Clinical Utility of Plasma Cell-Free DNA EGFR Mutation Analysis in Treatment-Naïve Stage IV Non-Small Cell Lung Cancer Patients
Abstract
:1. Introduction
2. Methods
2.1. Study Population and Data Collection
2.2. Methods of Analysis
2.3. Statistical Analyses
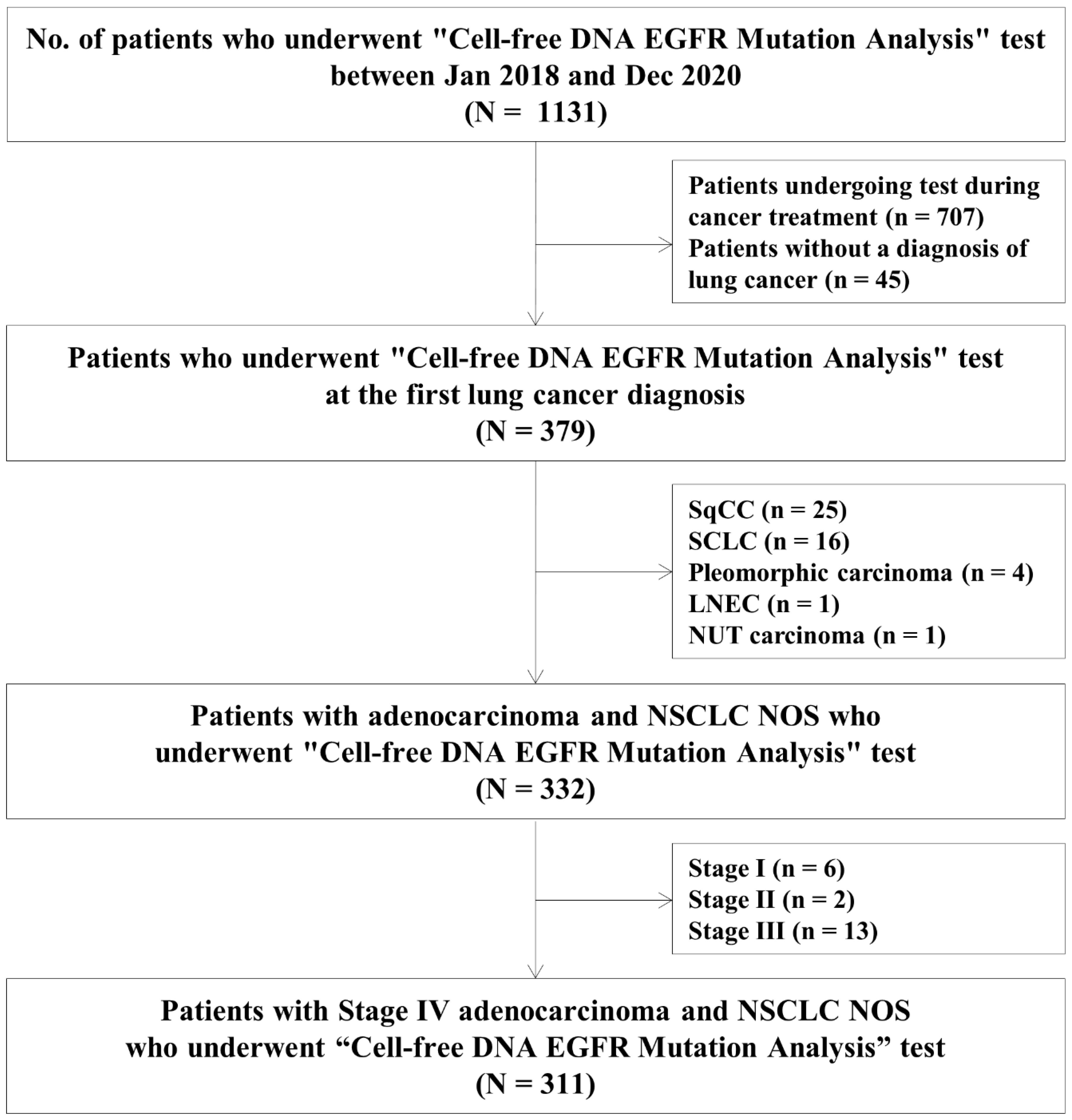
3. Results
3.1. Study Population
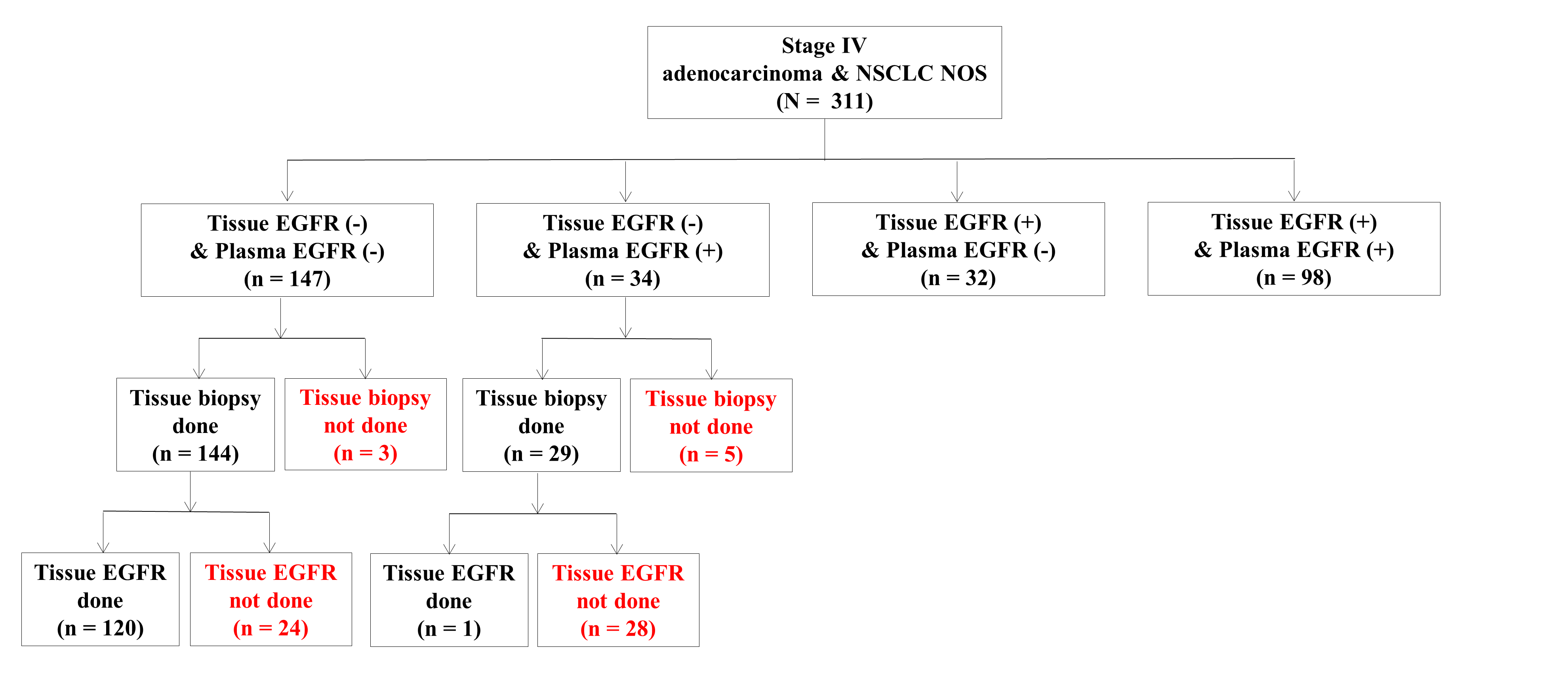
3.2. Diagnostic Performance of Tissue and Plasma EGFR Mutation Tests
3.3. Factors Associated with Positivity of the Plasma EGFR Mutation Assay
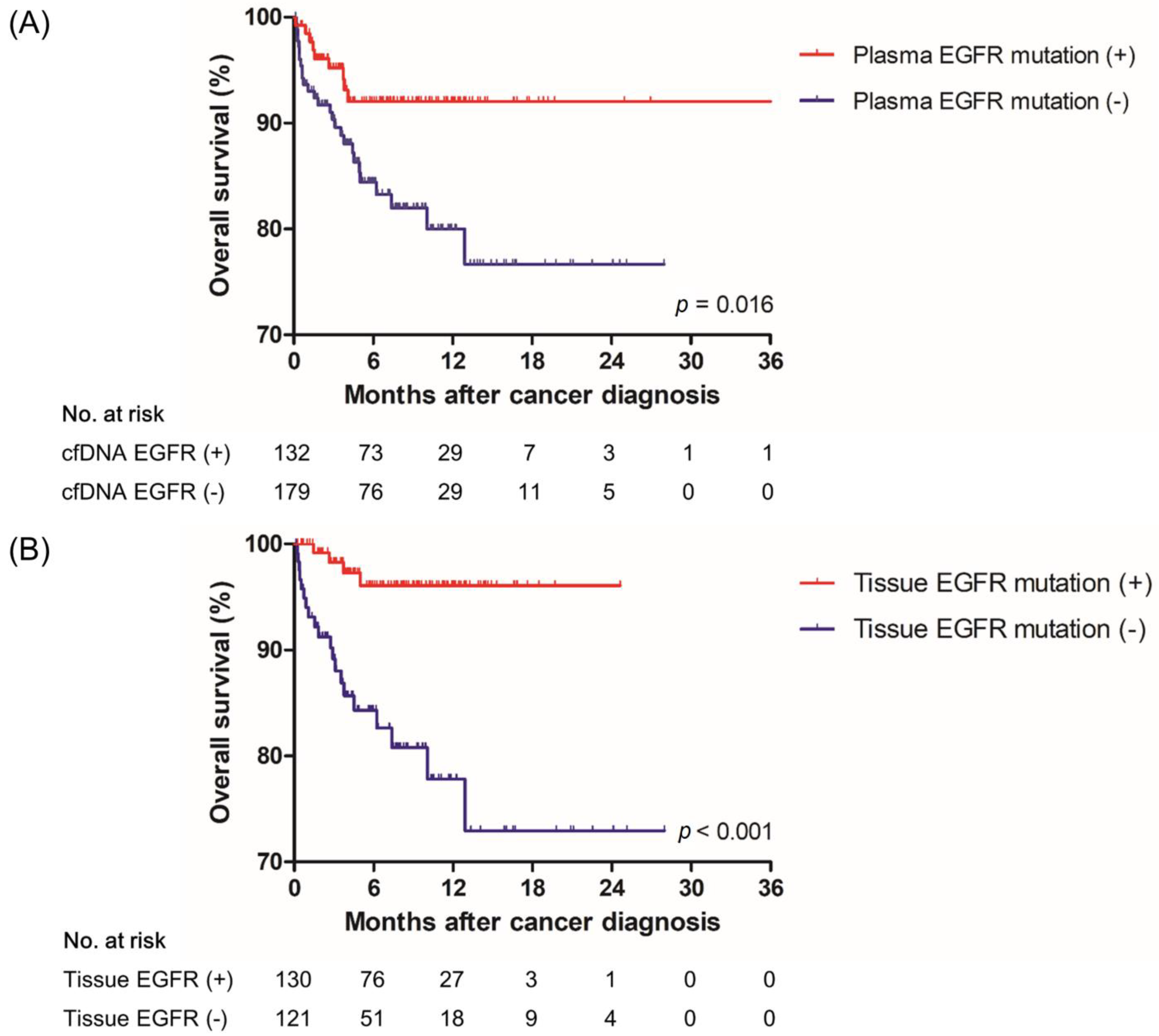
3.4. Effect of a Positive Result of the Plasma EGFR Mutation Test on Treatment and Overall Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| aOR | Adjusted odds ratio |
| BMI | Body mass inde |
| CEA | Carcinoembryonic antigen |
| cfDNA | Cell-free DNA |
| CI | Confidence interval |
| DNA | Deoxyribonucleic acid |
| ECLIA | Electro-chemiluminescence immunoassay |
| EGFR | Epidermal growth factor receptor |
| IRB | Institutional review board |
| IRMA | Immunoradiometric Assay |
| IQR | Interquartile range |
| NCCN | National Comprehensive Cancer Network |
| NGS | Next-generation sequencing |
| NPV | Negative predictive value |
| NSCLC | Non-small cell lung cancer |
| OS | Overall survival |
| PPV | Positive predictive value |
| RT-PCR | Real-time polymerase chain reaction |
| TAT | Turnaround time |
| TKI | Tyrosine kinase inhibitor |
| TTI | Time to treatment initiation |
References
- Bade, B.C.; Dela Cruz, C.S. Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clin. Chest Med. 2020, 41, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Fitzmaurice, C.; Allen, C.; Barber, R.M.; Barregard, L.; Bhutta, Z.A.; Brenner, H.; Dicker, D.J.; Chimed-Orchir, O.; Dandona, R.; Dandona, L.; et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar] [PubMed]
- Zhou, C.; Wu, Y.-L.; Chen, G.; Feng, J.; Liu, X.-Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef]
- Soria, J.-C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Network, T.C.G.A.R. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar]
- Liang, Z.; Cheng, Y.; Chen, Y.; Hu, Y.; Liu, W.-P.; Lu, Y.; Wang, J.; Wang, Y.; Wu, G.; Ying, J.-M.; et al. EGFR T790M ctDNA testing platforms and their role as companion diagnostics: Correlation with clinical outcomes to EGFR-TKIs. Cancer Lett. 2017, 403, 186–194. [Google Scholar] [CrossRef]
