Quantitative Assessment of Tumor-Infiltrating Lymphocytes Using Machine Learning Predicts Survival in Muscle-Invasive Bladder Cancer
Abstract
1. Introduction
2. Materials and Methods
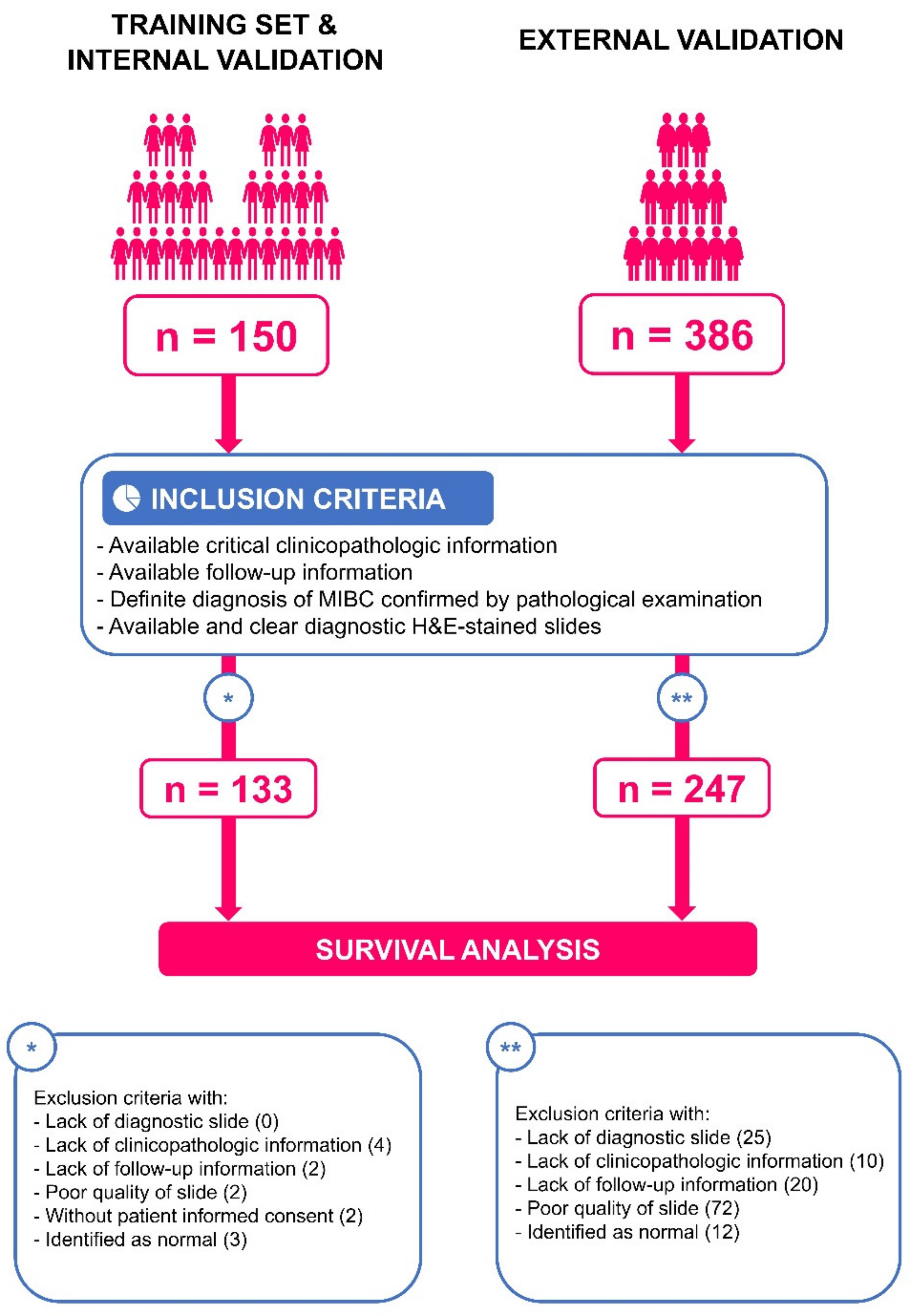
2.1. Clinical Cohorts
2.2. Ethics
2.3. Digital Scanning
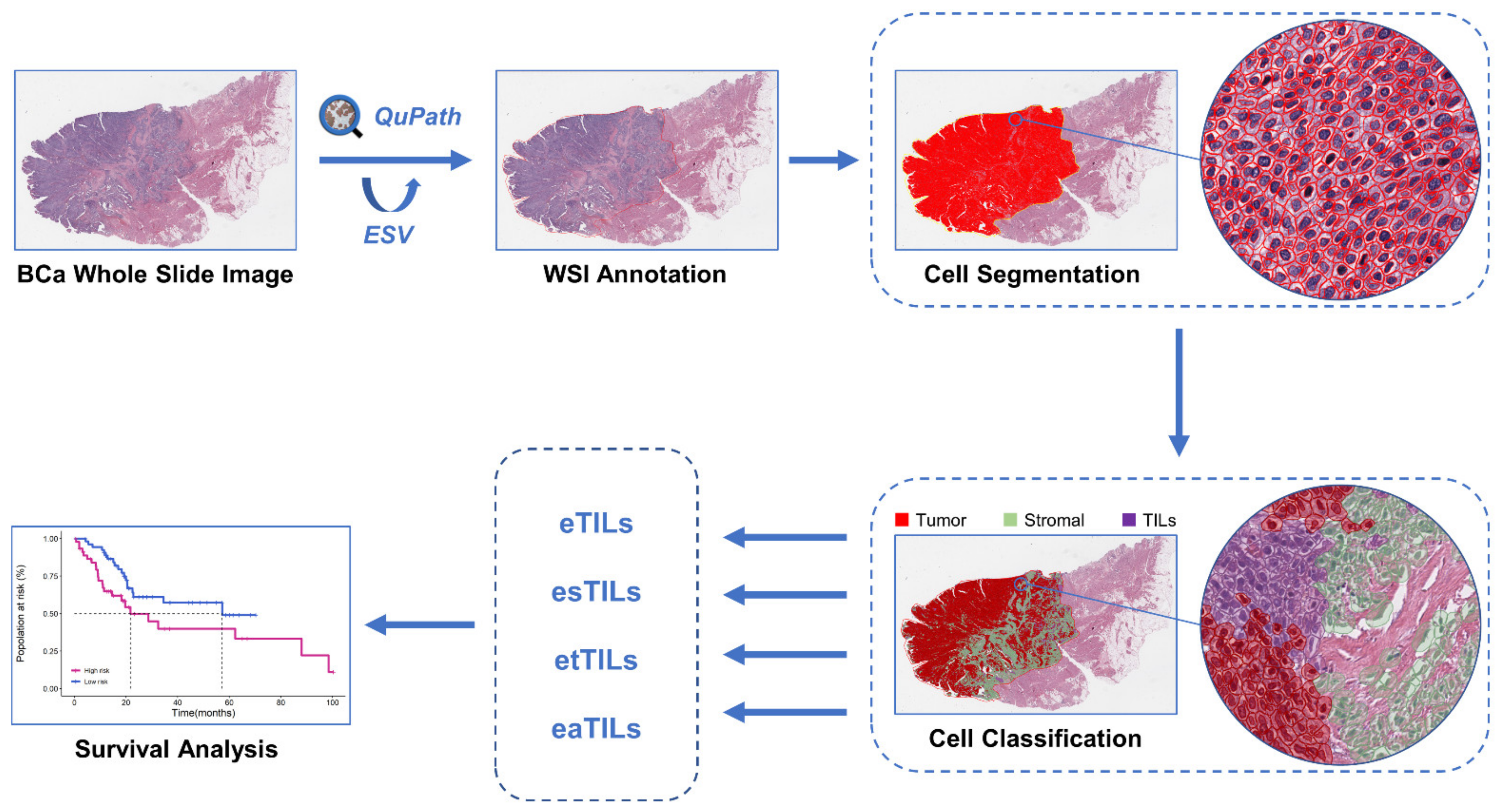
2.4. WSI Image Analysis
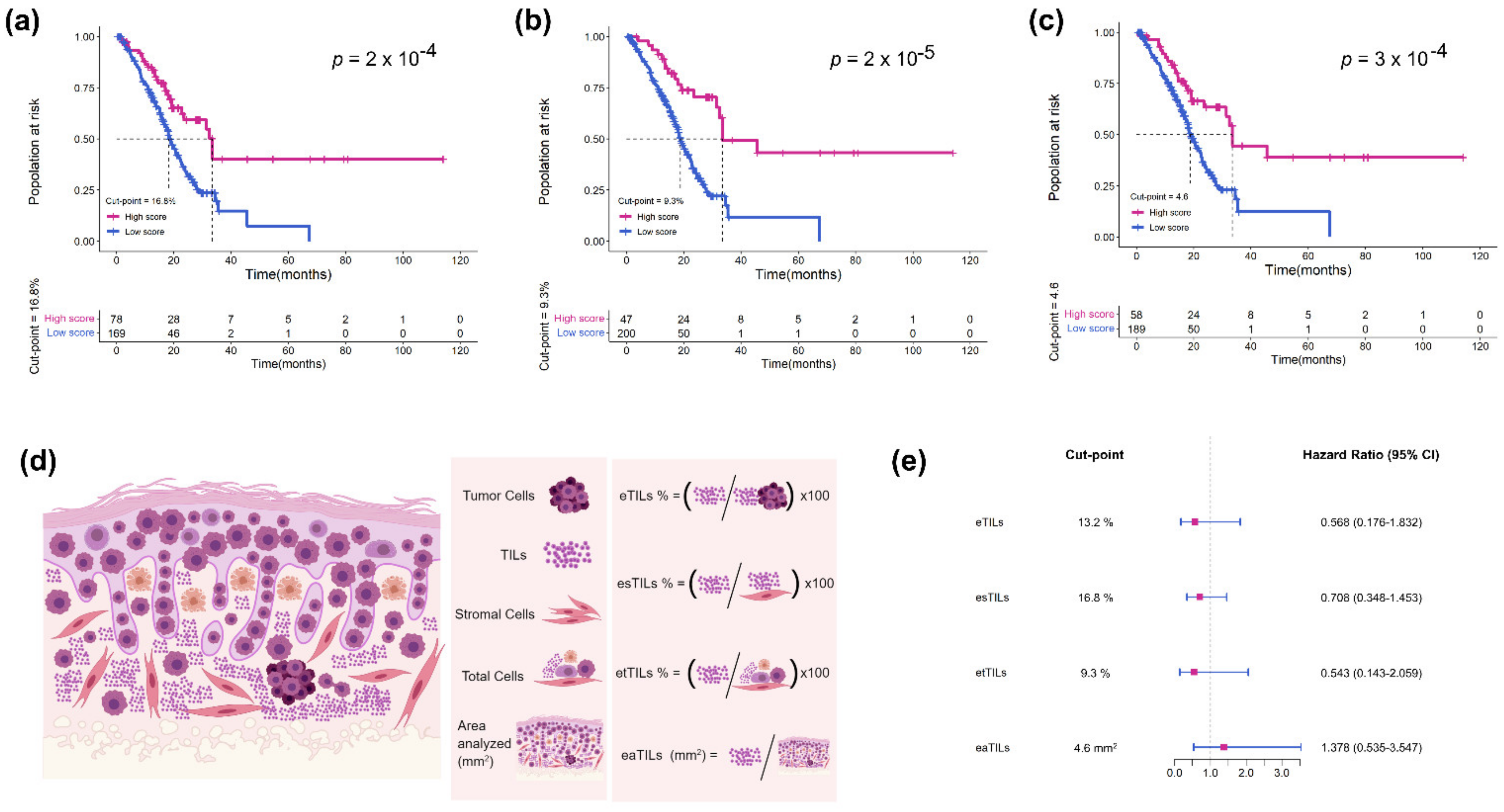
2.5. Assessment of TILs Using Four Variables
- (1)
- Calculate the proportion of TILs in tumor cells: eTILs% = (TILs/TILs + tumor cells) × 100
- (2)
- Calculate the proportion of TILs in stromal cells: esTILs% = (TILs/TILs + stromal cells) × 100
- (3)
- Calculate the proportion of TILs in total cells: etTILs% = (TILs/total cells) × 100
- (4)
- Calculate the infiltration density of TILs in the tumor region: eaTILs = TILs/tumor region areas analyzed (mm2)
2.6. Statistical Analyses
3. Results
3.1. Patient Characteristics
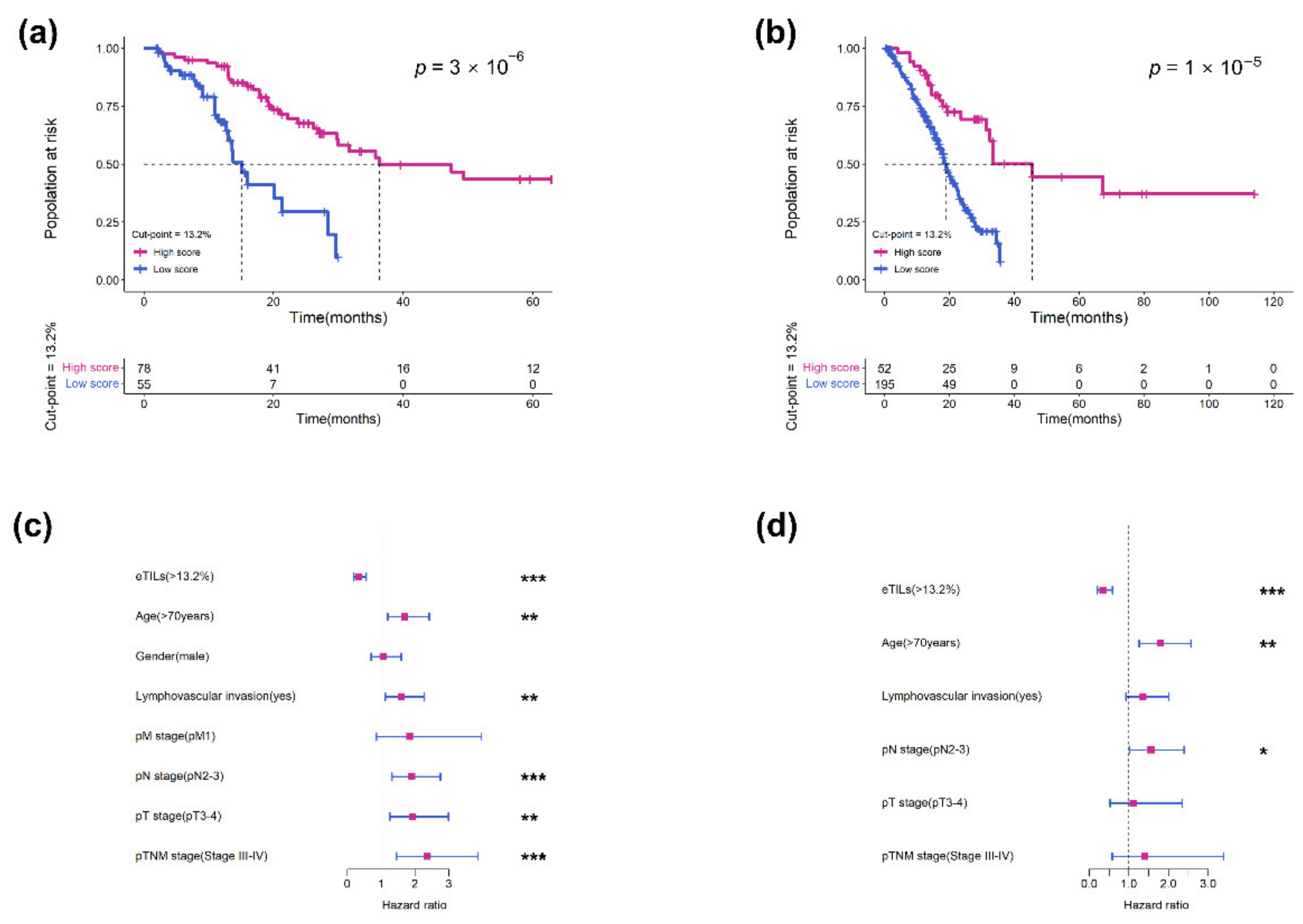
3.2. Measurement of eTILs as a Prognostic Variable in Two Cohorts
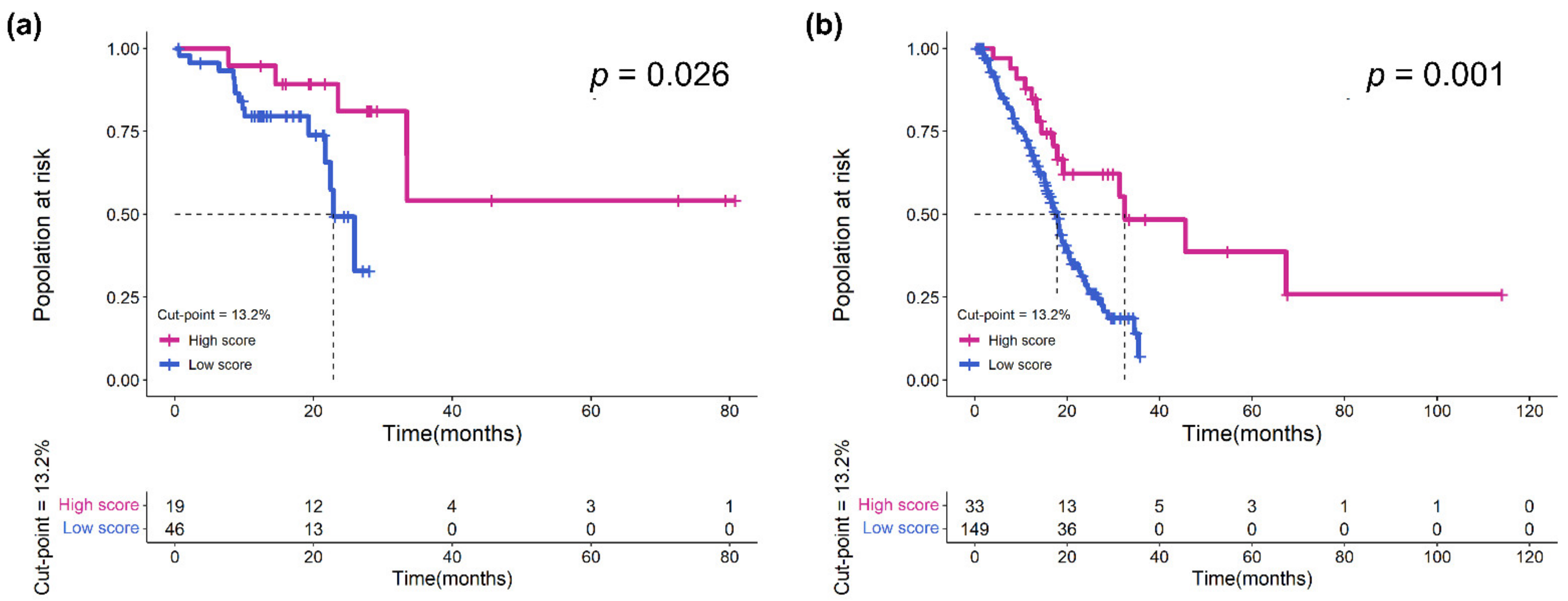
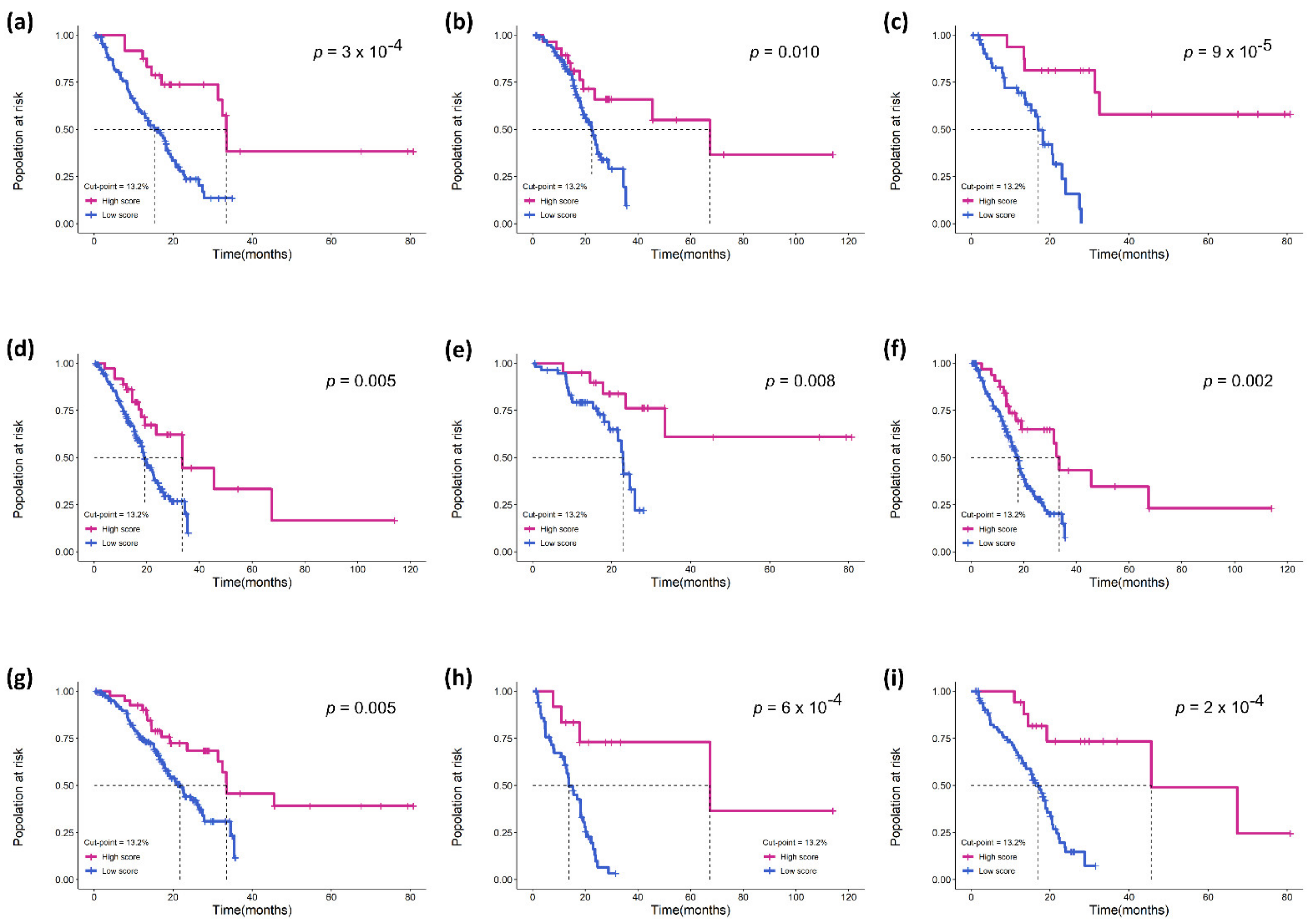
3.3. Validation of the Prognostic Effect of eTILs in Different Subgroups
3.4. Assessment of Four TIL Variables in TCGA Cohort
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Patel, V.G.; Oh, W.K.; Galsky, M.D. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J. Clin. 2020, 70, 404–423. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics, 2022. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, X.; Lin, T. Emerging strategies for the improvement of chemotherapy in bladder cancer: Current knowledge and future perspectives. J. Adv. Res. 2022, 39, 187–202. [Google Scholar] [CrossRef]
