Endometrial Cancer Patient-Derived Xenograft Models: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protocol and Registration
2.2. Information Sources and Search Strategies
2.3. Eligibility Criteria
2.4. Study Selection
2.5. Data Extraction and Synthesis
2.6. Quality Assessment
3. Results
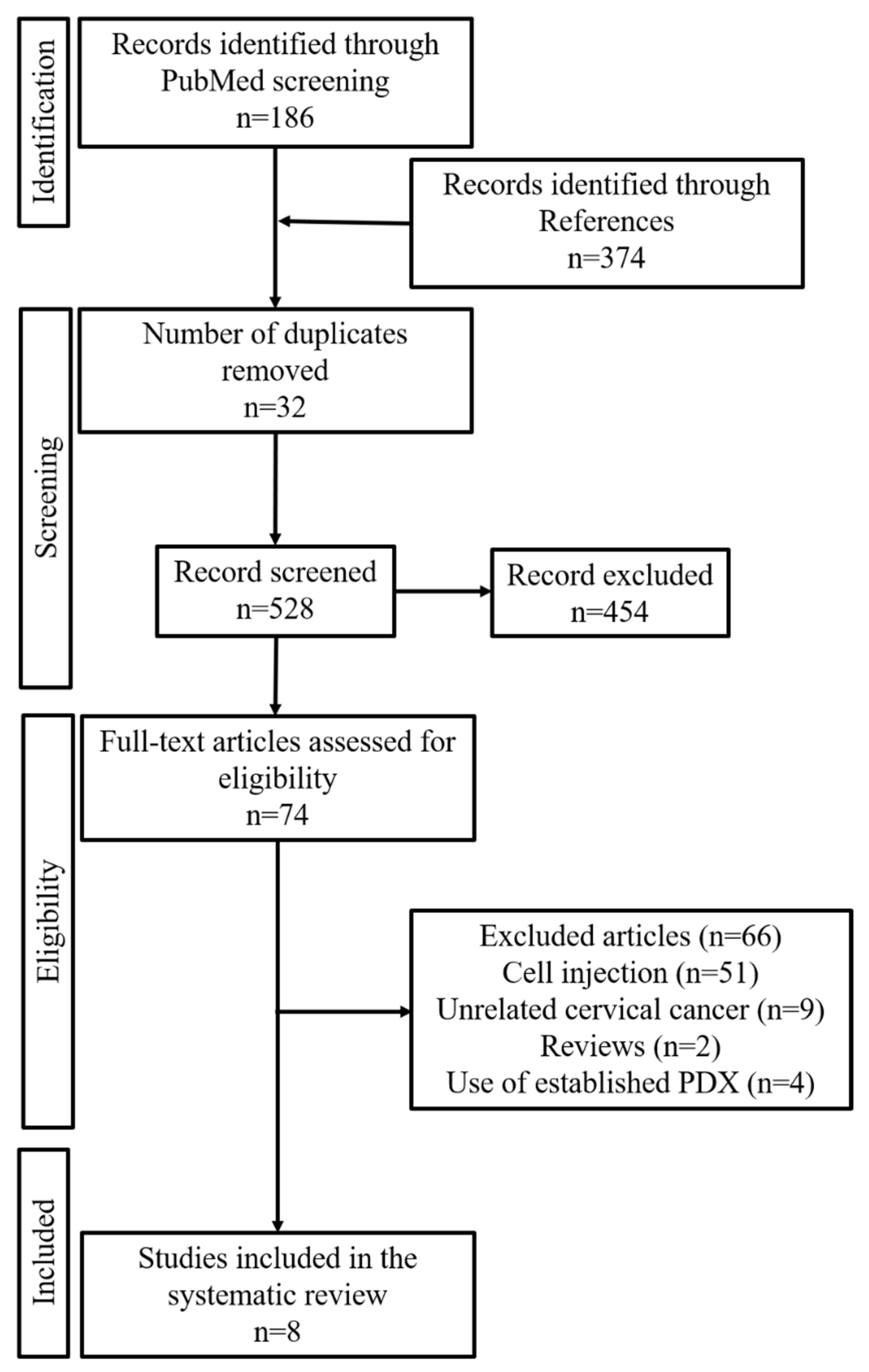
3.1. Study Selection
3.2. General Features of EC-PDX Models
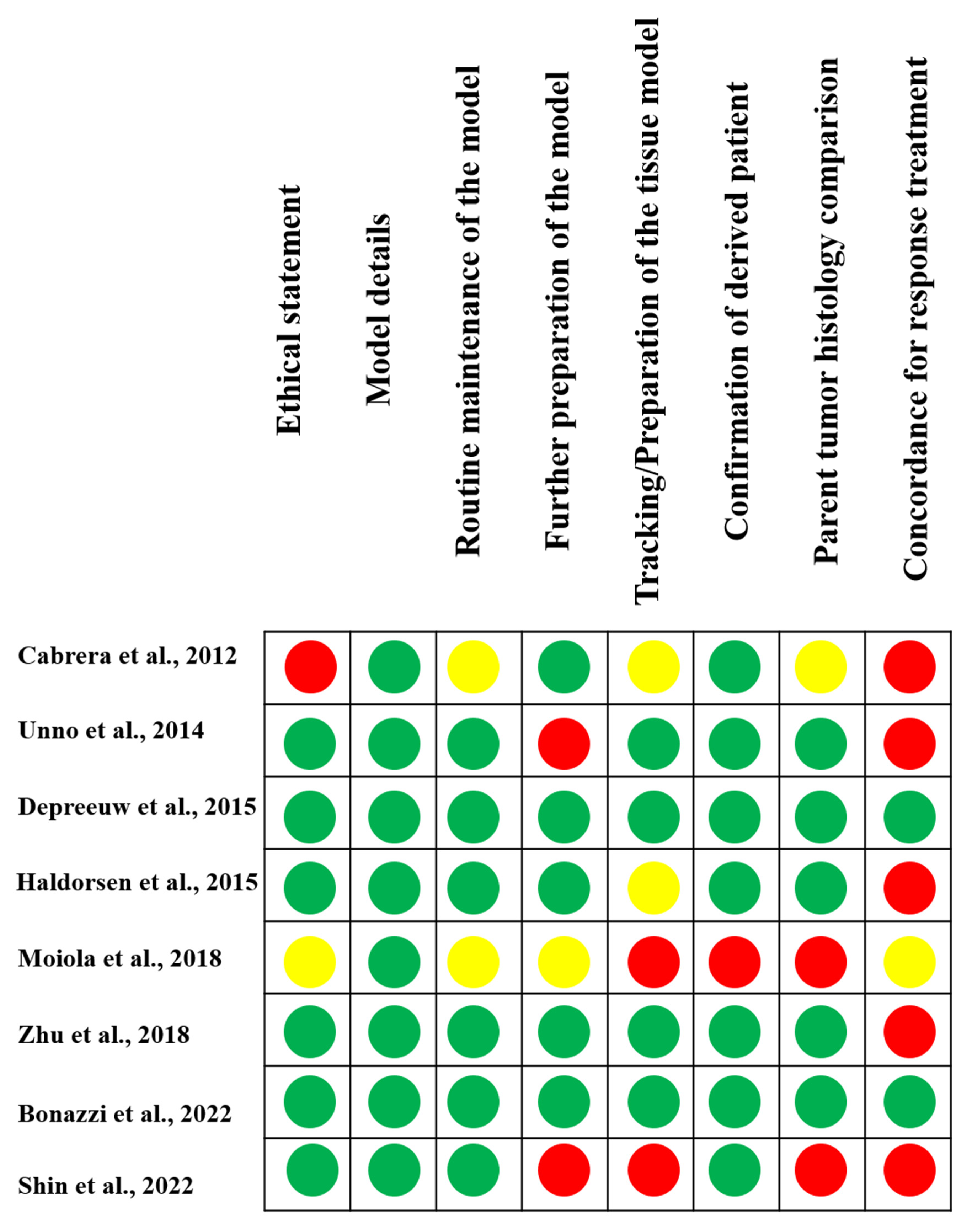
3.3. Quality Assessment
4. Discussion
4.1. Success Rate and Transplantation Method
4.2. Comparison of Original Tumors and PDXs
4.3. Implication for Further Research and Research Practice
4.3.1. Drug Repositioning
4.3.2. Precision Medicine
4.3.3. Mini-PDX Models
4.3.4. PDX Models and Co-Clinical Trials
4.3.5. Identifying Tumor Biomarkers
4.3.6. Humanized PDX Models for Immunotherapy
5. Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tanaka, T.; Ueda, S.; Miyamoto, S.; Terada, S.; Konishi, H.; Kogata, Y.; Fujiwara, S.; Tanaka, Y.; Taniguchi, K.; Komura, K.; et al. Oncologic outcomes for patients with endometrial cancer who received minimally invasive surgery: A retrospective observational study. Int. J. Clin. Oncol. 2020, 25, 1985–1994. [Google Scholar] [CrossRef]
- Tanaka, T.; Yamashita, S.; Kuroboshi, H.; Kamibayashi, J.; Sugiura, A.; Yoriki, K.; Mori, T.; Tanaka, K.; Nagashima, A.; Maeda, M.; et al. Oncologic outcomes in elderly patients who underwent hysterectomy for endometrial cancer: A multi-institutional survey in Kinki District, Japan. Int. J. Clin. Oncol. 2022. [Google Scholar] [CrossRef]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Qin, T.; Huang, Y.; Li, Y.; Chen, G.; Sun, C. Drug screening model meets cancer organoid technology. Transl. Oncol. 2020, 13, 100840. [Google Scholar] [CrossRef]
- Harrison, R.K. Phase II and phase III failures: 2013-2015. Nat. Rev. Drug Discov. 2016, 15, 817–818. [Google Scholar] [CrossRef]
- Tanaka, T.; Nishie, R.; Ueda, S.; Miyamoto, S.; Hashida, S.; Konishi, H.; Terada, S.; Kogata, Y.; Sasaki, H.; Tsunetoh, S.; et al. Patient-Derived Xenograft Models in Cervical Cancer: A Systematic Review. Int. J. Mol. Sci. 2021, 22, 9369. [Google Scholar] [CrossRef]