- Chen, J.; Yang, H.; Teo, A.S.M.; Amer, L.B.; Sherbaf, F.G.; Tan, C.Q.; Alvarez, J.J.S.; Lu, B.; Lim, J.Q.; Takano, A.; et al. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020, 52, 177–186. [Google Scholar] [CrossRef]
- Singh, A.P.; Li, S.; Cheng, H. Circulating DNA in EGFR-mutated lung cancer. Ann. Transl. Med. 2017, 5, 379. [Google Scholar] [CrossRef] [Green Version]
- Sacher, A.G.; Paweletz, C.; Dahlberg, S.E.; Alden, R.S.; O’Connell, A.; Feeney, N.; Mach, S.L.; Jänne, P.A.; Oxnard, G.R. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016, 2, 1014–1022. [Google Scholar] [CrossRef] [Green Version]
- Deng, Q.; Fang, Q.; Sun, H.; Singh, A.P.; Alexander, M.; Li, S.; Cheng, H.; Zhou, S. Detection of plasma EGFR mutations for personalized treatment of lung cancer patients without pathologic diagnosis. Cancer Med. 2020, 9, 2085–2095. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement from the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, D.S.; Wood, D.E.; Aisner, D.L.; Akerley, W.; Bauman, J.R.; Bharat, A.; Bruno, D.S.; Chang, J.Y.; Chirieac, L.R.; D’Amico, T.A.; et al. NCCN Guidelines Insights: Non–Small Cell Lung Cancer, Version 2.2021. J. Natl. Compr. Cancer Netw. 2021, 19, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. cobas EGFR Mutation Test v2. 2021. Available online: http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm504540.htm (accessed on 17 August 2021).
- Lee, J.; La Choi, Y.; Han, J.; Park, S.; Jung, H.A.; Sun, J.-M.; Lee, S.-H.; Ahn, J.S.; Park, K.; Ahn, M.-J. Osimertinib Improves Overall Survival in Patients with EGFR-Mutated NSCLC with Leptomeningeal Metastases Regardless of T790M Mutational Status. J. Thorac. Oncol. 2020, 15, 1758–1766. [Google Scholar] [CrossRef]
- Trevethan, R. Sensitivity, Specificity, and Predictive Values: Foundations, Pliabilities, and Pitfalls in Research and Practice. Front. Public Health 2017, 5, 307. [Google Scholar] [CrossRef]
- Cescon, D.W.; Bratman, S.V.; Chan, S.M.; Siu, L.L. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer 2020, 1, 276–290. [Google Scholar] [CrossRef]