- Motterle, G.; Andrews, J.R.; Morlacco, A.; Karnes, R.J. Predicting response to neoadjuvant chemotherapy in bladder cancer. Eur. Urol. Focus 2020, 6, 642–649. [Google Scholar] [CrossRef]
- Rouprêt, M.; Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Cowan, N.C.; Dominguez-Escrig, J.L.; Gontero, P.; Hugh, M.A.; et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2020 Update. Eur. Urol. 2021, 79, 62–79. [Google Scholar] [CrossRef]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef]
- Kumar, A.; Watkins, R.; Vilgelm, A.E. Cell therapy with TILs: Training and taming t cells to fight cancer. Front. Immunol. 2021, 12, 690499. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; Denardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; van de Vijver, K.; Estrada, M.V.; Gonzalez-Ericsson, P.I.; et al. Assessing tumor-infiltrating lymphocytes in solid tumors: A practical review for pathologists and proposal for a standardized method from the international immunooncology biomarkers working group: Part 1: Assessing the host immune response, TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv. Anat. Pathol. 2017, 24, 235–251. [Google Scholar]
- Liu, K.; Zhao, K.; Wang, L.; Sun, E. The prognostic values of tumor-infiltrating neutrophils, lymphocytes and neutrophil/lymphocyte rates in bladder urothelial cancer. Pathol. Res. Pract. 2018, 214, 1074–1080. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.J.; Meng, X.Y.; Wu, Q.J.; Zhou, X.H. High CD3D/CD4 ratio predicts better survival in muscle-invasive bladder cancer. Cancer Manag. Res. 2019, 11, 2987–2995. [Google Scholar] [CrossRef] [PubMed]