- Cho, S.Y.; Kang, W.; Han, J.Y.; Min, S.; Kang, J.; Lee, A.; Kwon, J.Y.; Lee, C.; Park, H. An Integrative Approach to Precision Cancer Medicine Using Patient-Derived Xenografts. Mol. Cells 2016, 39, 77–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabrera, S.; Llauradó, M.; Castellví, J.; Fernandez, Y.; Alameda, F.; Colás, E.; Ruiz, A.; Doll, A.; Schwartz, S., Jr.; Carreras, R.; et al. Generation and characterization of orthotopic murine models for endometrial cancer. Clin. Exp. Metastasis 2012, 29, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Unno, K.; Ono, M.; Winder, A.D.; Maniar, K.P.; Paintal, A.S.; Yu, Y.; Wei, J.J.; Lurain, J.R.; Kim, J.J. Establishment of human patient-derived endometrial cancer xenografts in NOD scid gamma mice for the study of invasion and metastasis. PLoS ONE 2014, 9, e116064. [Google Scholar] [CrossRef] [PubMed]
- Depreeuw, J.; Hermans, E.; Schrauwen, S.; Annibali, D.; Coenegrachts, L.; Thomas, D.; Luyckx, M.; Gutierrez-Roelens, I.; Debruyne, D.; Konings, K.; et al. Characterization of patient-derived tumor xenograft models of endometrial cancer for preclinical evaluation of targeted therapies. Gynecol. Oncol. 2015, 139, 118–126. [Google Scholar] [CrossRef]
- Haldorsen, I.S.; Popa, M.; Fonnes, T.; Brekke, N.; Kopperud, R.; Visser, N.C.; Rygh, C.B.; Pavlin, T.; Salvesen, H.B.; McCormack, E.; et al. Multimodal Imaging of Orthotopic Mouse Model of Endometrial Carcinoma. PLoS ONE 2015, 10, e0135220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moiola, C.P.; Lopez-Gil, C.; Cabrera, S.; Garcia, A.; Van Nyen, T.; Annibali, D.; Fonnes, T.; Vidal, A.; Villanueva, A.; Matias-Guiu, X.; et al. Patient-Derived Xenograft Models for Endometrial Cancer Research. Int. J. Mol. Sci. 2018, 19, 2431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Jia, N.; Nie, Y.; Chen, J.; Jiang, Y.; Lv, T.; Li, Y.; Yao, L.; Feng, W. Establishment of Patient-Derived Tumor Xenograft Models of High-Risk Endometrial Cancer. Int. J. Gynecol. Cancer 2018, 28, 1812–1820. [Google Scholar] [CrossRef] [PubMed]
- Bonazzi, V.F.; Kondrashova, O.; Smith, D.; Nones, K.; Sengal, A.T.; Ju, R.; Packer, L.M.; Koufariotis, L.T.; Kazakoff, S.H.; Davidson, A.L.; et al. Patient-derived xenograft models capture genomic heterogeneity in endometrial cancer. Genome Med. 2022, 14, 3. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Lee, E.J.; Yang, W.; Kim, H.S.; Chung, D.; Cho, H.; Kim, J.H. Identification of Prognostic Markers of Gynecologic Cancers Utilizing Patient-Derived Xenograft Mouse Models. Cancers 2022, 14, 829. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Lin, W.; Huang, Y.; Chen, X.; Wang, H.; Teng, L. The Essential Factors of Establishing Patient-derived Tumor Model. J. Cancer 2021, 12, 28–37. [Google Scholar] [CrossRef]
