WT1 Gene Mutations, rs16754 Variant, and WT1 Overexpression as Prognostic Factors in Acute Myeloid Leukemia Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Classical Cytogenetics and Fluorescence In Situ Hybridization (FISH)
2.2. DNA Isolation
2.3. Array Comparative Genomic Hybridization (aCGH)
2.4. WT1 Genotyping—Analysis of WT1 Mutations and rs16754 Variant
2.5. RNA Isolation and WT1 Expression
2.6. Analysis for Other Gene Mutations
2.7. Statistical Analysis
3. Results
3.1. WT1 rs16754 Variant
3.2. WT1 Mutations
- -
- In exon 7: c.1375G>A (p.A399V), c.1334C>A (p.R385R), c.1382A>T (p.P401P), c.1389delA (p.N404H); c.1320G>A (p.P381S), c.1314C>T (p.V379I), c.1324indelGTACAAGAG/GTACAAGAGGGTACAAGAG (frameshift variant);
- -
- In exon 9: c.1590delC (p.L491X), c.1567G>A (p.R463P), c.1557T>A/C (p.T460S/A).
3.3. WT1 Expression
3.4. FLT3, NPM1, and CEBPA Mutations
3.5. Cytogenetic Aberrations
- -
- losses of (5)(q23q32)—20%;
- -
- losses of (7)(p12.3q36.3)—13%;
- -
- gains of (8)(q12.1q24.3)—10%;
- -
- gains of (11)(q12.2q14.1)—12.2%;
- -
- losses of (11)(q22q23.3)—14.4%;
- -
- losses of (17)(p13.3p13.1)—16.7%;
- -
- losses of (18)(p11.32q23)—11%;
- -
- gains of (22)(q12.3q13.2)—5.5%.
4. Discussion
4.1. WT1 rs16754 Variant
4.2. WT1 Mutations
4.3. WT1 Expression
4.4. Cytogenetic Analysis
4.5. Clinical and Molecular Data
4.6. Limitations
4.7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Estey, E.; Döhner, H. Acute myeloid leukaemia. Lancet 2006, 368, 1894–1907. [Google Scholar] [CrossRef]
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Rostami, S.; Kazemi, A.; Chahardouli, B.; Mohammadi, S.; Nikbakht, M.; Alizadeh, N.; Mousavi, A.; Alimoghaddam, K.; Ardestani, M.T. The Prognostic Impact of WT1 Expression Levels, Mutations, and SNP rs16754 in AML Patients: A Retrospective Cohort Study. J. Adv. Med. Biomed. Res. 2021, 29, 109–117. [Google Scholar] [CrossRef]
- Sugiyama, H. Wilms’ tumor gene WT1: Its oncogenic function and clinical application. Int. J. Hematol. 2001, 73, 177–187. [Google Scholar] [CrossRef]
- Döhner, H.; Weisdorf, D.J.; Bloomfield, C.D. Acute Myeloid Leukemia. N. Engl. J. Med. 2015, 373, 1136–1152. [Google Scholar] [CrossRef]
- Kang, H.J.; Park, J.H.; Chen, W.; Kang, S.I.; Moroz, K.; Ladanyi, M.; Lee, S.B. EWS-WT1 oncoprotein activates neuronal reprogramming factor ASCL1 and promotes neural differentiation. Cancer Res. 2014, 74, 4526–4535. [Google Scholar] [CrossRef]
- Rampal, R.; Figueroa, M.E. Wilms tumor 1 mutations in the pathogenesis of acute myeloid leukemia. Haematologica 2016, 101, 672–679. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, J.-H.; Hur, E.-H.; Kang, M.J.; Kim, S.-D.; Lee, J.-H.; Kim, D.-Y.; Lim, S.-N.; Bae, K.-S.; Lim, H.-S.; et al. Single nucleotide polymorphism of Wilms’ tumor 1 gene rs16754 in Korean patients with cytogenetically normal acute myeloid leukemia. Ann. Hematol. 2011, 91, 671–677. [Google Scholar] [CrossRef]