- Salgado, R.; Denkert, C.; Demaria, S.; Sirtaine, N.; Klauschen, F.; Pruneri, G.; Wienert, S.; Van den Eynden, G.; Baehner, F.L.; Penault-Llorca, F.; et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: Recommendations by an International TILs Working Group 2014. Ann. Oncol. 2015, 26, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.J.; Elder, D.E.; Guerry, D.T.; Braitman, L.E.; Trock, B.J.; Schultz, D.; Synnestvedt, M.; Halpern, A.C. Model predicting survival in stage I melanoma based on tumor progression. J. Natl. Cancer Inst. 1989, 81, 1893–1904. [Google Scholar] [CrossRef] [PubMed]
- Roeke, T.; Sobral-Leite, M.; Dekker, T.; Wesseling, J.; Smit, V.; Tollenaar, R.; Schmidt, M.K.; Mesker, W.E. The prognostic value of the tumour-stroma ratio in primary operable invasive cancer of the breast: A validation study. Breast Cancer Res. Treat. 2017, 166, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Ledderose, S.; Rodler, S.; Eismann, L.; Ledderose, G.; Ledderose, C. Tumor-infiltrating lymphocytes predict survival in ≥ pT2 urothelial bladder cancer. Pathol. Res. Pract. 2022, 237, 154037. [Google Scholar] [CrossRef] [PubMed]
- Kos, Z.; Roblin, E.; Kim, R.S.; Michiels, S.; Gallas, B.D.; Chen, W.; van de Vijver, K.K.; Goel, S.; Adams, S.; Demaria, S.; et al. Pitfalls in assessing stromal tumor infiltrating lymphocytes (sTILs) in breast cancer. NPJ Breast Cancer 2020, 6, 17. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; Mcart, D.G.; Dunne, P.D.; Mcquaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Cole, K.; Martinez-Morilla, S.; Ahmed, F.S.; Zugazagoitia, J.; Staaf, J.; Bosch, A.; Ehinger, A.; Nimeus, E.; Hartman, J.; et al. An Open-Source, automated Tumor-Infiltrating lymphocyte algorithm for prognosis in Triple-Negative breast cancer. Clin. Cancer Res. 2021, 27, 5557–5565. [Google Scholar] [CrossRef]
