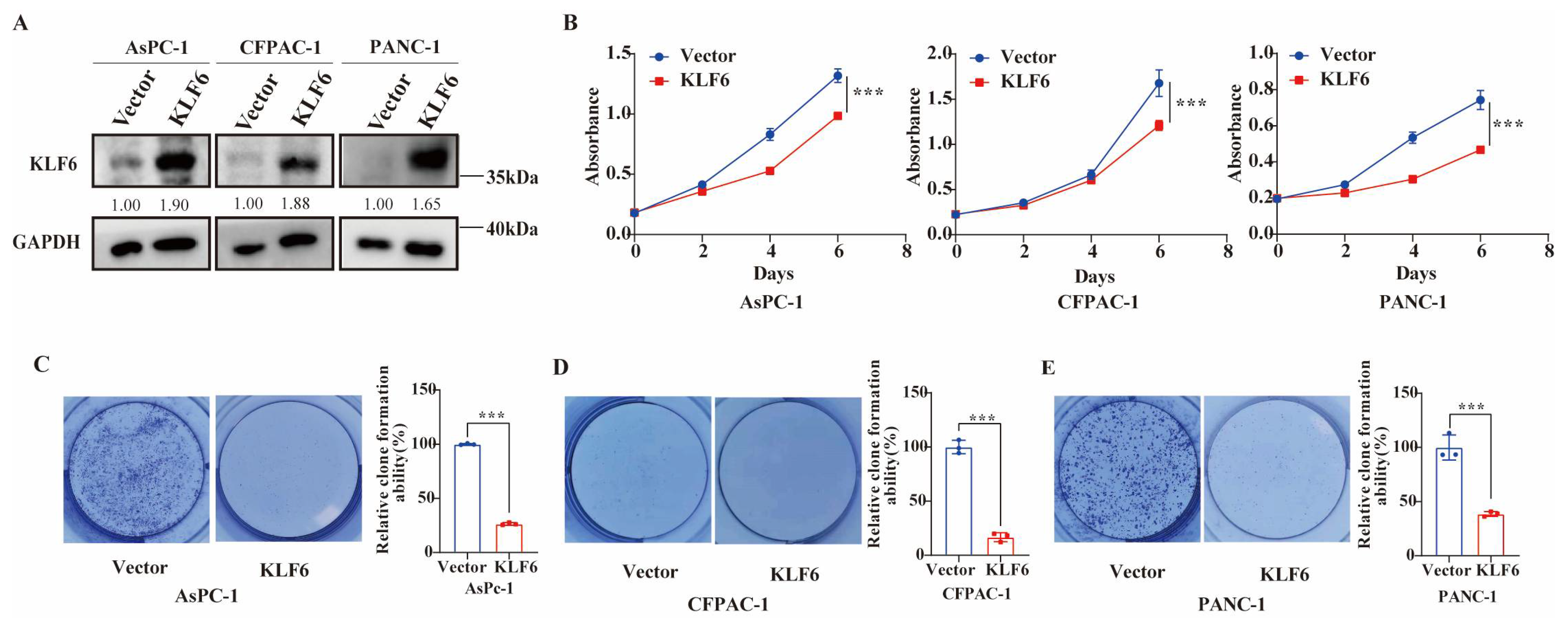
3.2. KLF6 Overexpression Inhibits Proliferation of Pancreatic Cancer Cells
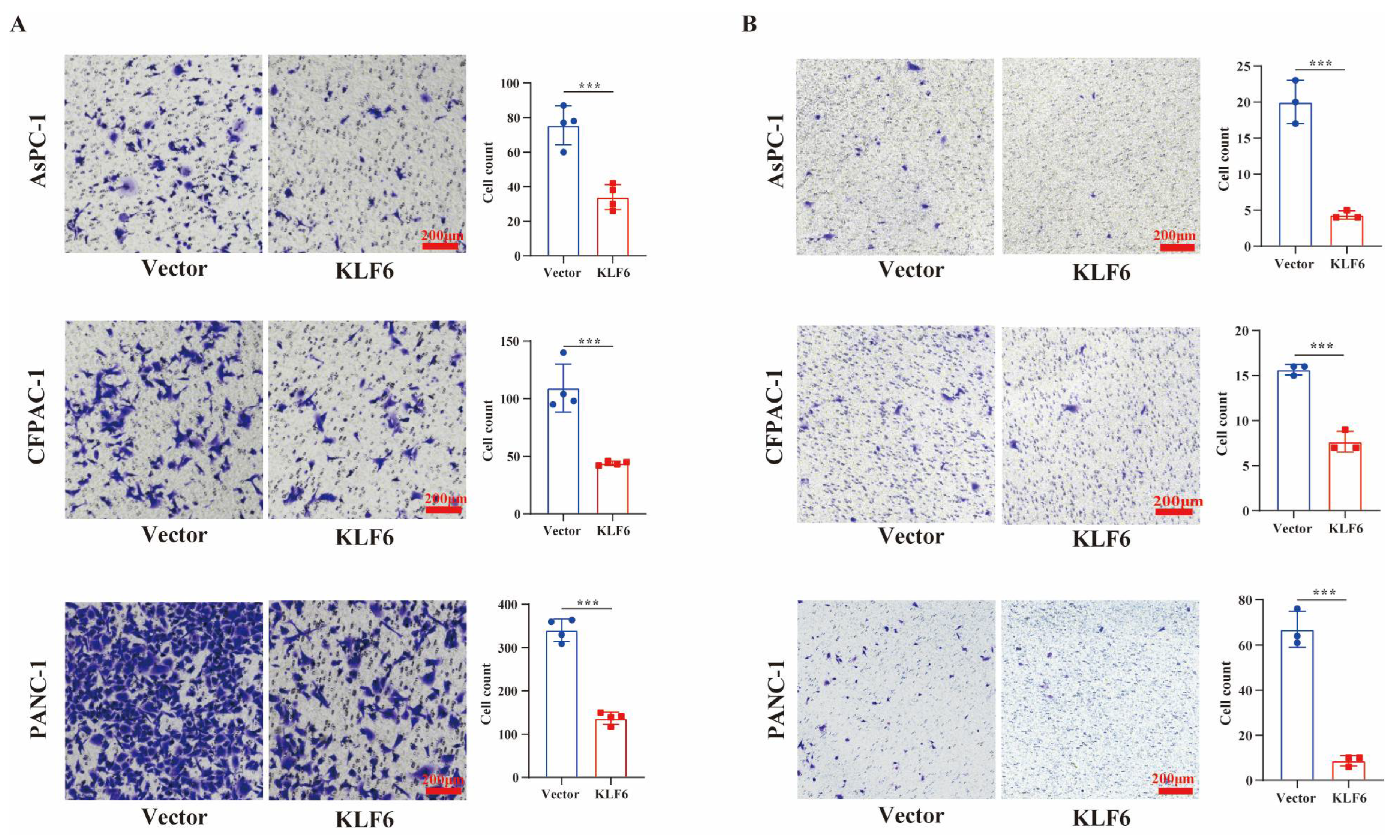
3.3. KLF6 Overexpression Inhibits Metastasis of Pancreatic Cancer Cells
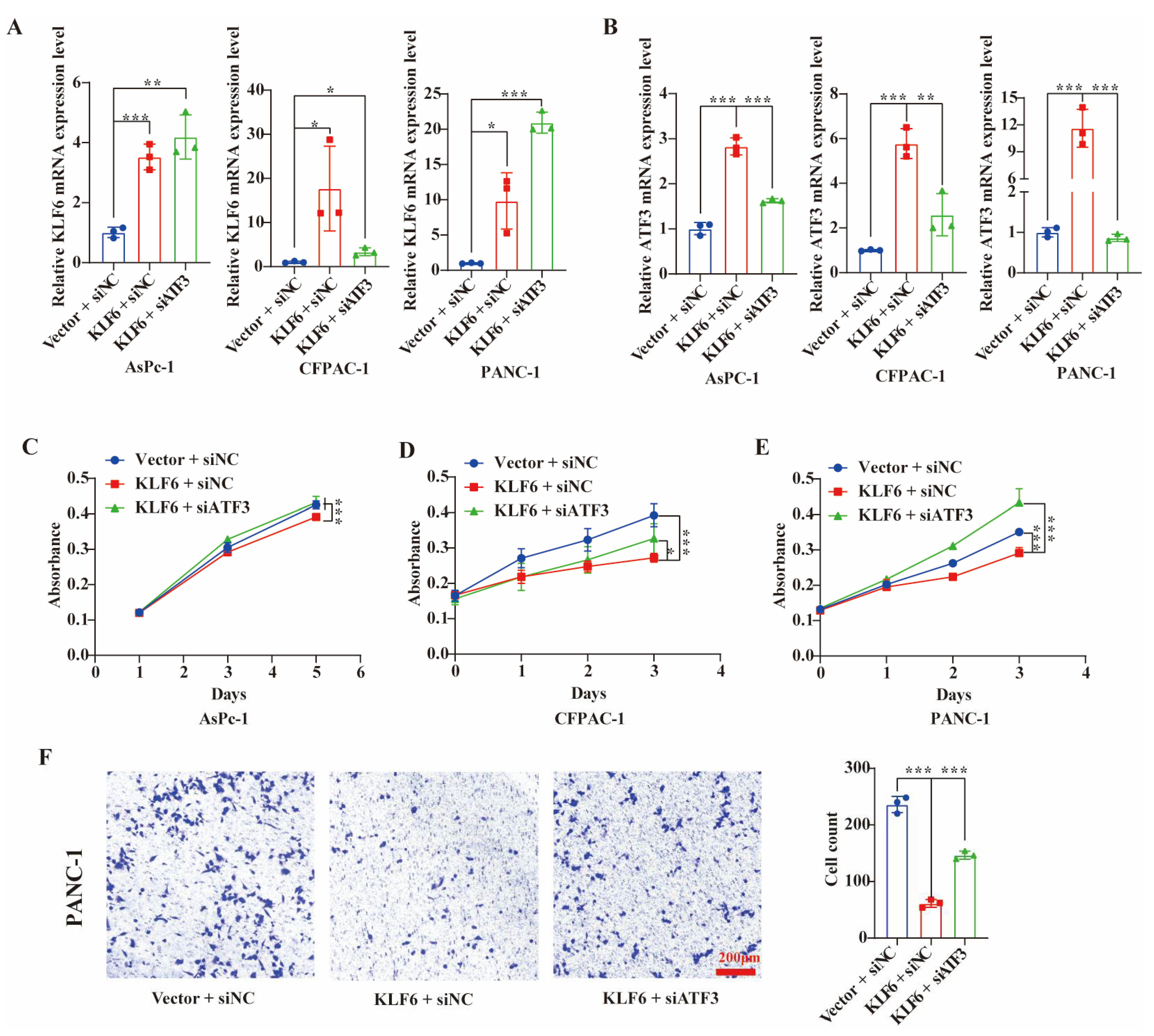
3.4. Overexpression of KLF6 Impairs EMT Progression and Works by Upregulating ATF3 in Pancreatic Cancer Cell Lines
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turner, J.; Crossley, M. Mammalian Kruppel-like transcription factors: More than just a pretty finger. Trends Biochem. Sci. 1999, 24, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Syafruddin, S.E.; Mohtar, M.A.; Nazarie, W.F.W.M.; Low, T.Y. Two Sides of the Same Coin: The Roles of KLF6 in Physiology and Pathophysiology. Biomolecules 2020, 10, 1378. [Google Scholar] [CrossRef] [PubMed]
- Leow, C.C.; Wang, B.E.; Ross, J.; Chan, S.M.; Zha, J.; Carano, R.A.; Frantz, G.; Shen, M.M.; de Sauvage, F.J.; Gao, W.Q. Prostate-specific Klf6 inactivation impairs anterior prostate branching morphogenesis through increased activation of the Shh pathway. J. Biol. Chem. 2009, 284, 21057–21065. [Google Scholar] [CrossRef] [PubMed]
- Laub, F.; Aldabe, R.; Ramirez, F.; Friedman, S. Embryonic expression of Kruppel-like factor 6 in neural and non-neural tissues. Mech. Dev. 2001, 106, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, N.; Kubo, A.; Liu, H.; Akita, K.; Laub, F.; Ramirez, F.; Keller, G.; Friedman, S.L. Developmental regulation of yolk sac hematopoiesis by Kruppel-like factor 6. Blood 2006, 107, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Date, D.; Das, R.; Narla, G.; Simon, D.I.; Jain, M.K.; Mahabeleshwar, G.H. Kruppel-like transcription factor 6 regulates inflammatory macrophage polarization. J. Biol. Chem. 2014, 289, 10318–10329. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.D.; Ng, H.P.; Chan, E.R.; Mahabeleshwar, G.H. Kruppel-like factor 6 promotes macrophage inflammatory and hypoxia response. FASEB J. 2020, 34, 3209–3223. [Google Scholar] [CrossRef]