- Palmirotta, R.; Lovero, D.; Cafforio, P.; Felici, C.; Mannavola, F.; Pellè, E.; Quaresmini, D.; Tucci, M.; Silvestris, F. Liquid biopsy of cancer: A multimodal diagnostic tool in clinical oncology. Ther. Adv. Med. Oncol. 2018, 10, 1758835918794630. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Venesio, T.; Marsoni, S.; Seoane, J.; Dive, C.; Papadopoulos, N.; Kopetz, S.; Corcoran, R.; Siu, L.; et al. How liquid biopsies can change clinical practice in oncology. Ann. Oncol. 2019, 30, 1580–1590. [Google Scholar] [CrossRef] [Green Version]
- Mok, T.; Wu, Y.-L.; Lee, J.S.; Yu, C.-J.; Sriuranpong, V.; Sandoval-Tan, J.; Ladrera, G.; Thongprasert, S.; Srimuninnimit, V.; Liao, M.; et al. Detection and Dynamic Changes of EGFR Mutations from Circulating Tumor DNA as a Predictor of Survival Outcomes in NSCLC Patients Treated with First-line Intercalated Erlotinib and Chemotherapy. Clin. Cancer Res. 2015, 21, 3196–3203. [Google Scholar] [CrossRef] [Green Version]
- Weber, B.; Meldgaard, P.; Hager, H.; Wu, L.; Wei, W.; Tsai, J.; Khalil, A.; Nexo, E.; Sorensen, B.S. Detection of EGFR mutations in plasma and biopsies from non-small cell lung cancer patients by allele-specific PCR assays. BMC Cancer 2014, 14, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thress, K.S.; Brant, R.; Carr, T.; Dearden, S.; Jenkins, S.; Brown, H.; Hammett, T.; Cantarini, M.; Barrett, J.C. EGFR mutation detection in ctDNA from NSCLC patient plasma: A cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer 2015, 90, 509–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, S.; Yang, J.C.-H.; Ramalingam, S.S.; Yu, K.; Patel, S.; Weston, S.; Hodge, R.; Cantarini, M.; Jänne, P.A.; Mitsudomi, T.; et al. Plasma ctDNA Analysis for Detection of the EGFR T790M Mutation in Patients with Advanced Non–Small Cell Lung Cancer. J. Thorac. Oncol. 2017, 12, 1061–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grunnet, M.; Sorensen, J. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer 2012, 76, 138–143. [Google Scholar] [CrossRef]
- Gao, X.C.; Wei, C.H.; Zhang, R.G.; Cai, Q.; He, Y.; Tong, F.; Dong, J.H.; Wu, G.; Dong, X.R. (18)F-FDG PET/CT SUV(max) and serum CEA levels as predictors for EGFR mutation state in Chinese patients with non-small cell lung cancer. Oncol. Lett. 2020, 20, 61. [Google Scholar] [CrossRef]
- Wang, W.-T.; Li, Y.; Ma, J.; Chen, X.-B.; Qin, J.-J. Serum carcinoembryonic antigen levels before initial treatment are associated with EGFR mutations and EML4- ALK fusion gene in lung adenocarcinoma patients. Asian Pac. J. Cancer Prev. 2014, 15, 3927–3932. [Google Scholar] [CrossRef] [Green Version]