- Long, J.; Fang, S.; Dai, Q.; Liu, X.; Zhu, W. The Wilms Tumor-1 (WT1) rs16754 polymorphism is a prognostic factor in acute myeloid leukemia (AML): A meta-analysis. Oncotarget 2016, 7, 32079–32087. [Google Scholar]
- Damm, F.; Heuser, M.; Morgan, M.; Yun, H.; Großhennig, A.; Göhring, G.; Schlegelberger, B.; Döhner, K.; Ottmann, O.; Lübbert, M.; et al. Single Nucleotide Polymorphism in the Mutational Hotspot of WT1 Predicts a Favorable Outcome in Patients With Cytogenetically Normal Acute Myeloid Leukemia. J. Clin. Oncol. 2010, 28, 578–585. [Google Scholar] [CrossRef]
- Bennett, J.M.; Catovsky, D.; Daniel, M.-T.; Flandrin, G.; Galton, D.A.G.; Gralnick, H.R.; Sultan, C. Proposals for the Classification of the Acute Leukaemias French-American-British (FAB) Co-operative Group. Br. J. Haematol. 1976, 33, 451–458. [Google Scholar] [CrossRef]
- King-Underwood, L.; Renshaw, J.; Pritchard-Jones, K. Mutations in the Wilms’ tumor gene WT1 in leukemias. Blood 1996, 87, 2171–2179. [Google Scholar] [CrossRef]
- Andersson, C.; Li, X.; Lorenz, F.; Golovleva, I.; Wahlin, A.; Li, A. Reduction in WT1 Gene Expression During Early Treatment Predicts the Outcome in Patients With Acute Myeloid Leukemia. Diagn. Mol. Pathol. 2012, 21, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Yu, K.; Yan, Q.-X.; Shen, Z.-J.; Wu, J.-B.; Chen, H.-M.; Gao, S.-M. Analysis of WT1 mutations, expression levels and single nucleotide polymorphism rs16754 inde novonon-M3 acute myeloid leukemia. Leuk. Lymphoma 2013, 55, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Green, C.L.; Koo, K.K.; Hills, R.; Burnett, A.K.; Linch, D.C.; Gale, R.E. Prognostic Significance of CEBPA Mutations in a Large Cohort of Younger Adult Patients With Acute Myeloid Leukemia: Impact of Double CEBPA Mutations and the Interaction With FLT3 and NPM1 Mutations. J. Clin. Oncol. 2010, 28, 2739–2747. [Google Scholar] [CrossRef]
- Paschka, P.; Marcucci, G.; Ruppert, A.S.; Whitman, S.P.; Mrózek, K.; Maharry, K.; Langer, C.; Baldus, C.D.; Zhao, W.; Powell, B.L.; et al. Wilms’ Tumor 1 Gene Mutations Independently Predict Poor Outcome in Adults With Cytogenetically Normal Acute Myeloid Leukemia: A Cancer and Leukemia Group B Study. J. Clin. Oncol. 2008, 26, 4595–4602. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, G.; Mrózek, K.; Ruppert, A.S.; Archer, K.J.; Pettenati, M.J.; Heerema, N.A.; Carroll, A.J.; Koduru, P.R.; Kolitz, J.E.; Sterling, L.J.; et al. Abnormal Cytogenetics at Date of Morphologic Complete Remission Predicts Short Overall and Disease-Free Survival, and Higher Relapse Rate in Adult Acute Myeloid Leukemia: Results From Cancer and Leukemia Group B Study 8461. J. Clin. Oncol. 2004, 22, 2410–2418. [Google Scholar] [CrossRef]
