Physicians and Machine-Learning Algorithm Performance in Predicting Left-Ventricular Systolic Dysfunction from a Standard 12-Lead-Electrocardiogram
Abstract
1. Introduction
2. Materials and Methods
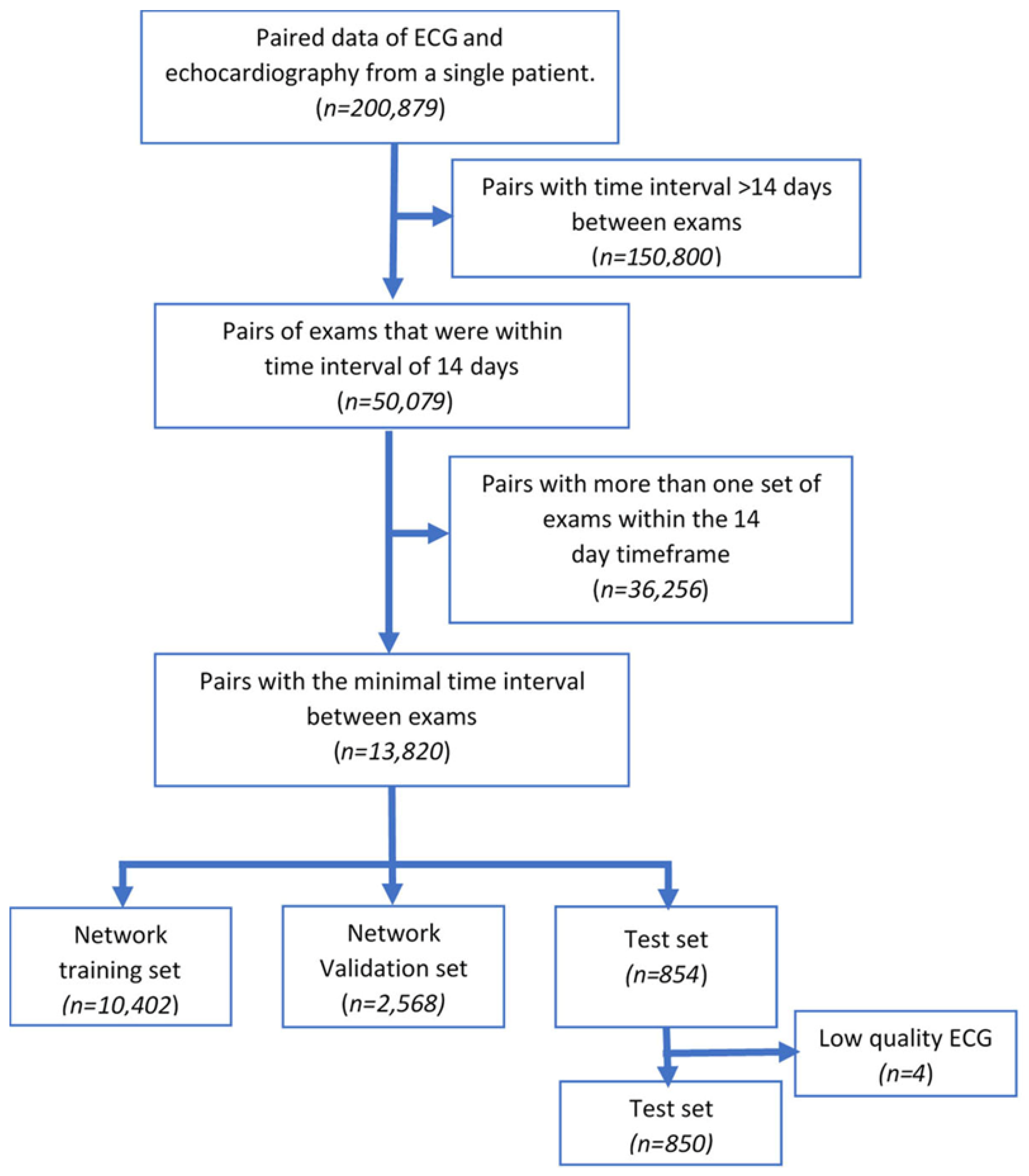
2.1. Study Population
2.2. Physician Predictions of EF by ECG
2.3. Deep Residual Convolutional Neural Network for EF Prediction
2.4. Statistical Analysis
3. Results
3.1. Baseline Characteristics
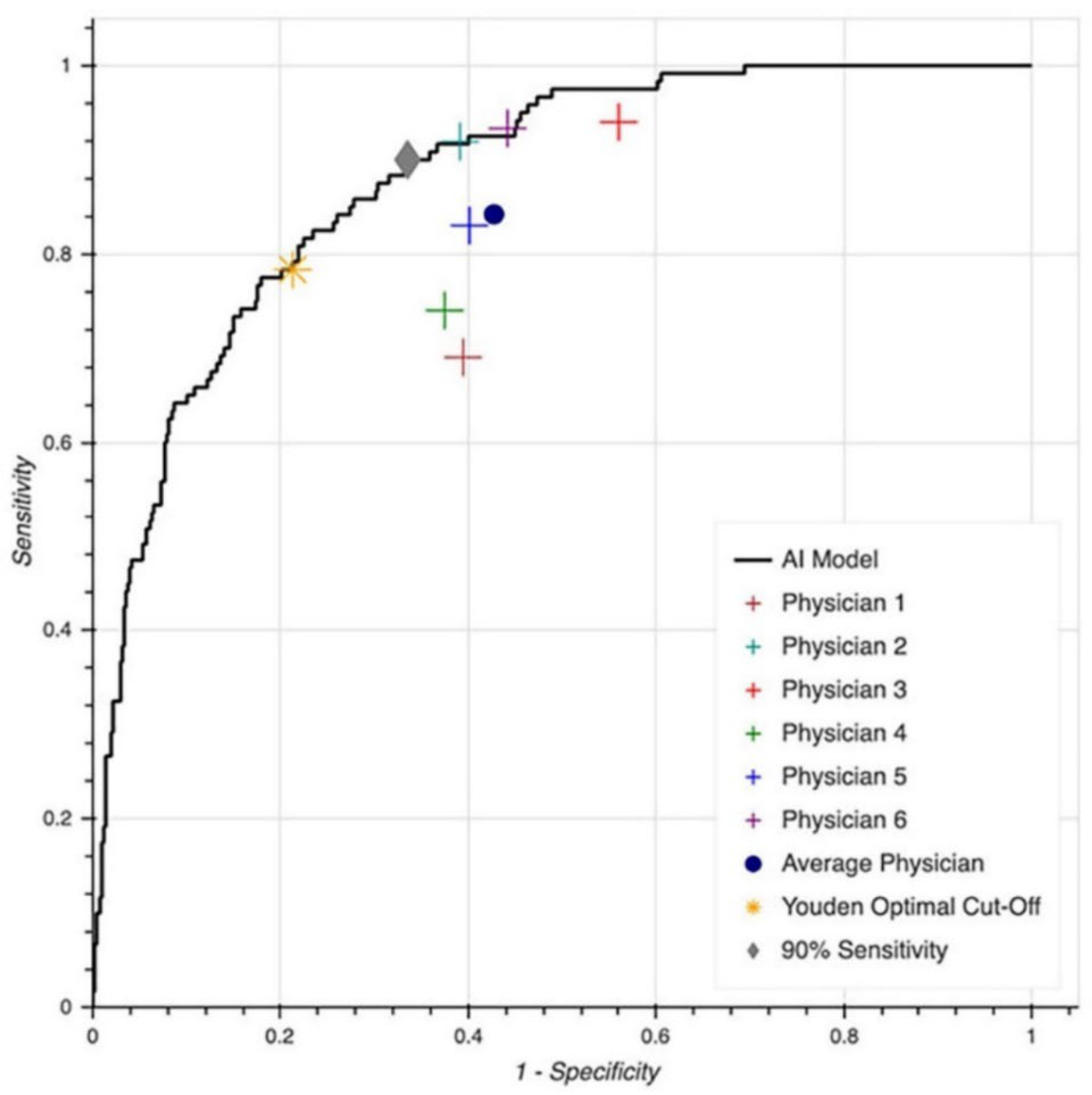
3.2. LVSD Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart Disease and Stroke Statistics-2020 Update: A Report from the American Heart Association. Circulation 2020, 141, e139–e596. [Google Scholar] [CrossRef] [PubMed]
- McDonagh, T.A.; Morrison, C.E.; Lawrence, A.; Ford, I.; Tunstall-Pedoe, H.; McMurray, J.J.; Dargie, H.J. Symptomatic and asymptomatic left-ventricular systolic dysfunction in an urban population. Lancet 1997, 350, 829–833. [Google Scholar] [CrossRef]
- Ammar, K.A.; Jacobsen, S.J.; Mahoney, D.W.; Kors, J.A.; Redfield, M.M.; Burnett, J.C., Jr.; Rodeheffer, R.J. Prevalence and prognostic significance of heart failure stages: Application of the American College of Cardiology/American Heart Association heart failure staging criteria in the community. Circulation 2007, 115, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Sara, J.D.; Toya, T.; Taher, R.; Lerman, A.; Gersh, B.; Anavekar, N.S. Asymptomatic Left Ventricle Systolic Dysfunction. Eur. Cardiol. 2020, 15, e13. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.J.; Evans, J.C.; Benjamin, E.J.; Levy, D.; LeRoy, E.C.; Vasan, R.S. Natural history of asymptomatic left ventricular systolic dysfunction in the community. Circulation 2003, 108, 977–982. [Google Scholar] [CrossRef]
- Investigators, S.; Yusuf, S.; Pitt, B.; Davis, C.E.; Hood, W.B., Jr.; Cohn, J.N. Effect of enalapril on mortality and the development of heart failure in asymptomatic patients with reduced left ventricular ejection fractions. N. Engl. J. Med. 1992, 327, 685–691. [Google Scholar] [CrossRef]
- Kober, L.; Torp-Pedersen, C.; Carlsen, J.E.; Bagger, H.; Eliasen, P.; Lyngborg, K.; Videbaek, J.; Cole, D.S.; Auclert, L.; Pauly, N.C. A clinical trial of the angiotensin-converting-enzyme inhibitor trandolapril in patients with left ventricular dysfunction after myocardial infarction. Trandolapril Cardiac Evaluation (TRACE) Study Group. N. Engl. J. Med. 1995, 333, 1670–1676. [Google Scholar] [CrossRef]