- Sholl, L.M.; Aisner, D.L.; Varella-Garcia, M.; Berry, L.D.; Dias-Santagata, D.; Wistuba, I.I.; Chen, H.; Fujimoto, J.; Kugler, K.; Franklin, W.A.; et al. Multi-institutional Oncogenic Driver Mutation Analysis in Lung Adenocarcinoma: The Lung Cancer Mutation Consortium Experience. J. Thorac. Oncol. 2015, 10, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.; Kim, E.Y.; Lee, S.H.; Kwon, D.S.; Kim, A.; Chang, Y.S. Presence of mEGFR ctDNA predicts a poor clinical outcome in lung adenocarcinoma. Thorac. Cancer 2019, 10, 2267–2273. [Google Scholar] [CrossRef]
- Guan, J.; Chen, M.; Xiao, N.; Li, L.; Zhang, Y.; Li, Q.; Yang, M.; Liu, L.; Chen, L. EGFR mutations are associated with higher incidence of distant metastases and smaller tumor size in patients with non-small-cell lung cancer based on PET/CT scan. Med. Oncol. 2015, 33, 1–8. [Google Scholar] [CrossRef]
- Yang, B.; Lee, H.; Um, S.-W.; Kim, K.; Zo, J.I.; Shim, Y.M.; Kwon, O.J.; Lee, K.S.; Ahn, M.-J.; Kim, H. Incidence of brain metastasis in lung adenocarcinoma at initial diagnosis on the basis of stage and genetic alterations. Lung Cancer 2019, 129, 28–34. [Google Scholar] [CrossRef]
- Doebele, R.C.; Lu, X.; Sumey, C.; Bs, D.A.M.; Weickhardt, A.J.; Oton, A.B.; Bunn, P.A.; Barón, A.E.; Franklin, W.A.; Aisner, D.L.; et al. Oncogene status predicts patterns of metastatic spread in treatment-naive nonsmall cell lung cancer. Cancer 2012, 118, 4502–4511. [Google Scholar] [CrossRef] [PubMed]
- Denis, M.G.; Lafourcade, M.-P.; Le Garff, G.; Dayen, C.; Falchero, L.; Thomas, P.; Locher, C.; Oliviero, G.; Licour, M.; Reck, M.; et al. Circulating free tumor-derived DNA to detect EGFR mutations in patients with advanced NSCLC: French subset analysis of the ASSESS study. J. Thorac. Dis. 2019, 11, 1370–1378. [Google Scholar] [CrossRef] [PubMed]
- Seki, Y.; Fujiwara, Y.; Kohno, T.; Yoshida, K.; Goto, Y.; Horinouchi, H.; Kanda, S.; Nokihara, H.; Yamamoto, N.; Kuwano, K.; et al. Circulating cell-free plasma tumour DNA shows a higher incidence of EGFR mutations in patients with extrathoracic disease progression. ESMO Open 2018, 3, e000292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikushima, H.; Sakatani, T.; Usui, K. Clinical Features of Patients with an Epidermal Growth Factor Receptor T790M Mutation Detected in Circulating Tumor DNA. Oncology 2019, 98, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Park, S.; Kim, W.S.; Lee, J.C.; Jang, S.J.; Choi, J.; Choi, C.