- Sydor, S.; Manka, P.; Best, J.; Jafoui, S.; Sowa, J.P.; Zoubek, M.E.; Hernandez-Gea, V.; Cubero, F.J.; Kalsch, J.; Vetter, D.; et al. Kruppel-like factor 6 is a transcriptional activator of autophagy in acute liver injury. Sci. Rep. 2017, 7, 8119. [Google Scholar] [CrossRef]
- Andreoli, V.; Gehrau, R.C.; Bocco, J.L. Biology of Krüppel-like factor 6 transcriptional regulator in cell life and death. IUBMB Life 2010, 62, 896–905. [Google Scholar] [CrossRef]
- Narla, G.; Heath, K.E.; Reeves, H.L.; Li, D.; Giono, L.E.; Kimmelman, A.C.; Glucksman, M.J.; Narla, J.; Eng, F.J.; Chan, A.M.; et al. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 2001, 294, 2563–2566. [Google Scholar] [CrossRef]
- Ito, G.; Uchiyama, M.; Kondo, M.; Mori, S.; Usami, N.; Maeda, O.; Kawabe, T.; Hasegawa, Y.; Shimokata, K.; Sekido, Y. Kruppel-like factor 6 is frequently down-regulated and induces apoptosis in non-small cell lung cancer cells. Cancer Res. 2004, 64, 3838–3843. [Google Scholar] [CrossRef] [PubMed]
- Reeves, H.L.; Narla, G.; Ogunbiyi, O.; Haq, A.I.; Katz, A.; Benzeno, S.; Hod, E.; Harpaz, N.; Goldberg, S.; Tal-Kremer, S.; et al. Kruppel-like factor 6 (KLF6) is a tumor-suppressor gene frequently inactivated in colorectal cancer. Gastroenterology 2004, 126, 1090–1103. [Google Scholar] [CrossRef] [PubMed]