- Betz, B.L.; Hess, J.L. Acute Myeloid Leukemia Diagnosis in the 21st Century. Arch. Pathol. Lab. Med. 2010, 134, 1427–1433. [Google Scholar] [CrossRef]
- Grimwade, D.; Walker, H.; Oliver, F.; Wheatley, K.; Harrison, C.; Harrison, G.; Rees, J.; Hann, I.; Stevens, R.; Burnett, A.; et al. The importance of diagnostic cytogenetics on outcome in AML: Analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children’s Leukaemia Working Parties. Blood 1998, 92, 2322–2333. Available online: http://www.ncbi.nlm.nih.gov/pubmed/9746770 (accessed on 10 February 2022). [CrossRef]
- Döhner, H.; Estey, E.H.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Burnett, A.K.; Dombret, H.; Fenaux, P.; Grimwade, D.; Larson, R.A.; et al. Diagnosis and management of acute myeloid leukemia in adults: Recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood 2010, 115, 453–474. [Google Scholar] [CrossRef]
- Summers, K.; Stevens, J.; Kakkas, I.; Smith, M.; Smith, L.L.; MacDougall, F.; Cavenagh, J.; Bonnet, D.; Young, B.D.; Lister, T.A.; et al. Wilms’ tumour 1 mutations are associated with FLT3-ITD and failure of standard induction chemotherapy in patients with normal karyotype AML. Leukemia 2007, 21, 550–551. [Google Scholar] [CrossRef][Green Version]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Cilloni, D.; Gottardi, E.; De Micheli, D.; Serra, A.; Volpe, G.; Messa, F.; Rege-Cambrin, G.; Guerrasio, A.; Divona, M.; Coco, F.L.; et al. Quantitative assessment of WT1 expression by real time quantitative PCR may be a useful tool for monitoring minimal residual disease in acute leukemia patients. Leukemia 2002, 16, 2115–2121. [Google Scholar] [CrossRef]
- Beillard, E.; Pallisgaard, N.; Van Der Velden, V.H.J.; Bi, W.; Dee, R.; Van Der Schoot, E.; Delabesse, E.; MacIntyre, E.; Gottardi, E.; Saglio, G.; et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)—A Europe against cancer program. Leukemia 2003, 17, 2474–2486. [Google Scholar] [CrossRef]
- De Marchi, F.; Candoni, A.; Zannier, M.E.; Haley, L.; Lau, B.W.Y.; Fanin, R. Concomitant monitoring of WT1 and FLT3-ITD expression in FLT3-ITD acute myeloid leukemia patients: Which should we trust as a minimal residual disease marker? Am. J. Hematol. 2017, 92, E72–E74. [Google Scholar] [CrossRef]
- Koczkodaj, D.; Zmorzyński, S.; Michalak-Wojnowska, M.; Wasik-Szczepanek, E.; Filip, A.A. Examination of the FLT3 and NPM1 mutational status in patients with acute myeloid leukemia from southeastern Poland. Arch. Med Sci. 2016, 1, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Benthaus, T.; Schneider, F.; Mellert, G.; Zellmeier, E.; Schneider, S.; Kakadia, P.M.; Hiddemann, W.; Bohlander, S.; Feuring-Buske, M.; Braess, J.; et al. Rapid and sensitive screening forCEBPAmutations in acute myeloid leukaemia. Br. J. Haematol. 2008, 143, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Preacher, K.J. Calculation for the Chi-Square Test: An Interactive Calculation Tool for Chi-Square Tests of Goodness of Fit and Independence. 2019. Available online: http://quantpsy.org/ (accessed on 21 January 2022).
