A Web-Based Automated Image Processing Research Platform for Cochlear Implantation-Related Studies
Abstract
1. Introduction
Background
2. Methods
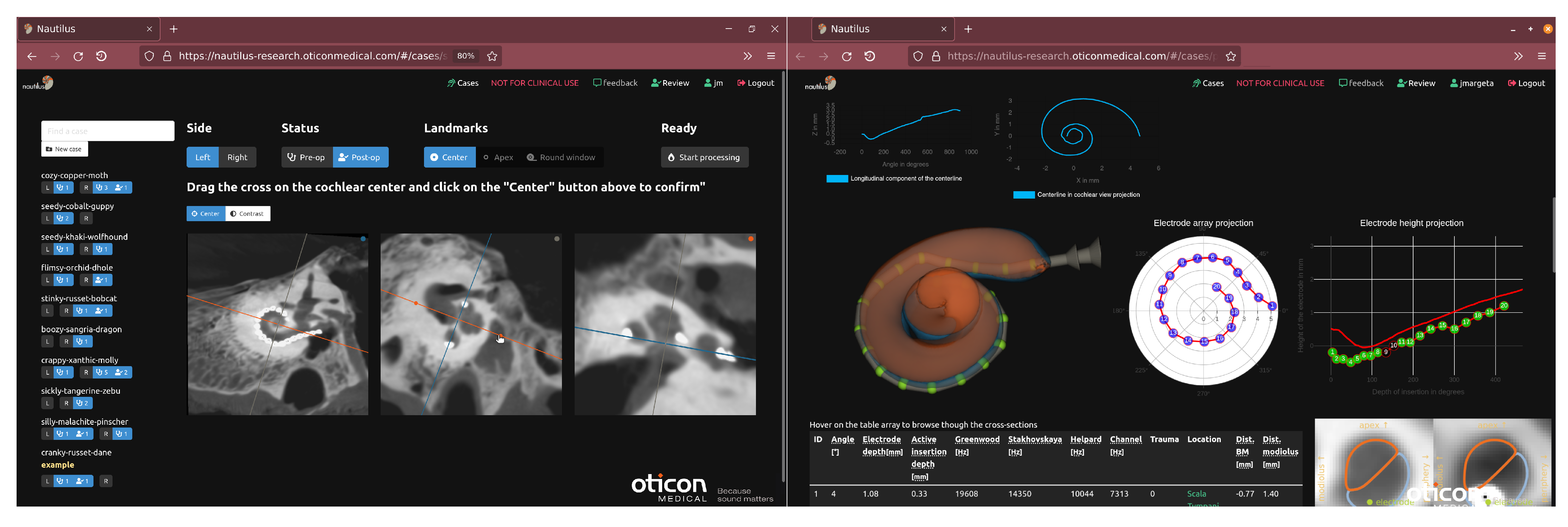
2.1. Data Upload and Pseudonymization via a Web-Based Frontend
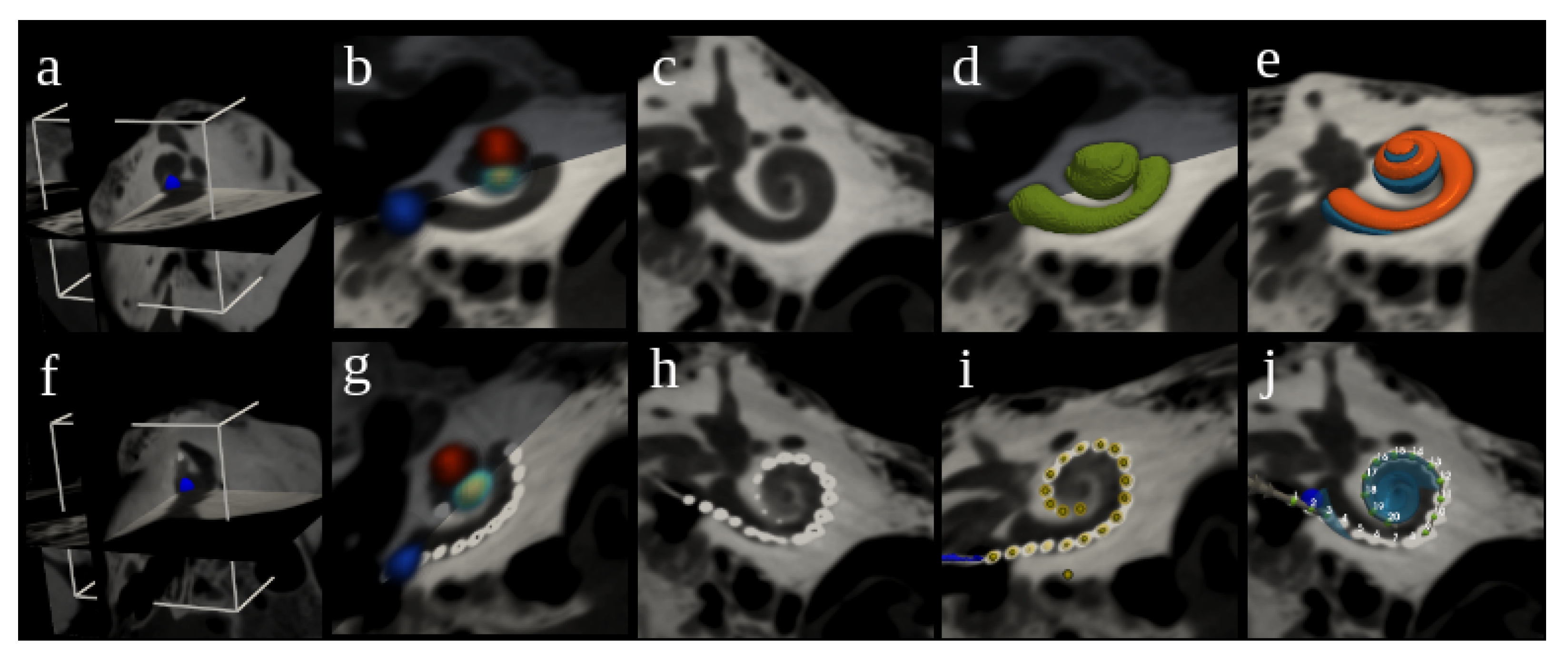
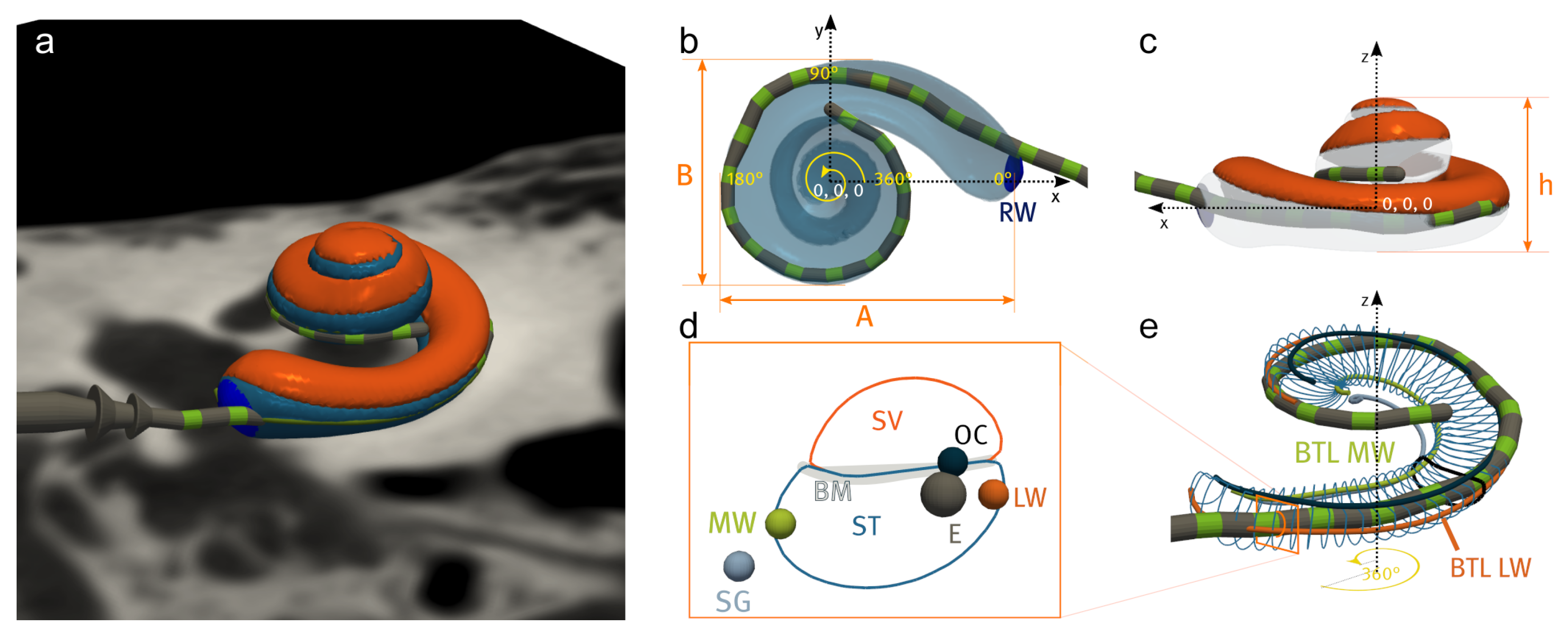
2.2. Cochlear Landmarks and Canonical Pose Estimation
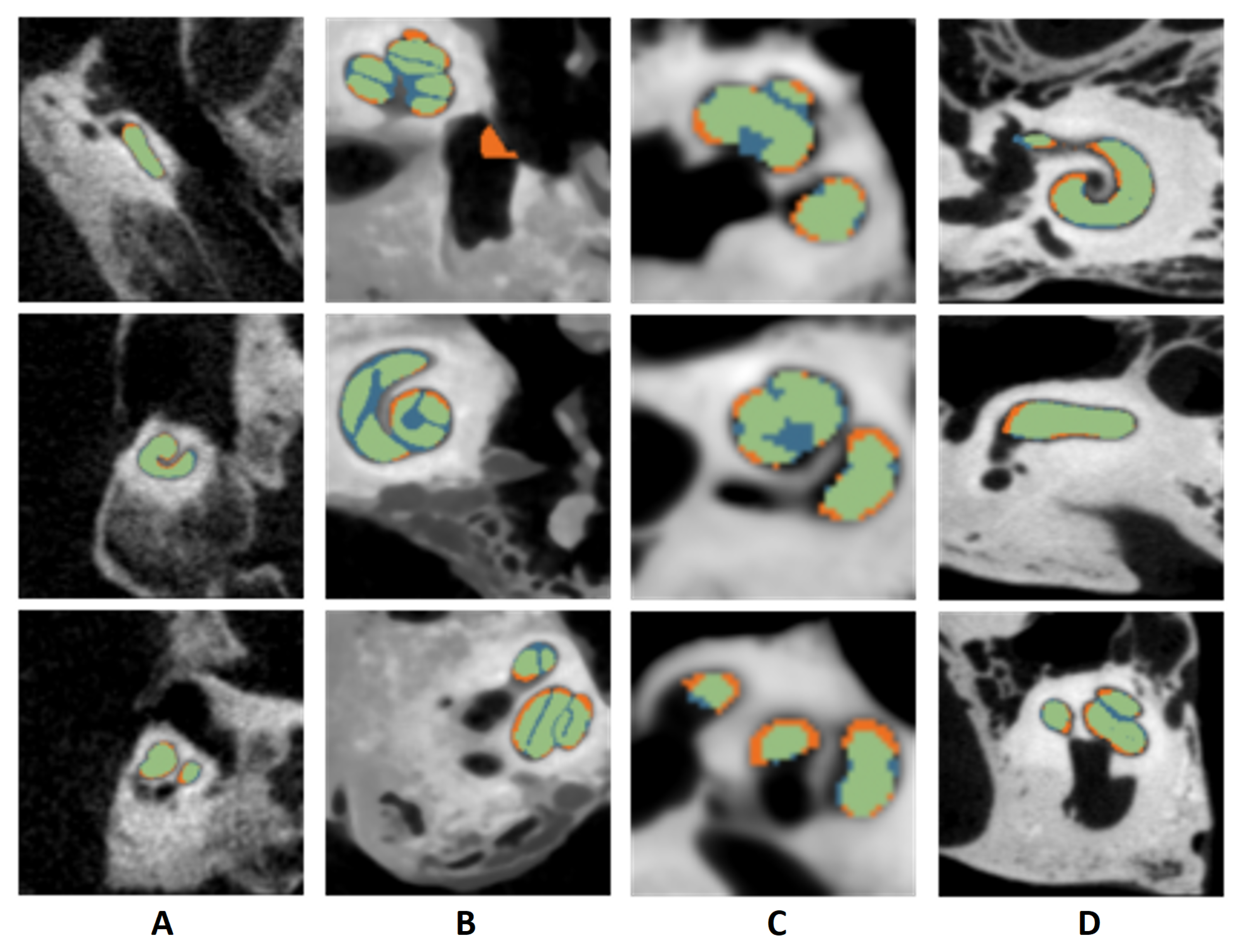
2.3. Segmentation of Cochlear Structures
2.4. Electrode Depth-to-Angular Coverage Prediction