- Kremer-Tal, S.; Narla, G.; Chen, Y.; Hod, E.; DiFeo, A.; Yea, S.; Lee, J.S.; Schwartz, M.; Thung, S.N.; Fiel, I.M.; et al. Downregulation of KLF6 is an early event in hepatocarcinogenesis, and stimulates proliferation while reducing differentiation. J. Hepatol. 2007, 46, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Vanegas, O.; Narla, G.; Teixeira, M.S.; DiFeo, A.; Misra, A.; Singh, G.; Chan, A.M.; Friedman, S.L.; Feuerstein, B.G.; Martignetti, J.A. Functional inactivation of the KLF6 tumor suppressor gene by loss of heterozygosity and increased alternative splicing in glioblastoma. Int. J. Cancer 2007, 121, 1390–1395. [Google Scholar] [CrossRef] [PubMed]
- DiFeo, A.; Narla, G.; Hirshfeld, J.; Camacho-Vanegas, O.; Narla, J.; Rose, S.L.; Kalir, T.; Yao, S.; Levine, A.; Birrer, M.J.; et al. Roles of KLF6 and KLF6-SV1 in ovarian cancer progression and intraperitoneal dissemination. Clin. Cancer Res. 2006, 12, 3730–3739. [Google Scholar] [CrossRef]
- Kimmelman, A.C.; Qiao, R.F.; Narla, G.; Banno, A.; Lau, N.; Bos, P.D.; Nunez Rodriguez, N.; Liang, B.C.; Guha, A.; Martignetti, J.A.; et al. Suppression of glioblastoma tumorigenicity by the Kruppel-like transcription factor KLF6. Oncogene 2004, 23, 5077–5083. [Google Scholar] [CrossRef]
- Sangodkar, J.; Shi, J.; DiFeo, A.; Schwartz, R.; Bromberg, R.; Choudhri, A.; McClinch, K.; Hatami, R.; Scheer, E.; Kremer-Tal, S.; et al. Functional role of the KLF6 tumour suppressor gene in gastric cancer. Eur. J. Cancer 2009, 45, 666–676. [Google Scholar] [CrossRef]
- Hsu, L.S.; Huang, R.H.; Lai, H.W.; Hsu, H.T.; Sung, W.W.; Hsieh, M.J.; Wu, C.Y.; Lin, Y.M.; Chen, M.K.; Lo, Y.S.; et al. KLF6 inhibited oral cancer migration and invasion via downregulation of mesenchymal markers and inhibition of MMP-9 activities. Int. J. Med. Sci. 2017, 14, 530–535. [Google Scholar] [CrossRef]
- DeKelver, R.C.; Lewin, B.; Lam, K.; Komeno, Y.; Yan, M.; Rundle, C.; Lo, M.C.; Zhang, D.E. Cooperation between RUNX1-ETO9a and novel transcriptional partner KLF6 in upregulation of Alox5 in acute myeloid leukemia. PLoS Genet. 2013, 9, e1003765. [Google Scholar] [CrossRef]