- Ono, A.; Terada, Y.; Kawata, T.; Serizawa, M.; Isaka, M.; Kawabata, T.; Imai, T.; Mori, K.; Muramatsu, K.; Hayashi, I.; et al. Assessment of associations between clinical and immune microenvironmental factors and tumor mutation burden in resected nonsmall cell lung cancer by applying machine learning to whole-slide images. Cancer Med. 2020, 9, 4864–4875. [Google Scholar] [CrossRef]
- Chou, M.; Illa-Bochaca, I.; Minxi, B.; Darvishian, F.; Johannet, P.; Moran, U.; Shapiro, R.L.; Berman, R.S.; Osman, I.; Jour, G.; et al. Optimization of an automated tumor-infiltrating lymphocyte algorithm for improved prognostication in primary melanoma. Mod. Pathol. 2021, 34, 562–571. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.Y.; Park, H.E.; Kim, J.H.; Wen, X.; Jeong, S.; Cho, N.Y.; Gwon, H.G.; Kim, K.; Lee, H.S.; Jeong, S.Y.; et al. Whole-Slide image analysis reveals quantitative landscape of Tumor-Immune microenvironment in colorectal cancers. Clin. Cancer Res. 2020, 26, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Paner, G.P.; Stadler, W.M.; Hansel, D.E.; Montironi, R.; Lin, D.W.; Amin, M.B. Updates in the eighth edition of the Tumor-Node-Metastasis staging classification for urologic cancers. Eur. Urol. 2018, 73, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Malpica, N.; De Solórzano, C.O.; Vaquero, J.J.; Santos, A.; Vallcorba, I.; García Sagredo, J.M.; Del Pozo, F. Applying watershed algorithms to the segmentation of clustered nuclei. Cytom. J. Int. Soc. Anal. Cytol. 