- Zmorzynski, S.; Szudy-Szczyrek, A.; Popek-Marciniec, S.; Korszen-Pilecka, I.; Wojcierowska-Litwin, M.; Luterek, M.; Chocholska, S.; Styk, W.; Swiderska-Kołacz, G.; Januszewska, J.; et al. ACE Insertion/Deletion Polymorphism (rs4646994) Is Associated With the Increased Risk of Multiple Myeloma. Front. Oncol. 2019, 9, 44. [Google Scholar] [CrossRef]
- Kim, N.; Kim, I.-S.; Chang, C.L.; Kang, J.-E.; Lee, E.Y.; Shin, H.-J. Risk-Reducing Genetic Variant of Wilms Tumor 1 Gene rs16754 in Korean Patients With BCR-ABL1-Negative Myeloproliferative Neoplasm. Ann. Lab. Med. 2015, 35, 348–351. [Google Scholar] [CrossRef]
- Zhang, D.-Y.; Yan, H.; Cao, S.; Zhang, W.; Li, X.-L.; Zeng, H.; Chen, X.-P. Wilms Tumor 1 rs16754 predicts favorable clinical outcomes for acute myeloid leukemia patients in South Chinese population. Leuk. Res. 2015, 39, 568–574. [Google Scholar] [CrossRef]
- Krauth, M.-T.; Alpermann, T.; Bacher, U.; Eder, C.; Dicker, F.; Ulke, M.; Kuznia, S.; Nadarajah, N.; Kern, W.; Haferlach, C.; et al. WT1 mutations are secondary events in AML, show varying frequencies and impact on prognosis between genetic subgroups. Leukemia 2014, 29, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Gaidzik, V.I.; Schlenk, R.F.; Moschny, S.; Becker, A.; Bullinger, L.; Corbacioglu, A.; Krauter, J.; Schlegelberger, B.; Ganser, A.; Döhner, H.; et al. Prognostic impact of WT1 mutations in cytogenetically normal acute myeloid leukemia: A study of the German-Austrian AML Study Group. Blood 2009, 113, 4505–4511. [Google Scholar] [CrossRef]
- Hou, H.-A.; Huang, T.-C.; Lin, L.-I.; Liu, C.-Y.; Chen, C.-Y.; Chou, W.-C.; Tang, J.-L.; Tseng, M.-H.; Huang, C.-F.; Chiang, Y.-C.; et al. WT1 mutation in 470 adult patients with acute myeloid leukemia: Stability during disease evolution and implication of its incorporation into a survival scoring system. Blood 2010, 115, 5222–5231. [Google Scholar] [CrossRef] [PubMed]
- Toogeh, G.; Ramzi, M.; Faranoush, M.; Amirizadeh, N.; Haghpanah, S.; Moghadam, M.; Cohan, N. Prevalence and Prognostic Impact of Wilms’ Tumor 1 (WT1) Gene, Including SNP rs16754 in Cytogenetically Normal Acute Myeloblastic Leukemia (CN-AML): An Iranian Experience. Clin. Lymphoma Myeloma Leuk. 2016, 16, e21–e26. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhang, Y.; Hu, J.; Ren, Y.; Wang, H. Clinical features and prognosis of normal karyotype acute myeloid leukemia pediatric patients with WT1 mutations: An analysis based on TCGA database. Hematology 2020, 25, 79–84. [Google Scholar] [CrossRef]