- Sirach, E.; Bureau, C.; Peron, J.M.; Pradayrol, L.; Vinel, J.P.; Buscail, L.; Cordelier, P. KLF6 transcription factor protects hepatocellular carcinoma-derived cells from apoptosis. Cell Death Differ. 2007, 14, 1202–1210. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.F.; Xiong, Q.L.; Yang, Y.; Weng, N.N.; Li, J.J.; Liu, J.L.; Yang, X.J.; Zeng, Z.; Zhang, Z.W.; Zhu, Q. Serum Nardilysin as a Prognostic Biomarker in Pancreatic Ductal Adenocarcinoma. J. Clin. Med. 2022, 11, 3101. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic. Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Miyaki, M.; Yamaguchi, T.; Iijima, T.; Funata, N.; Mori, T. Difference in the role of loss of heterozygosity at 10p15 (KLF6 locus) in colorectal carcinogenesis between sporadic and familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer patients. Oncology 2006, 71, 131–135. [Google Scholar] [CrossRef]
- Bureau, C.; Peron, J.M.; Bouisson, M.; Danjoux, M.; Selves, J.; Bioulac-Sage, P.; Balabaud, C.; Torrisani, J.; Cordelier, P.; Buscail, L.; et al. Hepatology—Expression of the transcription factor Klf6 in cirrhosis, macronodules, and hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2008, 23, 78–86. [Google Scholar] [CrossRef]
- Yea, S.; Narla, G.; Zhao, X.; Garg, R.; Tal-Kremer, S.; Hod, E.; Villanueva, A.; Loke, J.; Tarocchi, M.; Akita, K.; et al. Ras promotes growth by alternative splicing-mediated inactivation of the KLF6 tumor suppressor in hepatocellular carcinoma. Gastroenterology 2008, 134, 1521–1531. [Google Scholar] [CrossRef]
- Boyault, S.; Herault, A.; Balabaud, C.; Zucman-Rossi, J. Absence of KLF6 gene mutation in 71 hepatocellular carcinomas. Hepatology 2005, 41, 681–682. [Google Scholar] [CrossRef]