- Jin, K.T.; Du, W.L.; Lan, H.R.; Liu, Y.Y.; Mao, C.S.; Du, J.L.; Mou, X.Z. Development of humanized mouse with patient-derived xenografts for cancer immunotherapy studies: A comprehensive review. Cancer Sci. 2021, 112, 2592–2606. [Google Scholar] [CrossRef]
- Sajjad, H.; Imtiaz, S.; Noor, T.; Siddiqui, Y.H.; Sajjad, A.; Zia, M. Cancer models in preclinical research: A chronicle review of advancement in effective cancer research. Anim. Model. Exp. Med. 2021, 4, 87–103. [Google Scholar] [CrossRef]
- Bertotti, A.; Migliardi, G.; Galimi, F.; Sassi, F.; Torti, D.; Isella, C.; Corà, D.; Di Nicolantonio, F.; Buscarino, M.; Petti, C.; et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011, 1, 508–523. [Google Scholar] [CrossRef] [Green Version]
- Sartore-Bianchi, A.; Trusolino, L.; Martino, C.; Bencardino, K.; Lonardi, S.; Bergamo, F.; Zagonel, V.; Leone, F.; Depetris, I.; Martinelli, E.; et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 738–746. [Google Scholar] [CrossRef]
- Ma, D.; Hernandez, G.A.; Lefebvre, A.; Alshetaiwi, H.; Blake, K.; Dave, K.R.; Rauf, M.; Williams, J.W.; Davis, R.T.; Evans, K.T.; et al. Patient-derived xenograft culture-transplant system for investigation of human breast cancer metastasis. Commun. Biol. 2021, 4, 1268. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Chen, Y.; Li, N.; Sun, D.; Ju, H.; Chen, Z. Combination of GC-MS based metabolomics analysis with mouse xenograft models reveals a panel of dysregulated circulating metabolites and potential therapeutic targets for colorectal cancer. Transl. Cancer Res. 2021, 10, 1813–1825. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Maher, L.; Michaud, M.; Bae, S.W.; Kim, S.; Lee, H.S.; Im, S.A.; Yang, H.K.; Lee, C. Development of a Novel Orthotopic Gastric Cancer Mouse Model. Biol. Proced. Online 2021, 23, 1. [Google Scholar] [CrossRef]
- Chen, Q.; Wei, T.; Wang, J.; Zhang, Q.; Li, J.; Zhang, J.; Ni, L.; Wang, Y.; Bai, X.; Liang, T. Patient-derived xenograft model engraftment predicts poor prognosis after surgery in patients with pancreatic cancer. Pancreatology 2020, 20, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Cai, E.Y.; Garcia, J.; Liu, Y.; Vakar-Lopez, F.; Arora, S.; Nguyen, H.M.; Lakely, B.; Brown, L.; Wong, A.; Montgomery, B.; et al. A bladder cancer patient-derived xenograft displays aggressive growth dynamics in vivo and in organoid culture. Sci. Rep. 2021, 11, 4609. [Google Scholar] [CrossRef]