- Niktoreh, N.; Walter, C.; Zimmermann, M.; von Neuhoff, C.; von Neuhoff, N.; Rasche, M.; Waack, K.; Creutzig, U.; Hanenberg, H.; Reinhardt, D. Mutated WT1, FLT3-ITD, and NUP98-NSD1 Fusion in Various Combinations Define a Poor Prognostic Group in Pediatric Acute Myeloid Leukemia. J. Oncol. 2019, 2019, 1609128. [Google Scholar] [CrossRef]
- Di Stasi, A.; Jimenez, A.M.; Eminagawa, K.; Eal-Obaidi, M.; Erezvani, K. Review of the Results of WT1 Peptide Vaccination Strategies for Myelodysplastic Syndromes and Acute Myeloid Leukemia from Nine Different Studies. Front. Immunol. 2015, 6, 36. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Kim, H.-J.; Shin, S.-H.; Yahng, S.-A.; Lee, S.-E.; Cho, B.-S.; Eom, K.-S.; Kim, Y.-J.; Lee, S.; Min, C.-K.; et al. Serial Measurement of WT1 Expression and Decrement Ratio Until Hematopoietic Cell Transplantation as a Marker of Residual Disease in Patients with Cytogenetically Normal Acute Myelogenous Leukemia. Biol. Blood Marrow Transplant. 2013, 19, 958–966. [Google Scholar] [CrossRef]
- Shimada, A.; Taki, T.; Koga, D.; Tabuchi, K.; Tawa, A.; Hanada, R.; Tsuchida, M.; Horibe, K.; Tsukimoto, I.; Adachi, S.; et al. High WT1 mRNA expression after induction chemotherapy and FLT3-ITD have prognostic impact in pediatric acute myeloid leukemia: A study of the Japanese Childhood AML Cooperative Study Group. Int. J. Hematol. 2012, 96, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Suela, J.; Alvarez, S.; Cigudosa, J. DNA profiling by arrayCGH in acute myeloid leukemia and myelodysplastic syndromes. Cytogenet. Genome Res. 2007, 118, 304–309. [Google Scholar] [CrossRef]
- Evers, C.; Beier, M.; Poelitz, A.; Hildebrandt, B.; Servan, K.; Drechsler, M.; Germing, U.; Royer, H.-D.; Royer-Pokora, B. Molecular definition of chromosome arm 5q deletion end points and detection of hidden aberrations in patients with myelodysplastic syndromes and isolated del(5q) using oligonucleotide array CGH. Genes Chromosom. Cancer 2007, 46, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Rücker, F.G.; Bullinger, L.; Schwaenen, C.; Lipka, D.B.; Wessendorf, S.; Fröhling, S.; Bentz, M.; Miller, S.; Scholl, C.; Schlenk, R.F.; et al. Disclosure of Candidate Genes in Acute Myeloid Leukemia With Complex Karyotypes Using Microarray-Based Molecular Characterization. J. Clin. Oncol. 2006, 24, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.R.S.; Belangero, S.; Melaragno, M.I.; Chauffaille, M.D.L. Additional chromosomal abnormalities detected by array comparative genomic hybridization in AML. Med. Oncol. 2012, 29, 2083–2087. [Google Scholar] [CrossRef] [PubMed]