-M. Correlation between progression-free survival, tumor burden, and circulating tumor DNA in the initial diagnosis of advanced-stageEGFR-mutated non-small cell lung cancer. Thorac. Cancer 2018, 9, 1104–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, C.-Y.; Lee, M.-H.; Tsai, M.-J.; Yang, C.-J.; Hung, J.-Y.; Chong, I.-W. The Factors Predicting Concordant Epidermal Growth Factor Receptor (EGFR) Mutation Detected in Liquid/Tissue Biopsy and the Related Clinical Outcomes in Patients of Advanced Lung Adenocarcinoma with EGFR Mutations. J. Clin. Med. 2019, 8, 1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serdarevic, N.; Smajic, J. Comparison of chemiluminescent microparticle immunoassay (CMIA) with electrochemiluminescence immunoassay (ECLIA) for Carcinoembryonic antigen (CEA). J. Health Sci. 2018, 8, 94–100. [Google Scholar] [CrossRef] [Green Version]
- Valdes, R.; Stein, K.E. Comparison of RIA and IRMA methods for measurement of carcinoembryonic antigen (CEA). Clin. Biochem. 1982, 15, 241–247. [Google Scholar] [CrossRef]
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, C.; Rolfo, C.D.; Oxnard, G.R.; Gray, J.E.; Sholl, L.M.; Gandara, D.R. Strategies for the successful implementation of plasma-based NSCLC genotyping in clinical practice. Nat. Rev. Clin. Oncol. 2021, 18, 56–62. [Google Scholar] [CrossRef]
| Variables | Total (N = 311) | Tissue (−)/ Plasma (−) (N = 147) | Tissue (−)/ Plasma (+) (N = 34) | Tissue (+)/ Plasma (−) (N = 32) | Tissue (+)/ Plasma (+) (N = 98) | p |
|---|---|---|---|---|---|---|
| Age, years | 65 (57–74) | 65 (59–73) | 61 (55–79) | 66 (54–77) | 64 (55–72) | 0.732 |
| Sex, female | 176 (56.6) | 64 (43.5) | 19 (55.9) | 26 (81.3) | 67 (68.4) | <0.001 bc |
| Smoking history | <0.001 bc | |||||
| Never smoker | 195 (62.7) | 71 (48.3) | 21 (61.8) | 25 (78.1) | 78 (79.6) | |
| Ever smoker | 116 (37.3) | 76 (51.7) | 13 (38.2) | 7 (21.9) | 20 (20.4) | |
| ECOG at diagnosis | 0.079 | |||||
| ECOG 0–2 | 297 (95.5) | 136 (92.5) | 33 (97.1) | 31 (96.9) | 97 (99.0) | |
| ECOG 3–4 | 14 (4.5) | 11 (7.5) | 1 (2.9) | 1 (3.1) | 1 (1.0) | |