- Lundy, J.; Jenkins, B.J.; Saad, M.I. A Method for the Establishment of Human Lung Adenocarcinoma Patient-Derived Xenografts in Mice. Methods Mol. Biol. 2021, 2279, 165–173. [Google Scholar] [CrossRef]
- Sueyoshi, K.; Komura, D.; Katoh, H.; Yamamoto, A.; Onoyama, T.; Chijiwa, T.; Isagawa, T.; Tanaka, M.; Suemizu, H.; Nakamura, M.; et al. Multi-tumor analysis of cancer-stroma interactomes of patient-derived xenografts unveils the unique homeostatic process in renal cell carcinomas. iScience 2021, 24, 103322. [Google Scholar] [CrossRef]
- Wu, J.; Zheng, Y.; Tian, Q.; Yao, M.; Yi, X. Establishment of patient-derived xenograft model in ovarian cancer and its influence factors analysis. J. Obstet. Gynaecol. Res. 2019, 45, 2062–2073. [Google Scholar] [CrossRef]
- Cybula, M.; Wang, L.; Wang, L.; Drumond-Bock, A.L.; Moxley, K.M.; Benbrook, D.M.; Gunderson-Jackson, C.; Ruiz-Echevarria, M.J.; Bhattacharya, R.; Mukherjee, P.; et al. Patient-Derived Xenografts of High-Grade Serous Ovarian Cancer Subtype as a Powerful Tool in Pre-Clinical Research. Cancers 2021, 13, 6288. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef]
- Collins, A.; Ross, J.; Lang, S.H. A systematic review of the asymmetric inheritance of cellular organelles in eukaryotes: A critique of basic science validity and imprecision. PLoS ONE 2017, 12, e0178645. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Kim, H.; Lee, J.E.; Shin, S.J.; Oh, S.; Kwon, G.; Kim, H.; Choi, Y.Y.; White, M.A.; Paik, S.; et al. Selective Cytotoxicity of the NAMPT Inhibitor FK866 Toward Gastric Cancer Cells With Markers of the Epithelial-Mesenchymal Transition, Due to Loss of NAPRT. Gastroenterology 2018, 155, 799–814.e713. [Google Scholar] [CrossRef] [PubMed]
- Reddavid, R.; Corso, S.; Moya-Rull, D.; Giordano, S.; Degiuli, M. Patient-Derived Orthotopic Xenograft models in gastric cancer: A systematic review. Updates Surg. 2020, 72, 951–966. [Google Scholar] [CrossRef] [PubMed]
- Gillet, J.P.; Calcagno, A.M.; Varma, S.; Marino, M.; Green, L.J.; Vora, M.I.; Patel, C.; Orina, J.N.; Eliseeva, T.A.; Singal, V.; et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc. Natl. Acad. Sci. USA 2011, 108, 18708–18713. [Google Scholar] [CrossRef] [Green Version]
- Armando, R.G.; Mengual Gómez, D.L.; Gomez, D.E. New drugs are not enough-drug repositioning in oncology: An update. Int. J. Oncol. 2020, 56, 651–684. [Google Scholar] [CrossRef] [Green Version]