- Mehrotra, M.; Luthra, R.; Ravandi, F.; Sargent, R.L.; Barkoh, B.A.; Abraham, R.; Mishra, B.M.; Medeiros, L.J.; Patel, K.P. Identification of clinically important chromosomal aberrations in acute myeloid leukemia by array-based comparative genomic hybridization. Leuk. Lymphoma 2014, 55, 2538–2548. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Veigaard, C.; Nørgaard, J.M.; Kjeldsen, E. Genomic profiling in high hyperdiploid acute myeloid leukemia: A retrospective study of 19 cases. Cancer Genet. 2011, 204, 516–521. [Google Scholar] [CrossRef]
- Schoch, C.; Kern, W.; Kohlmann, A.; Hiddemann, W.; Schnittger, S.; Haferlach, T. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosom. Cancer 2005, 43, 227–238. [Google Scholar] [CrossRef]
- Itzhar, N.; Dessen, P.; Toujani, S.; Auger, N.; Preudhomme, C.; Richon, C.; Lazar, V.; Saada, V.; Bennaceur, A.; Bourhis, J.H.; et al. Chromosomal Minimal Critical Regions in Therapy-Related Leukemia Appear Different from Those of De Novo Leukemia by High-Resolution aCGH. PLoS ONE 2011, 6, e16623. [Google Scholar] [CrossRef][Green Version]
- Thiede, C.; Steudel, C.; Mohr, B.; Schaich, M.; Schäkel, U.; Platzbecker, U.; Wermke, M.; Bornhäuser, M.; Ritter, M.; Neubauer, A.; et al. Analysis of FLT3-activating mutations in 979 patients with acute myelogenous leukemia: Association with FAB subtypes and identification of subgroups with poor prognosis. Blood 2002, 99, 4326–4335. [Google Scholar] [CrossRef]
- Park, B.; Chi, H.-S.; Park, S.-J.; Min, S.K.; Jang, S.; Park, C.-J.; Kim, D.-Y.; Lee, J.-H.; Lee, J.-H.; Lee, K.-H. Clinical Implications of Non-A-Type NPM1 and FLT3 Mutations in Patients with Normal Karyotype Acute Myeloid Leukemia. Acta Haematol. 2012, 127, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Mrózek, K.; Marcucci, G.; Paschka, P.; Bloomfield, C.D. Advances in molecular genetics and treatment of core-binding factor acute myeloid leukemia. Curr. Opin. Oncol. 2008, 20, 711–718. [Google Scholar] [CrossRef]
- Fröhling, S.; Schlenk, R.F.; Breitruck, J.; Benner, A.; Kreitmeier, S.; Tobis, K.; Doöhner, H.; Doöhner, K. Prognostic significance of activating FLT3 mutations in younger adults (16 to 60 years) with acute myeloid leukemia and normal cytogenetics: A study of the AML Study Group Ulm. Blood 2002, 100, 4372–4380. [Google Scholar] [CrossRef]
| Age median (range) | 62.63 (18–85) |
| Gender (%) | |
| Male | 42 (47) |
| Female | 48 (53) |
| Laboratory parameters (range) | |
| HB g/dL | 9.26 (4.8–9.7) |
| WBC G/L | 55.15 (0.8–94.84) |
| PLT G/L | 89.06 (6.0–783.0) |
| FAB subtype: | |
| M0 | 6 |
| M1 | 7 |
| M2 | 7 |
| M3 | 12 |
| M4 | 42 |
| M5 | 15 |
| M6 | 1 |
| Risk category: | |
| favorable | 13 |
| intermediate | 34 |
| adverse | 43 |
| Induction therapy: | |
| DAC | 52 |
| Idarubicin + ATRA | 6 |
| AZA | 9 |
| reduced DA | 15 |
| low-dose cytarabine | 8 |