1997, 28, 289–297. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural networks for pattern recognition. Agric. Eng. Int. Cigr. J. Sci. Res. Dev. Manuscr. Pm 1995, 12, 1235–1242. [Google Scholar]
- Camp, R.L.; Dolled-Filhart, M.; Rimm, D.L. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin. Cancer Res. 2004, 10, 7252–7259. [Google Scholar] [CrossRef]
- Witjes, J.A.; Bruins, H.M.; Cathomas, R.; Compérat, E.M.; Cowan, N.C.; Gakis, G.; Hernández, V.; Linares, E.E.; Lorch, A.; Neuzillet, Y.; et al. European Association of Urology Guidelines on Muscle-invasive and Metastatic Bladder Cancer: Summary of the 2020 Guidelines. Eur. Urol. 2021, 79, 82–104. [Google Scholar] [CrossRef]
- Hsu, F.S.; Su, C.H.; Huang, K.H. A comprehensive review of US FDA-Approved immune checkpoint inhibitors in urothelial carcinoma. J. Immunol. Res. 2017, 2017, 6940546. [Google Scholar] [CrossRef]
- Cumberbatch, M.; Jubber, I.; Black, P.C.; Esperto, F.; Figueroa, J.D.; Kamat, A.M.; Kiemeney, L.; Lotan, Y.; Pang, K.; Silverman, D.T.; et al. Epidemiology of bladder cancer: A systematic review and contemporary update of risk factors in 2018. Eur. Urol. 2018, 74, 784–795. [Google Scholar] [CrossRef]
- Sharma, P.; Shen, Y.; Wen, S.; Yamada, S.; Jungbluth, A.A.; Gnjatic, S.; Bajorin, D.F.; Reuter, V.E.; Herr, H.; Old, L.J.; et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc. Natl. Acad. Sci. USA 2007, 104, 3967–3972. [Google Scholar] [CrossRef]