- Topp, M.D.; Hartley, L.; Cook, M.; Heong, V.; Boehm, E.; McShane, L.; Pyman, J.; McNally, O.; Ananda, S.; Harrell, M.; et al. Molecular correlates of platinum response in human high-grade serous ovarian cancer patient-derived xenografts. Mol. Oncol. 2014, 8, 656–668. [Google Scholar] [CrossRef]
- De Thaye, E.; Van de Vijver, K.; Van der Meulen, J.; Taminau, J.; Wagemans, G.; Denys, H.; Van Dorpe, J.; Berx, G.; Ceelen, W.; Van Bocxlaer, J.; et al. Establishment and characterization of a cell line and patient-derived xenograft (PDX) from peritoneal metastasis of low-grade serous ovarian carcinoma. Sci. Rep. 2020, 10, 6688. [Google Scholar] [CrossRef] [Green Version]
- Dong, R.; Qiang, W.; Guo, H.; Xu, X.; Kim, J.J.; Mazar, A.; Kong, B.; Wei, J.J. Histologic and molecular analysis of patient derived xenografts of high-grade serous ovarian carcinoma. J. Hematol. Oncol. 2016, 9, 92. [Google Scholar] [CrossRef] [Green Version]
- Ricci, F.; Bizzaro, F.; Cesca, M.; Guffanti, F.; Ganzinelli, M.; Decio, A.; Ghilardi, C.; Perego, P.; Fruscio, R.; Buda, A.; et al. Patient-derived ovarian tumor xenografts recapitulate human clinicopathology and genetic alterations. Cancer Res. 2014, 74, 6980–6990. [Google Scholar] [CrossRef] [Green Version]
- Weroha, S.J.; Becker, M.A.; Enderica-Gonzalez, S.; Harrington, S.C.; Oberg, A.L.; Maurer, M.J.; Perkins, S.E.; AlHilli, M.; Butler, K.A.; McKinstry, S.; et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin. Cancer Res. 2014, 20, 1288–1297. [Google Scholar] [CrossRef] [Green Version]
- Heo, E.J.; Cho, Y.J.; Cho, W.C.; Hong, J.E.; Jeon, H.K.; Oh, D.Y.; Choi, Y.L.; Song, S.Y.; Choi, J.J.; Bae, D.S.; et al. Patient-Derived Xenograft Models of Epithelial Ovarian Cancer for Preclinical Studies. Cancer Res. Treat. 2017, 49, 915–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cybulska, P.; Stewart, J.M.; Sayad, A.; Virtanen, C.; Shaw, P.A.; Clarke, B.; Stickle, N.; Bernardini, M.Q.; Neel, B.G. A Genomically Characterized Collection of High-Grade Serous Ovarian Cancer Xenografts for Preclinical Testing. Am. J. Pathol. 