| CEA, ng/mL (n = 248) | 0.006 c | |||||
| <3.2 ng/mL | 56 (22.6) | 39 (33.1) | 5 (20.8) | 5 (20.0) | 7 (8.6) | |
| 3.2–94.7 ng/mL | 124 (50.0) | 51 (43.2) | 13 (54.2) | 15 (60.0) | 45 (55.6) | |
| >94.7 ng/mL | 68 (27.4) | 28 (23.7) | 6 (25.0) | 5 (20.0) | 29 (35.8) | |
| Histology * | 0.022 a | |||||
| Adenocarcinoma | 309 (99.4) | 147 (100.0) | 32 (94.1) | 32 (100.0) | 98 (100.0) | |
| NSCLC NOS | 2 (0.6) | 0 (0.0) | 2 (5.9) | 0 (0.0) | 0 (0.0) | |
| Clinical stage at the diagnosis | ||||||
| T stage | 0.705 | |||||
| T1 | 67 (21.6) | 37 (25.2) | 8 (23.5) | 8 (25.0) | 14 (14.3) | |
| T2 | 94 (30.2) | 38 (25.8) | 11 (32.4) | 10 (31.3) | 35 (35.7) | |
| T3 | 57 (18.3) | 27 (18.4) | 7 (20.6) | 5 (15.6) | 18 (18.4) | |
| T4 | 93 (29.9) | 45 (30.6) | 8 (23.5) | 9 (28.1) | 31 (31.6) | |
| N stage | <0.001 bdf | |||||
| N0 | 35 (11.2) | 17 (11.6) | 2 (5.9) | 11 (34.4) | 5 (5.1) | |
| N1 | 22 (7.1) | 14 (9.5) | 0 (0.0) | 2 (6.3) | 6 (6.1) | |
| N2 | 77 (24.8) | 38 (25.8) | 10 (29.4) | 11 (34.4) | 18 (18.4) | |
| N3 | 177 (56.9) | 78 (53.1) | 22 (64.7) | 8 (25.0) | 69 (70.4) | |
| M stage | 0.004 f | |||||
| M1a | 91 (29.3) | 47 (32.0) | 10 (29.4) | 16 (50.0) | 18 (18.4) | |
| M1b | 31 (10.0) | 16 (10.9) | 3 (8.8) | 5 (15.6) | 7 (7.1) | |
| M1c | 189 (60.8) | 84 (57.1) | 21 (61.8) | 11 (34.4) | 73 (74.5) | |
| No. of metastatic sites | <0.001 acdf | |||||
| 1 | 101 (32.5) | 59 (40.1) | 9 (26.5) | 17 (53.1) | 16 (16.3) | |
| 2 | 73 (23.5) | 44 (29.9) | 4 (11.8) | 9 (28.1) | 16 (16.3) | |
| 3 | 37 (11.9) | 23 (15.6) | 2 (5.9) | 5 (15.6) | 7 (7.1) | |
| ≥4 | 100 (32.2) | 21 (14.3) | 19 (55.9) | 1 (3.1) | 59 (60.2) | |
| Location of metastasis ** | ||||||
| Brain | 94 (30.2) | 34 (23.1) | 12 (35.3) | 8 (25.0) | 40 (40.8) | 0.023 c |
| Bone | 139 (44.7) | 67 (45.6) | 12 (35.3) | 10 (31.3) | 50 (51.0) | 0.159 |
| Intrathoracic metastasis † | 192 (61.7) | 79 (53.7) | 22 (64.7) | 23 (71.9) | 68 (69.4) | 0.050 |
| Intraabdominal metastasis ‡ | 74 (23.8) | 38 (25.9) | 6 (17.6) | 3 (9.4) | 27 (27.6) | 0.140 |
| Others § | 14 (4.5) | 8 (5.5) | 0 (0.0) | 0 (0.0) | 6 (6.1) | 0.337 |
| Tissue EGFR mutation type ∥ | ||||||
| Exon 19 deletion | 68 (21.9) | 0 (0.0) | 0 (0.0) | 13 (40.6) | 55 (56.1) | <0.001 bcde |
| L858R (exon 21) | 54 (17.4) | 0 (0.0) | 0 (0.0) | 12 (37.5) | 42 (42.9) | <0.001 bcde |