- Authors/Task Force, M.; McDonagh, T.A.; Metra, M.; Adamo, M.; Gardner, R.S.; Baumbach, A.; Bohm, M.; Burri, H.; Butler, J.; Celutkiene, J.; et al. 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: Developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. J. Heart Fail. 2022, 24, 4–131. [Google Scholar] [CrossRef]
- Rutten, F.H.; Grobbee, D.E.; Hoes, A.W. Diagnosis and management of heart failure: A questionnaire among general practitioners and cardiologists. Eur. J. Heart Fail. 2003, 5, 345–348. [Google Scholar] [CrossRef]
- Clarke, K.W.; Gray, D.; Hampton, J.R. Evidence of inadequate investigation and treatment of patients with heart failure. Br. Heart J. 1994, 71, 584–587. [Google Scholar] [CrossRef]
- Hobbs, F.D.; Jones, M.I.; Allan, T.F.; Wilson, S.; Tobias, R. European survey of primary care physician perceptions on heart failure diagnosis and management (Euro-HF). Eur. Heart J. 2000, 21, 1877–1887. [Google Scholar] [CrossRef]
- Vasan, R.S.; Benjamin, E.J.; Larson, M.G.; Leip, E.P.; Wang, T.J.; Wilson, P.W.; Levy, D. Plasma natriuretic peptides for community screening for left ventricular hypertrophy and systolic dysfunction: The Framingham heart study. JAMA 2002, 288, 1252–1259. [Google Scholar] [CrossRef]
- Ledwidge, M.; Gallagher, J.; Conlon, C.; Tallon, E.; O’Connell, E.; Dawkins, I.; Watson, C.; O’Hanlon, R.; Bermingham, M.; Patle, A.; et al. Natriuretic peptide-based screening and collaborative care for heart failure: The STOP-HF randomized trial. JAMA 2013, 310, 66–74. [Google Scholar] [CrossRef]
- Huelsmann, M.; Neuhold, S.; Resl, M.; Strunk, G.; Brath, H.; Francesconi, C.; Adlbrecht, C.; Prager, R.; Luger, A.; Pacher, R.; et al. PONTIAC (NT-proBNP selected prevention of cardiac events in a population of diabetic patients without a history of cardiac disease): A prospective randomized controlled trial. J. Am. Coll. Cardiol. 2013, 62, 1365–1372. [Google Scholar] [CrossRef]