- Koivisto, P.A.; Zhang, X.H.; Sallinen, S.L.; Sallinen, P.; Helin, H.J.; Dong, J.T.; Van Meir, E.G.; Haapasalo, H.; Hyytinen, E.R. Absence of KLF6 gene mutations in human astrocytic tumors and cell lines. Int. J. Cancer 2004, 111, 642–643. [Google Scholar] [CrossRef] [PubMed]
- Lievre, A.; Landi, B.; Cote, J.F.; Veyrie, N.; Zucman-Rossi, J.; Berger, A.; Laurent-Puig, P. Absence of mutation in the putative tumor-suppressor gene KLF6 in colorectal cancers. Oncogene 2005, 24, 7253–7256. [Google Scholar] [CrossRef] [PubMed]
- Kohler, B.; Wolter, M.; Blaschke, B.; Reifenberger, G. Absence of mutations in the putative tumor suppressor gene KLF6 in glioblastomas and meningiomas. Int. J. Cancer 2004, 111, 644–645. [Google Scholar] [CrossRef] [PubMed]
- Agell, L.; Hernandez, S.; de Muga, S.; Lorente, J.A.; Juanpere, N.; Esgueva, R.; Serrano, S.; Gelabert, A.; Lloreta, J. KLF6 and TP53 mutations are a rare event in prostate cancer: Distinguishing between Taq polymerase artifacts and true mutations. Mod. Pathol. 2008, 21, 1470–1478. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Narla, G.; DiFeo, A.; Yao, S.; Banno, A.; Hod, E.; Reeves, H.L.; Qiao, R.F.; Camacho-Vanegas, O.; Levine, A.; Kirschenbaum, A.; et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005, 65, 5761–5768. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, J.Y.; Lu, L.J.; Wang, C.H.; Wang, L.H. MiR-630 promotes epithelial ovarian cancer proliferation and invasion via targeting KLF6. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4542–4547. [Google Scholar]
- Britschgi, A.; Trinh, E.; Rizzi, M.; Jenal, M.; Ress, A.; Tobler, A.; Fey, M.F.; Helin, K.; Tschan, M.P. DAPK2 is a novel E2F1/KLF6 target gene involved in their proapoptotic function. Oncogene 2008, 27, 5706–5716. [Google Scholar] [CrossRef]