| Stem cell transplantation: | |
| no | 42 |
| allogeneic | 37 |
| autologous | 11 |
| Cytogenetic: | |
| Normal karyotype | 45 |
| Abnormal karyotype | 45 (22 *) |
| FISH (%): | |
| del(13)(q14.3) | 51 (56.6) |
| del(17)(p13.1) | 15 (16.7) |
| del(11)(q23) | 13 (14.4) |
| del(6)(q23) | 5 (5.5) |
| t(15;17)(q22;q21.1) | 6 (6.6) |
| BCR/ABL | 2 (2.2) |
| MLL amplification | 1 (1.1) |
| MYC amplification | 1 (1.1) |
| EVI1 amplification | 1 (1.1) |
| Immunophenotype—negativity/positivity | |
| CD34 | 51/39 |
| CD33 | 19/71 |
| CD14 | 66/24 |
| Molecular variants—present/absent | |
| FLT3-ITD | 8/82 |
| NPM1 mutation | 11/79 |
| CEBPA mutation | 7/83 |
| GROUPS | GENOTYPES | Total | HWE p Value and χ2 * | ||
|---|---|---|---|---|---|
| WT1 rs16754 variant | |||||
| - | AA | GA | GG | - | - |
| CONTROL | |||||
| E | 79.21 | 19.58 | 1.21 | 100 | p = 0.44, χ2 = 0.57 |
| O | 80 | 18 | 2 | 100 | |
| CASE | |||||
| E | 70.2 | 18.55 | 1.25 | 90 | p = 0.0007, χ2 = 11.5 |
| O | 74 | 11 | 5 | 90 | |
| Gene Variants and Alleles | AML n (%) | Controls n (%) | Odds Ratio | 95% CI | p Values |
|---|---|---|---|---|---|
| Codominant model | |||||
| AA | 74 (82.2%) | 80 (80%) | 1 | - | - |
| GA | 11 (12.2%) | 18 (18%) | 1.51 | 0.67–3.41 | 0.31 |
| GG | 5 (5.5%) | 2 (2%) | 0.37 | 0.06–1.96 | 0.41 |
| Dominant model | |||||
| AA | 74 (82.2%) | 80 (80%) | 1 | - | - |
| GA + GG | 16 (17.7%) | 20 (20%) | 1.15 | 0.55–2.40 | 0.69 |
| Recessive model | |||||
| AA + GA | 85 (94.4%) | 98 (98%) | 1 | - | - |
| GG | 5 (5.5%) | 2 (2%) | 0.34 | 0.06–1.83 | 0.36 |
| Total: | 90 (100%) | 100 (100%) | |||
| Alleles | |||||
| A | 159 (88.3%) | 178 (89%) | 1 | - | - |
| G | 21 (11.7%) | 22 (11%) | 0.93 | 0.49–1.76 | 0.84 |
| Total: | 180 (100%) | 200 (100%) | |||
| Features | WT1 AA Genotype | WT1 GA + GG Genotype | p Value | WT1 Mutated | WT1 Wild Type | p Value | WT1 Expression * | WT1 Expression ** | p Value |
|---|---|---|---|---|---|---|---|---|---|
| Gender | |||||||||
| Male | 34 | 8 | 0.76 | 18 | 38 | 0.38 | 37 | 12 | 0.61 |
| Female | 40 | 8 | 8 | 26 | 29 | 12 | |||
| Age | |||||||||
| Age < 65 years | 62 | 12 | 0.63 | 23 | 13 | <0.0001 | 55 | 19 | 0.64 |
| Age ≥ 65 years | 12 | 4 | 3 | 51 | 11 | 5 | |||
| Cytogenetics | |||||||||
| Normal karyotype | 35 | 10 | 0.27 | 16 | 29 | 0.16 | 32 | 13 | 0.63 |
| Abnormal karyotype | 39 | 6 | 10 | 35 | 34 | 11 | |||
| Point mutations | |||||||||
| NPM1 wild type | 66 | 13 | 0.64 | 18 | 61 | 0.002 | 58 | 21 | 0.75 |
| NPM1 mutated | 8 | 3 | 8 | 3 | 8 | 3 | |||
| FLT3 wild type | 67 | 15 | 0.93 | 21 | 61 | 0.07 | 62 | 20 | 0.25 |
| FLT3 mutated | 7 | 1 | 5 | 3 | 4 | 4 | |||
| CEBPA wild type | 69 | 14 | 0.80 | 20 | 63 | 0.002 | 60 | 23 | 0.74 |
| CEBPA mutated | 5 | 2 | 6 | 1 | 6 | 1 | |||
| Clinical values | |||||||||