2018, 188, 1120–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.F.; Palakurthi, S.; Zeng, Q.; Zhou, S.; Ivanova, E.; Huang, W.; Zervantonakis, I.K.; Selfors, L.M.; Shen, Y.; Pritchard, C.C.; et al. Establishment of Patient-Derived Tumor Xenograft Models of Epithelial Ovarian Cancer for Preclinical Evaluation of Novel Therapeutics. Clin. Cancer Res. 2017, 23, 1263–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- George, E.; Kim, H.; Krepler, C.; Wenz, B.; Makvandi, M.; Tanyi, J.L.; Brown, E.; Zhang, R.; Brafford, P.; Jean, S.; et al. A patient-derived-xenograft platform to study BRCA-deficient ovarian cancers. JCI Insight 2017, 2, e89760. [Google Scholar] [CrossRef]
- Colombo, P.E.; du Manoir, S.; Orsett, B.; Bras-Gonçalves, R.; Lambros, M.B.; MacKay, A.; Nguyen, T.T.; Boissière, F.; Pourquier, D.; Bibeau, F.; et al. Ovarian carcinoma patient derived xenografts reproduce their tumor of origin and preserve an oligoclonal structure. Oncotarget 2015, 6, 28327–28340. [Google Scholar] [CrossRef]
- Ricci, F.; Guffanti, F.; Affatato, R.; Brunelli, L.; Roberta, P.; Fruscio, R.; Perego, P.; Bani, M.R.; Chiorino, G.; Rinaldi, A.; et al. Establishment of patient-derived tumor xenograft models of mucinous ovarian cancer. Am. J. Cancer Res. 2020, 10, 572–580. [Google Scholar]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, W.; Long, Y.; Liu, H.; Cheng, J.; Guo, L.; Li, R.; Meng, C.; Yu, S.; Zhao, Q.; et al. Characterization of drug responses of mini patient-derived xenografts in mice for predicting cancer patient clinical therapeutic response. Cancer Commun. 2018, 38, 60. [Google Scholar] [CrossRef] [Green Version]
- Malaney, P.; Nicosia, S.V.; Davé, V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer Lett. 2014, 344, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sanmamed, M.F.; Rodriguez, I.; Schalper, K.A.; Oñate, C.; Azpilikueta, A.; Rodriguez-Ruiz, M.E.; Morales-Kastresana, A.; Labiano, S.; Pérez-Gracia, J.L.; Martín-Algarra, S.; et al. Nivolumab and Urelumab Enhance Antitumor Activity of Human T Lymphocytes Engrafted in Rag2-/-IL2Rγnull Immunodeficient Mice. Cancer Res. 2015, 75, 3466–3478. [Google Scholar] [CrossRef] [Green Version]
- Rongvaux, A.; Willinger, T.; Martinek, J.; Strowig, T.; Gearty, S.V.; Teichmann, L.L.; Saito, Y.; Marches, F.; Halene, S.; Palucka, A.K.; et al. Development and function of human innate immune cells in a humanized mouse model. Nature Biotechnol. 2014, 32, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Brainard, D.M.; Seung, E.; Frahm, N.; Cariappa, A.; Bailey, C.C.; Hart, W.K.; Shin, H.S.; Brooks, S.F.; Knight, H.L.; Eichbaum, Q.; et al. Induction of robust cellular and humoral virus-specific adaptive immune responses in human immunodeficiency virus-infected humanized BLT mice. J. Virol. 2009, 83, 7305–7321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Author, Year | Country | Animal Model | Histology | Type of Procedure for Obtaining the Tumor | Aim of the Study |