- Li, D.; Yea, S.; Li, S.; Chen, Z.; Narla, G.; Banck, M.; Laborda, J.; Tan, S.; Friedman, J.M.; Friedman, S.L.; et al. Krüppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J. Biol. Chem. 2005, 280, 26941–26952. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.; Idelman, G.; Plymate, S.R.; Narla, G.; Friedman, S.L.; Werner, H. Transcriptional activation of the insulin-like growth factor I receptor gene by the Kruppel-like factor 6 (KLF6) tumor suppressor protein: Potential interactions between KLF6 and p53. Endocrinology 2004, 145, 3769–3777. [Google Scholar] [CrossRef]
- Hai, T.; Wolfgang, C.D.; Marsee, D.K.; Allen, A.E.; Sivaprasad, U. ATF3 and stress responses. Gene Expr. 1999, 7, 321–335. [Google Scholar]
- Huang, X.; Li, X.; Guo, B. KLF6 induces apoptosis in prostate cancer cells through up-regulation of ATF3. J. Biol. Chem. 2008, 283, 29795–29801. [Google Scholar] [CrossRef] [PubMed]

| Parameters | n | Low Expression | High Expression | p Value | |
|---|---|---|---|---|---|
| Number of patients | 57 | 28 | 29 | ||
| Age | ≤60 | 28 | 14 | 14 | 0.896 |
| >60 | 29 | 14 | 15 | ||
| Sex | Male | 36 | 19 | 17 | 0.470 |
| Female | 21 | 9 | 12 | ||
| TNM stage | I-II | 54 | 26 | 28 | 0.611 |
| III-IV | 3 | 2 | 1 | ||
| Grade | Well and moderate | 25 | 10 | 15 | 0.223 |
| Poor | 32 | 18 | 14 | ||
| Diabetes | Yes | 11 | 5 | 6 | 0.786 |
| No | 46 | 23 | 23 | ||
| Smoke | Yes | 22 | 13 | 9 | 0.233 |
| No | 35 | 15 | 20 | ||
| Drinking | Yes | 17 | 12 | 5 | 0.035 * |
| No | 40 | 16 | 24 | ||
| Preoperative CA19-9 value | ≥37 | 47 | 24 | 23 | 0.525 |
| <37 | 10 | 4 | 6 | ||
| Preoperative CEA value | ≥5 | 18 | 8 | 10 | 0.631 |
| <5 | 39 | 20 | 19 | ||
| Variable | Hazard Ratio | 95% Confidence Interval | p Value |
|---|---|---|---|
| KLF6 expression (low/high) | 2.431 | 1.166–5.070 | 0.017 * |