- Sikic, D.; Weyerer, V.; Geppert, C.I.; Bertz, S.; Lange, F.; Taubert, H.; Wach, S.; Schmitz-Draeger, B.J.; Wullich, B.; Hartmann, A.; et al. Utility of stromal tumor infiltrating lymphocyte scoring (sTILs) for risk stratification of patients with muscle-invasive urothelial bladder cancer after radical cystectomy. Urol. Oncol. 2022, 40, 19–63. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.R.; Friesner, I.D.; Rizk, E.M.; Fullerton, B.T.; Mondal, M.; Trager, M.H.; Mendelson, K.; Chikeka, I.; Kurc, T.; Gupta, R.; et al. Automated digital TIL analysis (ADTA) adds prognostic value to standard assessment of depth and ulceration in primary melanoma. Sci. Rep. 2021, 11, 2809. [Google Scholar] [CrossRef] [PubMed]
- Shvetsov, N.; Grønnesby, M.; Pedersen, E.; Møllersen, K.; Busund, L.R.; Schwienbacher, R.; Bongo, L.A.; Kilvaer, T.K. A pragmatic machine learning approach to quantify Tumor-Infiltrating lymphocytes in whole slide images. Cancers 2022, 14, 2974. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, X.; He, T.F.; Gruosso, T.; Zuo, D.; Souleimanova, M.; Ramos, V.M.; Omeroglu, A.; Meterissian, S.; Guiot, M.C.; et al. Predicting relapse in patients with triple negative breast cancer (TNBC) using a Deep-Learning approach. Front. Physiol. 2020, 11, 511071. [Google Scholar] [CrossRef] [PubMed]
- Acs, B.; Ahmed, F.S.; Gupta, S.; Wong, P.F.; Gartrell, R.D.; Sarin, P.J.; Rizk, E.M.; Gould, R.B.; Saenger, Y.M.; Rimm, D.L. An open source automated tumor infiltrating lymphocyte algorithm for prognosis in melanoma. Nat. Commun. 2019, 10, 5440. [Google Scholar] [CrossRef] [PubMed]
| RHWU (N = 133) | TCGA (N = 247) | |
|---|---|---|
| Age (years) | 66 (26, 87) | 69 (37, 90) |
| Sex | ||
| female | 20 (15.04%) | 60 (24.29%) |
| male | 113 (84.96%) | 187 (75.71%) |