- Betti, I.; Castelli, G.; Barchielli, A.; Beligni, C.; Boscherini, V.; De Luca, L.; Messeri, G.; Gheorghiade, M.; Maisel, A.; Zuppiroli, A. The role of N-terminal PRO-brain natriuretic peptide and echocardiography for screening asymptomatic left ventricular dysfunction in a population at high risk for heart failure. The PROBE-HF study. J. Card. Fail. 2009, 15, 377–384. [Google Scholar] [CrossRef]
- Redfield, M.M.; Rodeheffer, R.J.; Jacobsen, S.J.; Mahoney, D.W.; Bailey, K.R.; Burnett, J.C., Jr. Plasma brain natriuretic peptide to detect preclinical ventricular systolic or diastolic dysfunction: A community-based study. Circulation 2004, 109, 3176–3181. [Google Scholar] [CrossRef]
- Attia, Z.I.; Kapa, S.; Lopez-Jimenez, F.; McKie, P.M.; Ladewig, D.J.; Satam, G.; Pellikka, P.A.; Enriquez-Sarano, M.; Noseworthy, P.A.; Munger, T.M.; et al. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat. Med. 2019, 25, 70–74. [Google Scholar] [CrossRef]
- Kwon, J.M.; Kim, K.H.; Jeon, K.H.; Kim, H.M.; Kim, M.J.; Lim, S.M.; Song, P.S.; Park, J.; Choi, R.K.; Oh, B.H. Development and Validation of Deep-Learning Algorithm for Electrocardiography-Based Heart Failure Identification. Korean Circ. J. 2019, 49, 629–639. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ribeiro, A.H.; Ribeiro, M.H.; Paixao, G.M.M.; Oliveira, D.M.; Gomes, P.R.; Canazart, J.A.; Ferreira, M.P.S.; Andersson, C.R.; Macfarlane, P.W.; Meira, W., Jr.; et al. Automatic diagnosis of the 12-lead ECG using a deep neural network. Nat. Commun. 2020, 11, 1760. [Google Scholar] [CrossRef]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Agarap, A.F. Deep Learning using Rectified Linear Units (ReLU). arXiv 2018, arXiv:1803.08375. [Google Scholar]
- Dunne, R.; Campbell, N.A. On the pairing of the softmax activation and cross-entropy penalty functions and the derivation of the softmax activation function. In Proceedings of the 8th Australian Conference on the Neural Networks, Melbourne, Australia, 11–13 June 1997; p. 185. [Google Scholar]
- Kline, D.M.; Berardi, V.L. Revisiting squared-error and cross-entropy functions for training neural network classifiers. Neural Comput. Appl. 2005, 14, 310–318. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2015, arXiv:1412.6980. [Google Scholar]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. In Neural Networks for Machine Learning; Coursera: Elmira, NY, USA, 2012; Volume 2, pp. 21–26. [Google Scholar]