|---|---|---|---|---|---|
| Cabrera et al., 2012 [8] | Spain | Nude | EEC | Surgery | Evaluate the PDX method |
| Unno et al., 2014 [9] | USA | NSG | EEC, SEC, CCEC, and UCS | Surgery | Evaluate the PDX method |
| Depreeuw et al., 2015 [10] | Belgium | Nude | EEC, SEC, CCEC, and UDC | Surgery | Evaluate the PDX model |
| Haldorsen et al., 2015 [11] | Norway | NSG | EEC | Biopsy | Imaging evaluation using PDX model |
| Moiola et al., 2018 [12] | Spain | Nude or NSG | EEC, SEC, CCEC, UCS, and others | Surgery | PDX cohort |
| Zhu et al., 2018 [13] | China | NOD/SCID | EEC, SEC, CCEC, and UCS | Surgery | Evaluate the PDX model and drug evaluation |
| Bonazzi et al., 2022 [14] | Australia | NSG | EEC, SEC, CCEC, and UCS | Surgery | Evaluate the PDX model and drug evaluation |
| Shin et al., 2022 [15] | Korea | Nude | EEC, SEC, CCEC, and UCS | Surgery | Evaluate the PDX model |
| Author, Year | Time between Surgery and Implantation | Fragment Size | Site of Transplantation | Method of Graft | Mean Latency | Number of Donor Patients | Engraftment Rate (%) |
|---|---|---|---|---|---|---|---|
| Cabrera et al., 2012 [8] | Immediately | 1 mm3 | Subcutaneous | Direct | N.I. | 2 | 100 (2/2) |
| Immediately | Crumbled | Uterine cavity | Injection | 62.7 d | 2 | 100 (2/2) | |
| Unno et al., 2014 [9] | N.I. | 1.5 mm × 1.5 mm | Renal capsule | Direct | N.I. | 11 | 36.4 (4/11) |
| Depreeuw et al., 2015 [10] | Within 4 h | 8–10 mm3 | Subcutaneous | Direct | 1.5–9 mo | 40 | 60 (24/40) |
| Haldorsen et al., 2015 [11] | N.I. | Cell suspension | Uterine cavity | Injection | 3–4 mo | 1 | 100 (1/1) |
| Moiola et al., 2018 [12] | N.I. | Small tissue fragment | Orthotopic | Direct | 1–5 mo | 64 | N.I. |
| N.I. | 5–10 mm3 | Subcutaneous | Direct | 2–3 mo | 40 | N.I. | |
| N.I. | 8–10 mm3 | Subcutaneous | Direct | 3–5 mo | 15 | N.I. | |
| N.I. | Cell suspension | Orthotopic | Direct | 3–13 mo | 5 | N.I. | |
| Zhu et al., 2018 [13] | Within 5 h | 1 × 1.5 × 1.5 mm3 | Subcutaneous | Direct | 2–11 wk | 18 | 50 (9/18) |
| Within 5 h | 1 × 1.5 × 1.5 mm3 | Renal capsule | Direct | 4–10 wk | 16 | 62.5 (10/16) | |
| Bonazzi et al., 2022 [14] | Within 4 h | 1–2 mm3 | Subcutaneous | Direct | N.I. | 32 | 61 (13/32) |