| Others ¶ | 17 (5.5) | 0 (0.0) | 0 (0.0) | 6 (18.8) | 11 (11.2) | <0.001 bc |
| Plasma EGFR mutation type ∥ | ||||||
| Exon 19 deletion | 71 (22.8) | 0 (0.0) | 17 (50.0) | 0 (0.0) | 54 (55.1) | <0.001 acdf |
| L858R (exon 21) | 58 (18.6) | 0 (0.0) | 16 (47.1) | 0 (0.0) | 42 (42.9) | <0.001 acdf |
| Others ++ | 6 (1.9) | 0 (0.0) | 1 (2.9) | 0 (0.0) | 5 (5.1) | 0.019 c |
| Use of EGFR-TKI | 154 (49.5) | 0 (0.0) | 32 (94.1) | 26 (81.3) | 96 (98.0) | <0.001 abcf |
| Gefitinib | 46 (14.8) | 0 (0.0) | 11 (32.3) | 12 (37.5) | 23 (23.5) | <0.001 abc |
| Erlotinib | 23 (7.4) | 0 (0.0) | 4 (11.8) | 2 (6.3) | 17 (17.3) | <0.001 ac |
| Afatinib | 77 (24.7) | 0 (0.0) | 14 (41.2) | 10 (31.2) | 53 (54.1) | <0.001 abc |
| Osimertinib | 8 (2.6) | 0 (0.0) | 3 (8.8) | 2 (6.3) | 3 (3.1) | 0.014 a |
| Use of EGFR-TKI as 1st line treatment | 147 (47.3) | 0 (0.0) | 29 (85.3) | 25 (78.1) | 93 (94.9) | <0.001 abc |
| Variables | Univariable | Multivariable | ||
|---|---|---|---|---|
| Unadjusted OR (95% CI) | p-Value | Adjusted OR (95% CI) | p-Value | |
| Age, years | 0.99 (0.97–1.01) | 0.242 | 0.98 (0.95–1.00) | 0.086 |
| Sex, female | 1.85 (1.16–2.94) | 0.009 | ||
| Smoking history | ||||
| Ever smoker | Reference | Reference | ||
| Never smoker | 2.59 (1.59–4.24) | < 0.001 | 2.83 (1.55–5.20) | 0.001 |
| CEA level | ||||
| <3.2 ng/mL | Reference | Reference | ||
| 3.2–94.7 ng/mL | 3.22 (1.55–6.68) | 0.002 | 2.61 (1.16–5.84) | 0.020 |
| >94.7 ng/mL | 3.89 (1.75–8.62) | 0.001 | 2.98 (1.21–7.35) | 0.018 |
| N stage | ||||
| N0 | Reference | Reference | ||
| N1 | 1.50 (0.43–5.24) | 0.525 | 1.65 (0.38–7.17) | 0.501 |
| N2 | 2.29 (0.88–5.91) | 0.088 | 2.52 (0.78–8.17) | 0.124 |
| N3 | 4.23 (1.76–10.20) | 0.001 | 4.22 (1.41–12.62) | 0.010 |
| M stage | ||||
| M1a | Reference | |||
| M1b | 1.07 (0.45–2.57) | 0.877 | ||
| M1c | 2.23 (1.31–3.78) | 0.003 | ||
| Type of metastatic organs * | ||||
| Brain | 2.12 (1.30–3.47) | 0.003 | 2.73 (1.39–5.36) | 0.003 |
| Bone | 1.14 (0.72–1.79) | 0.575 | ||
| Intrathoracic metastasis ** | 1.62 (1.01–2.59) | 0.045 | 2.61 (1.38–4.96) | 0.003 |
| Intraabdominal metastasis † | 1.12 (0.66–1.90) | 0.668 | 1.85 (0.93–3.68) | 0.079 |
| Others ‡ | 1.01 (0.34–2.99) | 0.983 | ||
| Variables | N = 311 |
|---|---|
| Tissue biopsy | |
| Yes | 303 (97.4) |
| No | 8 (2.6) |
| Biopsy methods | |