- Bottou, L. Large-Scale Machine Learning with Stochastic Gradient Descent. In Proceedings of the COMPSTAT’2010, Paris France, 22–27 August 2010; Lechevallier, Y., Saporta, G., Eds.; Physica-Verlag HD: Berlin/Heidelberg, Germany, 2010; pp. 177–186. [Google Scholar]
- Oord, A.V.D.; Dieleman, S.; Zen, H.; Simonyan, K.; Vinyals, O.; Graves, A.; Kalchbrenner, N.; Senior, A.; Kavukcuoglu, K. WaveNet: A Generative Model for Raw Audio. arXiv 2016, arXiv:1609.03499. [Google Scholar]
- Sundermeyer, M.; Schlüter, R.; Ney, H. LSTM Neural Networks for Language Modeling. In Proceedings of the Thirteenth Annual Conference of the International Speech Communication Association, Portland, OR, USA, 9–13 September 2012. [Google Scholar]
- O’Neal, W.T.; Mazur, M.; Bertoni, A.G.; Bluemke, D.A.; Al-Mallah, M.H.; Lima, J.A.C.; Kitzman, D.; Soliman, E.Z. Electrocardiographic Predictors of Heart Failure with Reduced Versus Preserved Ejection Fraction: The Multi-Ethnic Study of Atherosclerosis. J. Am. Heart Assoc. 2017, 6, e006023. [Google Scholar] [CrossRef]
- Zhang, Z.M.; Rautaharju, P.M.; Prineas, R.J.; Loehr, L.; Rosamond, W.; Soliman, E.Z. Ventricular conduction defects and the risk of incident heart failure in the Atherosclerosis Risk in Communities (ARIC) Study. J. Card. Fail. 2015, 21, 307–312. [Google Scholar] [CrossRef]
- Rautaharju, P.M.; Kooperberg, C.; Larson, J.C.; LaCroix, A. Electrocardiographic predictors of incident congestive heart failure and all-cause mortality in postmenopausal women: The Women’s Health Initiative. Circulation 2006, 113, 481–489. [Google Scholar] [CrossRef]
- Rautaharju, P.M.; Prineas, R.J.; Wood, J.; Zhang, Z.M.; Crow, R.; Heiss, G. Electrocardiographic predictors of new-onset heart failure in men and in women free of coronary heart disease (from the Atherosclerosis in Communities [ARIC] Study). Am. J. Cardiol. 2007, 100, 1437–1441. [Google Scholar] [CrossRef]
- Schlegel, T.T.; Kulecz, W.B.; Feiveson, A.H.; Greco, E.C.; DePalma, J.L.; Starc, V.; Vrtovec, B.; Rahman, M.A.; Bungo, M.W.; Hayat, M.J.; et al. Accuracy of advanced versus strictly conventional 12-lead ECG for detection and screening of coronary artery disease, left ventricular hypertrophy and left ventricular systolic dysfunction. BMC Cardiovasc. Disord. 2010, 10, 28. [Google Scholar] [CrossRef]