| pT stage | ||
| pT2 | 52 (39.10%) | 70 (28.34%) |
| pT3 | 63 (47.37%) | 131 (53.04%) |
| pT4 | 18 (13.53%) | 40 (16.19%) |
| pTx | 0 (0%) | 6 (2.43%) |
| pN stage | ||
| pN0 | 66 (49.62%) | 141 (57.09%) |
| pN1 | 34 (25.56%) | 29 (11.74%) |
| pN2 | 18 (13.53%) | 58 (23.48%) |
| pN3 | 15 (11.28%) | 5 (2.02%) |
| pNx | 0 (0%) | 14 (5.67%) |
| pM stage | ||
| pM0 | 129 (96.99%) | 100 (40.49%) |
| pM1 | 4 (3.01%) | 7 (2.83%) |
| pMx | 0 (0%) | 140 (56.68%) |
| pTNM stage | ||
| Stage II | 38 (28.57%) | 65 (16.31%) |
| Stage III | 74 (55.64%) | 86 (34.82%) |
| Stage IV | 21 (15.79%) | 96 (38.87%) |
| Lymphovascular invasion | ||
| No | 83 (62.41%) | 90 (33.21%) |
| Yes | 50 (37.59%) | 99 (36.53%) |
| Missing | 0 (0%) | 82 (30.26%) |
| Survival status | ||
| Alive | 77 (57.89%) | 121 (48.99%) |
| Dead | 56 (42.11%) | 126 (51.01%) |
| OS time (months) | 15.3 (1.9, 66.0) | 17.5 (0.5, 114.0) |
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| HR (95%CI) | p Value | HR (95%CI) | p Value | |
| Age | ||||
| <70 | Ref. | Ref. | ||
| ≥70 | 1.702 (1.195, 2.424) | <0.001 | 1.798 (1.259, 2.567) | 0.001 |
| Gender | ||||
| female | Ref. | |||
| male | 1.058 (0.703, 1.591) | 0.788 | ||
| pT stage | ||||
| pT1–2 | Ref. | Ref. | ||
| pT3–4 | 1.938 (1.258, 2.985) | 0.003 | 1.111 (0.526, 2.345) | 0.783 |
| pN stage | ||||
| pN0–1 | Ref. | Ref. | ||
| pN2–3 | 1.907 (1.323, 2.749) | <0.001 | 1.557 (1.014, 2.391) | 0.043 |
| pM stage | ||||
| pM0 | Ref. | |||
| pM1 | 1.849 (0.859, 3.981) | 0.116 | ||
| pTNM stage | ||||
| Stage II | Ref. | Ref. | ||
| Stage III–IV | 2.438 (1.591, 3.876) | <0.001 | 1.407 (0.581, 3.403) | 0.449 |