2.5. Registration of the Pre- and Post-Operative Images
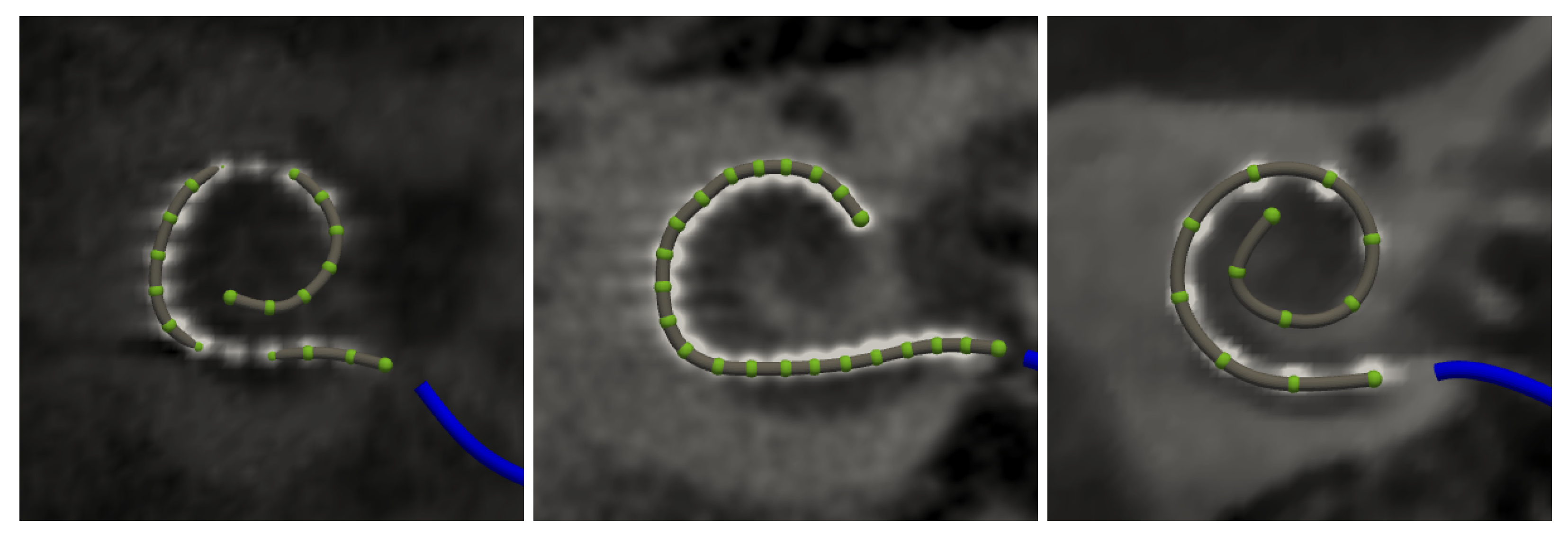
2.6. Electrode Array Detection
2.7. Extracted Measurements
2.7.1. Global Pre-Operative Metrics
2.7.2. Local Pre- and Post-Operative Metrics
2.8. Failure Flagging Mechanisms
2.9. Data Export
3. Results
3.1. Evaluation Datasets
3.2. Accuracy
3.2.1. Landmark Detection
3.2.2. Segmentation
3.2.3. Registration
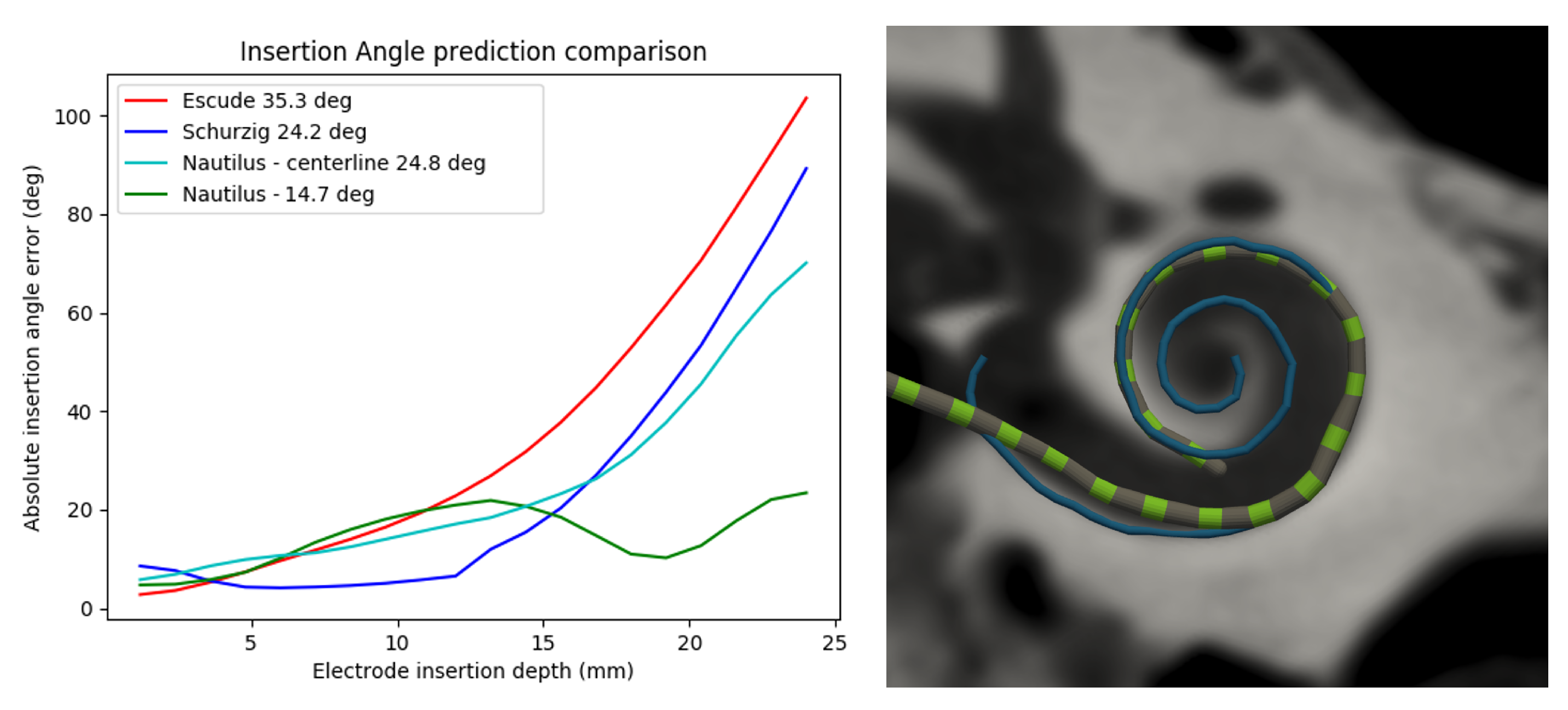
3.2.4. Electrode Detection
3.3. Robustness