- Gladding, P.A.; Loader, S.; Smith, K.; Zarate, E.; Green, S.; Villas-Boas, S.; Shepherd, P.; Kakadiya, P.; Hewitt, W.; Thorstensen, E.; et al. Multiomics, virtual reality and artificial intelligence in heart failure. Future Cardiol. 2021, 17, 1335–1347. [Google Scholar] [CrossRef]
- Johnson, K.; Neilson, S.; To, A.; Amir, N.; Cave, A.; Scott, T.; Orr, M.; Parata, M.; Day, V.; Gladding, P. Advanced Electrocardiography Identifies Left Ventricular Systolic Dysfunction in Non-Ischemic Cardiomyopathy and Tracks Serial Change over Time. J. Cardiovasc. Dev. Dis. 2015, 2, 93–107. [Google Scholar] [CrossRef]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 2018, 20, 65. [Google Scholar] [CrossRef]
- Zhang, J.; Gajjala, S.; Agrawal, P.; Tison, G.H.; Hallock, L.A.; Beussink-Nelson, L.; Lassen, M.H.; Fan, E.; Aras, M.A.; Jordan, C.; et al. Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation 2018, 138, 1623–1635. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, S.; Yuan, X.; Zhang, P. Interpretable deep learning for automatic diagnosis of 12-lead electrocardiogram. iScience 2021, 24, 102373. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef]
- Bleijendaal, H.; Ramos, L.A.; Lopes, R.R.; Verstraelen, T.E.; Baalman, S.W.E.; Oudkerk Pool, M.D.; Tjong, F.V.Y.; Melgarejo-Meseguer, F.M.; Gimeno-Blanes, F.J.; Gimeno-Blanes, J.R.; et al. Computer versus cardiologist: Is a machine learning algorithm able to outperform an expert in diagnosing a phospholamban p.Arg14del mutation on the electrocardiogram? Heart Rhythm 2021, 18, 79–87. [Google Scholar] [CrossRef]
- Zhu, H.; Cheng, C.; Yin, H.; Li, X.; Zuo, P.; Ding, J.; Lin, F.; Wang, J.; Zhou, B.; Li, Y.; et al. Automatic multilabel electrocardiogram diagnosis of heart rhythm or conduction abnormalities with deep learning: A cohort study. Lancet Digit. Health 2020, 2, e348–e357. [Google Scholar] [CrossRef]
- Adedinsewo, D.; Carter, R.E.; Attia, Z.; Johnson, P.; Kashou, A.H.; Dugan, J.L.; Albus, M.; Sheele, J.M.; Bellolio, F.; Friedman, P.A.; et al. Artificial Intelligence-Enabled ECG Algorithm to Identify Patients with Left Ventricular Systolic Dysfunction Presenting to the Emergency Department with Dyspnea. Circ. Arrhythm Electrophysiol. 2020, 13, e008437. [Google Scholar] [CrossRef]