| EBUS-TBNA | 146 (46.9) |
| Percutaneous core needle biopsy | 99 (31.8) |
| TBLB | 38 (12.2) |
| VATS | 16 (5.1) |
| Others * | 4 (1.3) |
| Biopsy sites | |
| Mediastinal lymph nodes | 146 (46.9) |
| Lung | 103 (33.1) |
| SCN | 33 (10.6) |
| Others † | 21 (6.8) |
| Tissue EGFR mutation test | |
| Performed | 251 (80.7) |
| Time taken from the first hospital visit to the test, days | 7 (4–12) |
| EGFR gene, mutation real-time PCR | 236 (75.9) |
| Turnaround time, days | 11 (9–13) |
| NGS | 15 (4.8) |
| Turnaround time, days | 41 (34–49) |
| Not performed | 60 (19.3) |
| Result of tissue EGFR mutation test (N = 251) | |
| Positive | 130 (51.8) |
| Negative | 121 (48.2) |
| Plasma EGFR mutation | |
| Performed | 311 (100) |
| Time taken from the first hospital visit to the test, days | 1 (0–5) |
| Turnaround time, days | 5 (4–6) |
| Result of plasma EGFR mutation test (N = 311) | |
| Positive | 132 (42.4) |
| Negative | 179 (57.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, B.-G.; Jang, J.-H.; Kim, J.-W.; Shin, S.H.; Jeong, B.-H.; Lee, K.; Kim, H.; Kwon, O.J.; Ahn, M.-J.; Um, S.-W. Clinical Utility of Plasma Cell-Free DNA EGFR Mutation Analysis in Treatment-Naïve Stage IV Non-Small Cell Lung Cancer Patients. J. Clin. Med. 2022, 11, 1144. https://doi.org/10.3390/jcm11041144
Kim B-G, Jang J-H, Kim J-W, Shin SH, Jeong B-H, Lee K, Kim H, Kwon OJ, Ahn M-J, Um S-W. Clinical Utility of Plasma Cell-Free DNA EGFR Mutation Analysis in Treatment-Naïve Stage IV Non-Small Cell Lung Cancer Patients. Journal of Clinical Medicine. 2022; 11(4):1144. https://doi.org/10.3390/jcm11041144
Chicago/Turabian StyleKim, Bo-Guen, Ja-Hyun Jang, Jong-Won Kim, Sun Hye Shin, Byeong-Ho Jeong, Kyungjong Lee, Hojoong Kim, O Jung Kwon, Myung-Ju Ahn, and Sang-Won Um. 2022. "Clinical Utility of Plasma Cell-Free DNA EGFR Mutation Analysis in Treatment-Naïve Stage IV Non-Small Cell Lung Cancer Patients" Journal of Clinical Medicine 11, no. 4: 1144. https://doi.org/10.3390/jcm11041144
APA StyleKim, B.-G., Jang, J.-H., Kim, J.-W., Shin, S. H., Jeong, B.-H., Lee, K., Kim, H., Kwon, O. J., Ahn, M.-J., & Um, S.-W. (2022). Clinical Utility of Plasma Cell-Free DNA EGFR Mutation Analysis in Treatment-Naïve Stage IV Non-Small Cell Lung Cancer Patients. Journal of Clinical Medicine, 11(4), 1144. https://doi.org/10.3390/jcm11041144