| Lymphovascular invasion | ||||
| No | Ref. | Ref. | ||
| Yes | 1.603 (1.128, 2.277) | 0.008 | 1.360 (0.924, 2.003) | 0.119 |
| eTILs | ||||
| Low eTILs | Ref. | Ref. | ||
| High eTILs | 0.333 (0.199, 0.558) | <0.001 | 0.345 (0.203, 0.586) | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Q.; Yang, R.; Ni, X.; Yang, S.; Jiao, P.; Wu, J.; Xiong, L.; Wang, J.; Jian, J.; Jiang, Z.; et al. Quantitative Assessment of Tumor-Infiltrating Lymphocytes Using Machine Learning Predicts Survival in Muscle-Invasive Bladder Cancer. J. Clin. Med. 2022, 11, 7081. https://doi.org/10.3390/jcm11237081
Zheng Q, Yang R, Ni X, Yang S, Jiao P, Wu J, Xiong L, Wang J, Jian J, Jiang Z, et al. Quantitative Assessment of Tumor-Infiltrating Lymphocytes Using Machine Learning Predicts Survival in Muscle-Invasive Bladder Cancer. Journal of Clinical Medicine. 2022; 11(23):7081. https://doi.org/10.3390/jcm11237081
Chicago/Turabian StyleZheng, Qingyuan, Rui Yang, Xinmiao Ni, Song Yang, Panpan Jiao, Jiejun Wu, Lin Xiong, Jingsong Wang, Jun Jian, Zhengyu Jiang, and et al. 2022. "Quantitative Assessment of Tumor-Infiltrating Lymphocytes Using Machine Learning Predicts Survival in Muscle-Invasive Bladder Cancer" Journal of Clinical Medicine 11, no. 23: 7081. https://doi.org/10.3390/jcm11237081
APA StyleZheng, Q., Yang, R., Ni, X., Yang, S., Jiao, P., Wu, J., Xiong, L., Wang, J., Jian, J., Jiang, Z., Wang, L., Chen, Z., & Liu, X. (2022). Quantitative Assessment of Tumor-Infiltrating Lymphocytes Using Machine Learning Predicts Survival in Muscle-Invasive Bladder Cancer. Journal of Clinical Medicine, 11(23), 7081. https://doi.org/10.3390/jcm11237081