- Cho, J.; Lee, B.; Kwon, J.M.; Lee, Y.; Park, H.; Oh, B.H.; Jeon, K.H.; Park, J.; Kim, K.H. Artificial Intelligence Algorithm for Screening Heart Failure with Reduced Ejection Fraction Using Electrocardiography. ASAIO J. 2021, 67, 314–321. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yao, X.; Rushlow, D.R.; Inselman, J.W.; McCoy, R.G.; Thacher, T.D.; Behnken, E.M.; Bernard, M.E.; Rosas, S.L.; Akfaly, A.; Misra, A.; et al. Artificial intelligence-enabled electrocardiograms for identification of patients with low ejection fraction: A pragmatic, randomized clinical trial. Nat. Med. 2021, 27, 815–819. [Google Scholar] [CrossRef] [PubMed]
- Siontis, K.C.; Noseworthy, P.A.; Arghami, A.; Weston, S.A.; Attia, Z.I.; Crestanello, J.A.; Friedman, P.A.; Chamberlain, A.M.; Gersh, B.J. Use of Artificial Intelligence Tools Across Different Clinical Settings: A Cautionary Tale. Circ. Cardiovasc. Qual. Outcomes 2021, 14, e008153. [Google Scholar] [CrossRef] [PubMed]
- Brisk, R.; Bond, R.; Finlay, D.; McLaughlin, J.; Piadlo, A.; Leslie, S.J.; Gossman, D.E.; Menown, I.B.; McEneaney, D.J.; Warren, S. The effect of confounding data features on a deep learning algorithm to predict complete coronary occlusion in a retrospective observational setting. Eur. Heart J.—Digit. Health 2021, 2, 127–134. [Google Scholar] [CrossRef]
- The Lancet Respiratory, M. Opening the black box of machine learning. Lancet Respir. Med. 2018, 6, 801. [Google Scholar] [CrossRef]


| Index | Training Dataset (n = 10,402) | Validation Dataset (n = 2568) | Test Dataset (n = 850) | p-Value |
|---|---|---|---|---|
| Age (years [IQR]) | 69.8 [59.6–80.1] | 70.0 [59.4–80.2] | 71.3 [61.1–81.4] | 0.07 |
| Male (n; %) | 6347; 61.0% | 1569; 61.1% | 560; 65.9% | 0.02 |
| Family history of CAD | 2591; 27.1% | 590; 25.1% | 224; 29.2% | 0.04 |
| Diabetes mellitus (n; %) | 5254; 54.9% | 1272; 54.2% | 423; 54.8% | 0.80 |
| Hypertension (n; %) | 7236; 75.8% | 1804; 76.9% | 587; 76.6% | 0.50 |
| Hyperlipidemia (n; %) | 6007; 63.9% | 1509; 64.3% | 511; 66.7% | 0.68 |
| Chronic kidney disease (n; %) | 3426; 35.9% | 806; 34.4% | 310; 40.5% | 0.09 |