| WBC median | 42.16 | 113.59 | 0.06 | 13.74 | 61.19 | 0.64 | 50.82 | 74.63 | 0.96 |
| Hb median | 9.18 | 9.74 | 0.33 | 8.85 | 9.33 | 0.27 | 9.28 | 9.17 | 0.17 |
| PLT median | 78.37 | 137.18 | 0.02 | 86 | 90.28 | 0.19 | 90.79 | 84.45 | 0.45 |
| Feature | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| p Value | HR | 95% CI | p Value | HR | 95% CI | |
| OS | ||||||
| Age | <0.01 | 2.52 | 1.33–4,81 | 0.01 | 0.41 | 0.22–0.81 |
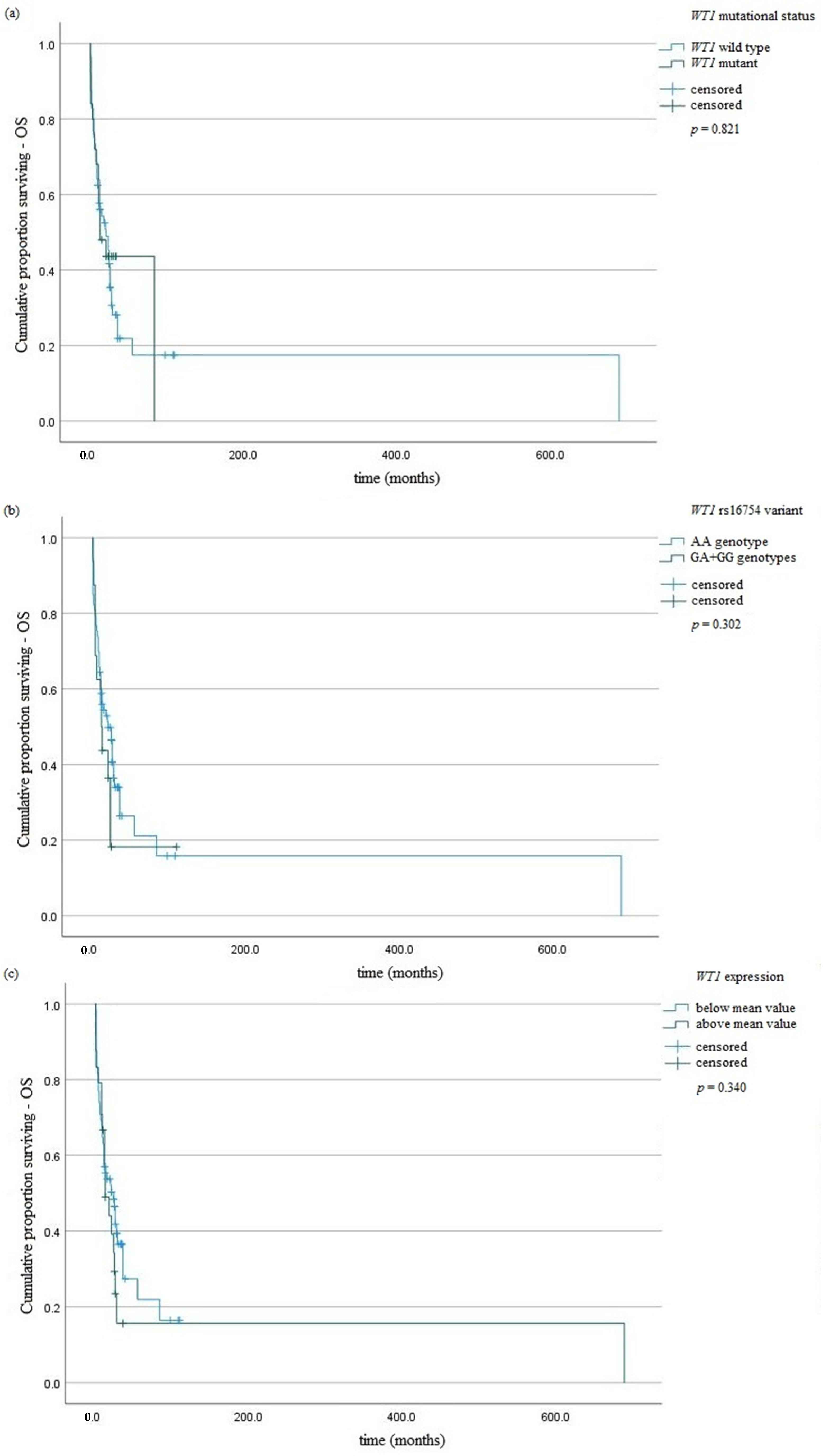
| WT1 mutation | 0.82 | 1.07 | 0.59–2.33 | 0.74 | 0.89 | 0.48–1.52 |
| WT1 overexpression | 0.35 | 0.76 | 0.44–1.34 | 0.54 | 0.84 | 0.45–1.82 |
| NPM1 mutation | 0.13 | 2.03 | 0.81–5.10 | 0.31 | 1.73 | 0.61–4.93 |
| FLT3-ITD | 0.76 | 0.86 | 0.35–2.17 | 0.61 | 0.78 | 0.29–1.93 |
| CEBPA mutation | 0.54 | 1.44 | 0.45–4.62 | 0.96 | 0.97 | 0.27–3.27 |
| Abnormal karyotype | 0.64 | 0.89 | 0.53–1.47 | 0.77 | 0.92 | 0.51–1.56 |
| RFS | ||||||
| Age | 0.07 | 2.08 | 0.92–4.65 | 0.24 | 0.59 | 0.27–1.52 |
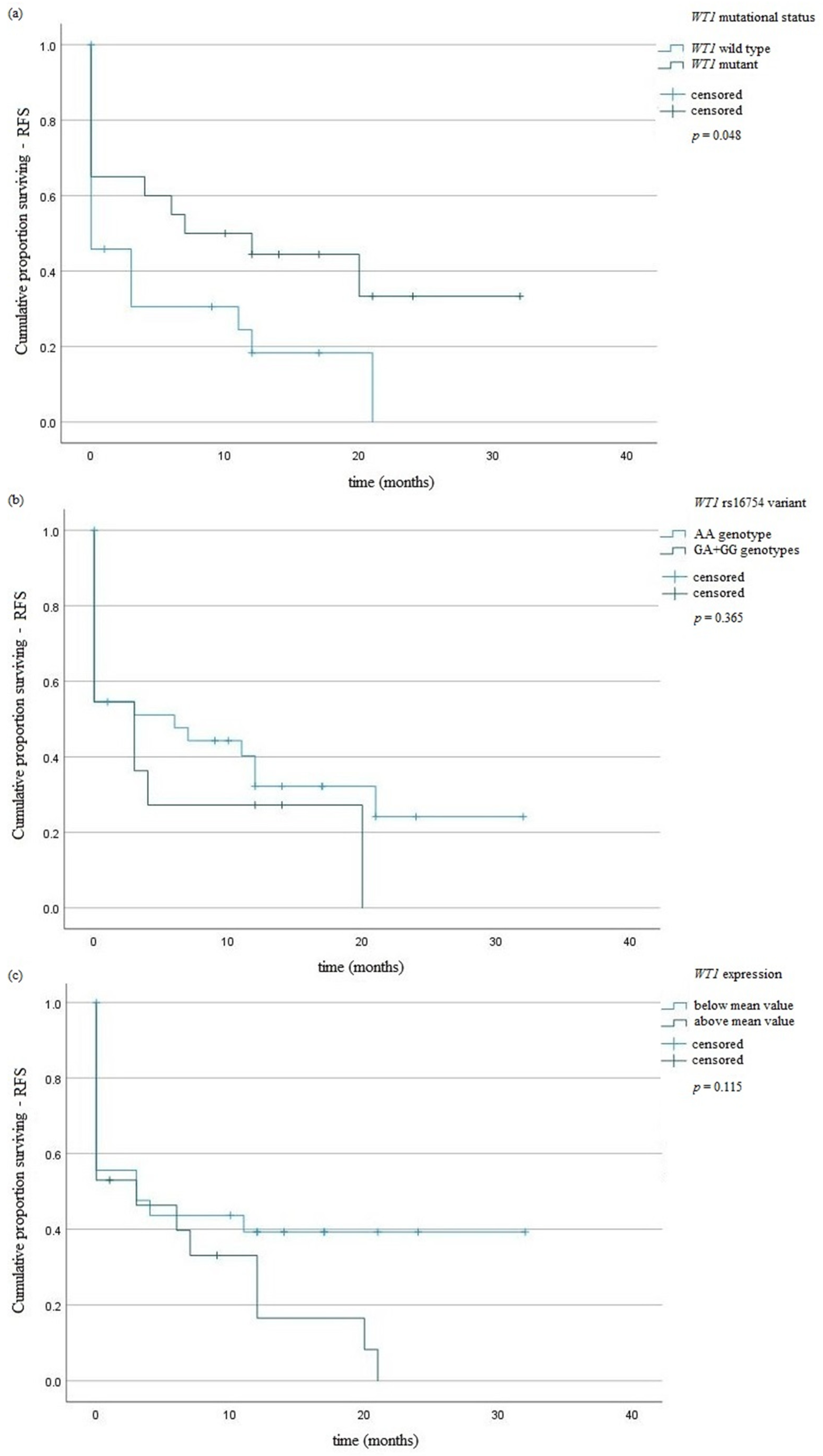
| WT1 mutation | 0.11 | 1.84 | 0.59–3.25 | 0.76 | 1.13 | 0.36–2.18 |
| WT1 overexpression | 0.19 | 0.63 | 0.31–1.27 | 0.59 | 0.79 | 0.33–1.83 |
| NPM1 mutation | 0.04 | 3.46 | 1.03–6.55 | 0.17 | 2.75 | 0.92–5.45 |
| FLT3-ITD | 0.58 | 0.74 | 0.26–2.13 | 0.38 | 0.57 | 0.15–1.99 |
| CEBPA mutation | 0.16 | 2.37 | 0.70–7.95 | 0.62 | 1.42 | 0.39–5.75 |
| Abnormal karyotype | 0.51 | 0.78 | 0.37–1.62 | 0.69 | 0.84 | 0.35–1.87 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koczkodaj, D.; Zmorzyński, S.; Grygalewicz, B.; Pieńkowska-Grela, B.; Styk, W.; Popek-Marciniec, S.; Filip, A.A. WT1 Gene Mutations, rs16754 Variant, and WT1 Overexpression as Prognostic Factors in Acute Myeloid Leukemia Patients. J. Clin. Med. 2022, 11, 1873. https://doi.org/10.3390/jcm11071873
Koczkodaj D, Zmorzyński S, Grygalewicz B, Pieńkowska-Grela B, Styk W, Popek-Marciniec S, Filip AA. WT1 Gene Mutations, rs16754 Variant, and WT1 Overexpression as Prognostic Factors in Acute Myeloid Leukemia Patients. Journal of Clinical Medicine. 2022; 11(7):1873. https://doi.org/10.3390/jcm11071873
Chicago/Turabian StyleKoczkodaj, Dorota, Szymon Zmorzyński, Beata Grygalewicz, Barbara Pieńkowska-Grela, Wojciech Styk, Sylwia Popek-Marciniec, and Agata Anna Filip. 2022. "WT1 Gene Mutations, rs16754 Variant, and WT1 Overexpression as Prognostic Factors in Acute Myeloid Leukemia Patients" Journal of Clinical Medicine 11, no. 7: 1873. https://doi.org/10.3390/jcm11071873
APA StyleKoczkodaj, D., Zmorzyński, S., Grygalewicz, B., Pieńkowska-Grela, B., Styk, W., Popek-Marciniec, S., & Filip, A. A. (2022). WT1 Gene Mutations, rs16754 Variant, and WT1 Overexpression as Prognostic Factors in Acute Myeloid Leukemia Patients. Journal of Clinical Medicine, 11(7), 1873. https://doi.org/10.3390/jcm11071873