| 4 °C overnight | 1–2 mm3 | Subcutaneous | Direct | N.I. | 11 | 27 (3/11) | |
| Viably Frozen | 1–2 mm3 | Subcutaneous | Direct | N.I. | 11 | 18 (2/11) | |
| Shin et al., 2022 [15] | Immediately | 3 mm3 | Subcutaneous | Direct | 6 mo | 31 | 56 (17/31) |
| Author, Year of Publication | Histology | Driver Gene Mutation | Gene Expression | Copy Number Variation | Proteomics | Immunohistochemistry | Other |
|---|---|---|---|---|---|---|---|
| Cabrera et al., 2012 [8] | Yes | No | No | No | No | p53, ER, PR, Ki67, E-cadherin, MSH2, MLH1, MSH6 | No |
| Unno et al., 2014 [9] | Yes | No | No | No | No | p53, ER, PR, Ki67, CD31, cytokeratin, vimentin, E-cadherin, PTEN, uPA, uPAR | No |
| Depreeuw et al., 2015 [10] | Yes | Yes | Yes | Yes | No | ER, PR, vimentin, MLH, MSH2, cytokeratin | PI3K/mTOR and MEK inhibitor |
| Haldorsen et al., 2015 [11] | Yes | No | No | No | No | No | No |
| Moiola et al., 2018 [12] | Yes | No | No | No | No | No | No |
| Zhu et al., 2018 [13] | Yes | Yes | Yes | No | No | No | No |
| Bonazzi et al., 2022 [14] | Yes | Yes | Yes | Yes | No | No | POLE, MMRd, p53 and HRD |
| Shin et al., 2022 [15] | No | No | No | No | No | No | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, T.; Nishie, R.; Ueda, S.; Miyamoto, S.; Hashida, S.; Konishi, H.; Terada, S.; Kogata, Y.; Sasaki, H.; Tsunetoh, S.; et al. Endometrial Cancer Patient-Derived Xenograft Models: A Systematic Review. J. Clin. Med. 2022, 11, 2606. https://doi.org/10.3390/jcm11092606
Tanaka T, Nishie R, Ueda S, Miyamoto S, Hashida S, Konishi H, Terada S, Kogata Y, Sasaki H, Tsunetoh S, et al. Endometrial Cancer Patient-Derived Xenograft Models: A Systematic Review. Journal of Clinical Medicine. 2022; 11(9):2606. https://doi.org/10.3390/jcm11092606
Chicago/Turabian StyleTanaka, Tomohito, Ruri Nishie, Shoko Ueda, Shunsuke Miyamoto, Sousuke Hashida, Hiromi Konishi, Shinichi Terada, Yuhei Kogata, Hiroshi Sasaki, Satoshi Tsunetoh, and et al. 2022. "Endometrial Cancer Patient-Derived Xenograft Models: A Systematic Review" Journal of Clinical Medicine 11, no. 9: 2606. https://doi.org/10.3390/jcm11092606
APA StyleTanaka, T., Nishie, R., Ueda, S., Miyamoto, S., Hashida, S., Konishi, H., Terada, S., Kogata, Y., Sasaki, H., Tsunetoh, S., Taniguchi, K., Komura, K., & Ohmichi, M. (2022). Endometrial Cancer Patient-Derived Xenograft Models: A Systematic Review. Journal of Clinical Medicine, 11(9), 2606. https://doi.org/10.3390/jcm11092606