| Chronic dialysis | 2490; 26.1% | 571; 24.3% | 215; 28.1% | 0.08 |
| Peripheral vascular disease | 2780; 29.1% | 658; 28.0% | 256; 33.4% | 0.02 |
| Obesity (n; %) | 2569; 37.4% | 856; 36.9% | 283; 36.9% | 0.88 |
| Atrial fibrillation/flutter | 3653; 38.3% | 839; 35.8% | 314; 41.0% | 0.02 |
| Pacemaker | 2607; 27.3% | 592; 25.2% | 233; 30.4% | 0.01 |
| Smoking history | 4351; 45.6% | 1052; 44.8% | 367; 47.9% | 0.33 |
| COPD | 2977; 31.2% | 716; 30.5% | 263; 34.3% | 0.13 |
| Stroke/TIA | 2983; 31.3% | 673; 28,7% | 244; 31.9% | 0.04 |
| Ischemic heart disease | 4479; 46.9% | 1084; 46.2% | 408; 53.3% | <0.001 |
| Coronary artery bypass surgery | 2729; 28.6% | 632; 26.9% | 247; 32.2% | 0.02 |
| PCI | 2880; 30.2% | 675; 28.8% | 251;32.8% | 0.10 |
| EF (%) median [IQR] | 55 [40–60] | 55 [40–60] | 50 [40–60] | <0.001 |
| EF ≥ 50% (n; %) | 6729; 64.7% | 1642; 63.9% | 427; 50.2% | <0.001 |
| Index | EF ≥ 50% (n = 8798) | EF < 50% (n = 5022) | p-Value |
|---|---|---|---|
| Age (median years [IQR]) | 70.1 [58.8–80.6] | 69.9 [60.9–79.7] | 0.26 |
| Male (n; %) | 4797; 54.5% | 3679; 73.3% | <0.001 |
| Family history of coronary artery disease (n; %) | 1640; 20.9% | 1765; 36.8% | <0.001 |
| Diabetes Mellitus (n; %) | 3855; 49.0% | 3082; 64.3% | <0.001 |
| Hypertension (n; %) | 5833; 74.2% | 3794; 79.2% | <0.001 |
| Chronic kidney disease (n; %) | 2301; 29.3% | 2241; 46.8% | <0.001 |
| Dialysis (n; %) | 1558; 19.8% | 1720; 35.9% | <0.001 |
| Peripheral vascular disease (n; %) | 1810; 23.0% | 1885; 39.3% | <0.001 |
| Hyperlipidemia (n; %) | 4698; 59.7% | 3329; 69.5% | <0.001 |
| Obesity (n; %) | 2568; 32.9% | 2131; 44.5% | <0.001 |
| Atrial fibrillation/flutter (n; %) | 2591; 32.9% | 2215; 46.2% | <0.001 |
| Pacemaker (n; %) | 1639; 20.8% | 1793; 37.4% | <0.001 |
| Chronic obstructive pulmonary disease (n; %) | 2034; 25.9% | 1922; 40.1% | <0.001 |
| Smoking history | 2984; 37.9% | 2786; 58.1% | <0.001 |
| Stroke/Transient ischemia accident (n; %) | 1971; 25.1% | 1929; 40.2% | <0.001 |
| Ischemic heart disease | 2924; 37.2% | 3047; 63.6% | <0.001 |
| Coronary artery bypass surgery (n; %) | 1683; 21.4% | 1925; 40.2% | <0.001 |
| Percutaneous coronary intervention (n; %) | 1824; 23.2% | 1982; 41.4% | <0.001 |
| Ejection fraction (median % [IQR]) | 60 [55–60] | 40 [30–40] | <0.001 |