| Age (≤60/>60) | 0.819 | 0.424–1.581 | 0.552 |
| Gender (female/male) | 0.605 | 0.296–1.236 | 0.168 |
| TNM stage (I–II/III–IV) | 0.503 | 0.150–1.689 | 0.266 |
| Grade (poor/moderate and well) | 2.316 | 1.138–4.711 | 0.020 * |
| Preoperative CA19-9 value (<37/≥37) | 0.556 | 0.213–1.450 | 0.230 |
| Preoperative CEA value (<5/≥5) | 0.570 | 0.272–1.192 | 0.135 |
| Variables | Hazard Ratio | 95.0% CI | p Value |
|---|---|---|---|
| TNM stage | 0.330 | 0.094–1.163 | 0.085 |
| Grade | 2.313 | 1.103–4.850 | 0.026 * |
| KLF6 expression | 2.254 | 1.068–4.757 | 0.033 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Q.; Zhang, Z.; Yang, Y.; Xu, Y.; Zhou, Y.; Zhang, S.; Liu, J.; Zheng, Y.; Zhu, Q. Krüppel-like Factor 6 Suppresses the Progression of Pancreatic Cancer by Upregulating Activating Transcription Factor 3. J. Clin. Med. 2023, 12, 200. https://doi.org/10.3390/jcm12010200
Xiong Q, Zhang Z, Yang Y, Xu Y, Zhou Y, Zhang S, Liu J, Zheng Y, Zhu Q. Krüppel-like Factor 6 Suppresses the Progression of Pancreatic Cancer by Upregulating Activating Transcription Factor 3. Journal of Clinical Medicine. 2023; 12(1):200. https://doi.org/10.3390/jcm12010200
Chicago/Turabian StyleXiong, Qunli, Zhiwei Zhang, Yang Yang, Yongfeng Xu, Ying Zhou, Su Zhang, Jinlu Liu, Ying Zheng, and Qing Zhu. 2023. "Krüppel-like Factor 6 Suppresses the Progression of Pancreatic Cancer by Upregulating Activating Transcription Factor 3" Journal of Clinical Medicine 12, no. 1: 200. https://doi.org/10.3390/jcm12010200
APA StyleXiong, Q., Zhang, Z., Yang, Y., Xu, Y., Zhou, Y., Zhang, S., Liu, J., Zheng, Y., & Zhu, Q. (2023). Krüppel-like Factor 6 Suppresses the Progression of Pancreatic Cancer by Upregulating Activating Transcription Factor 3. Journal of Clinical Medicine, 12(1), 200. https://doi.org/10.3390/jcm12010200