| Predicting EF < 50% | Predicting EF ≤ 35% | |||
|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | |
| Physician 1 | 59.4% | 67.1% | 69.6% | 59.5% |
| Physician 2 | 76.1% | 76.1% | 90.0% | 60.0% |
| Physician 3 | 80.8% | 53.3% | 93.2% | 43.4% |
| Physician 4 | 63.2% | 73.9% | 75.8% | 62.9% |
| Physician 5 | 69.2% | 69.7% | 83.9% | 58.5% |
| Physician 6 | 82.7% | 71.1% | 95.0% | 53.8% |
| Average physician | 71.9% | 68.5% | 84.6% | 56.3% |
| Average senior physician | 77.6% | 64.7% | 90.7% | 51.9% |
| MLA performance based on Youden cut point | 78.3% | 78.1% | 78% | 79% |
| MLA performance based on 90% sensitivity | 90% | 55.7% | 90% | 66% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Golany, T.; Radinsky, K.; Kofman, N.; Litovchik, I.; Young, R.; Monayer, A.; Love, I.; Tziporin, F.; Minha, I.; Yehuda, Y.; et al. Physicians and Machine-Learning Algorithm Performance in Predicting Left-Ventricular Systolic Dysfunction from a Standard 12-Lead-Electrocardiogram. J. Clin. Med. 2022, 11, 6767. https://doi.org/10.3390/jcm11226767
Golany T, Radinsky K, Kofman N, Litovchik I, Young R, Monayer A, Love I, Tziporin F, Minha I, Yehuda Y, et al. Physicians and Machine-Learning Algorithm Performance in Predicting Left-Ventricular Systolic Dysfunction from a Standard 12-Lead-Electrocardiogram. Journal of Clinical Medicine. 2022; 11(22):6767. https://doi.org/10.3390/jcm11226767
Chicago/Turabian StyleGolany, Tomer, Kira Radinsky, Natalia Kofman, Ilya Litovchik, Revital Young, Antoinette Monayer, Itamar Love, Faina Tziporin, Ido Minha, Yakir Yehuda, and et al. 2022. "Physicians and Machine-Learning Algorithm Performance in Predicting Left-Ventricular Systolic Dysfunction from a Standard 12-Lead-Electrocardiogram" Journal of Clinical Medicine 11, no. 22: 6767. https://doi.org/10.3390/jcm11226767
APA StyleGolany, T., Radinsky, K., Kofman, N., Litovchik, I., Young, R., Monayer, A., Love, I., Tziporin, F., Minha, I., Yehuda, Y., Ziv-Baran, T., Fuchs, S., & Minha, S. (2022). Physicians and Machine-Learning Algorithm Performance in Predicting Left-Ventricular Systolic Dysfunction from a Standard 12-Lead-Electrocardiogram. Journal of Clinical Medicine, 11(22), 6767. https://doi.org/10.3390/jcm11226767