3.4. Failure Detection
3.5. Computational Performances
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ASSD | Average symmetric surface distance |
| BM | Basilar membrane |
| BTL | Basal turn length |
| CBCT | Cone-beam computed tomography |
| CDL | Cochlear duct length |
| HD95 | Hausdorff distance at the 95th percentile |
| MRI | Magnetic resonance imaging |
| MW | Modiolar wall |
| µCT | Micro computed tomography |
| LW | Lateral wall |
| OC | Organ of Corti |
| RAVD | Relative absolute volume difference |
| ROC | Receiver operating characteristic curve |
| RW | Round window |
| SG | Spiral ganglion |
| ST | Scala tympani |
| SV | Scala vestibuli |
| TB | Temporal bone |
References
- Carlson, M.L. Cochlear Implantation in Adults. N. Engl. J. Med. 2020, 382, 1531–1542. [Google Scholar] [CrossRef] [PubMed]
- NIDCD. Cochlear Implants—Who Gets Cochlear Implants? 2021. Available online: https://www.nidcd.nih.gov/health/cochlear-implants (accessed on 22 July 2022).
- Kan, A.; Stoelb, C.; Litovsky, R.Y.; Goupell, M.J. Effect of Mismatched Place-of-Stimulation on Binaural Fusion and Lateralization in Bilateral Cochlear-Implant Usersa. J. Acoust. Soc. Am. 2013, 134, 2923. [Google Scholar] [CrossRef] [PubMed]
- Goupell, M.J.; Stakhovskaya, O.A.; Bernstein, J.G.W. Contralateral Interference Caused by Binaurally Presented Competing Speech in Adult Bilateral Cochlear-Implant Users. Ear Hear. 2018, 39, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.E.; Litovsky, R.Y. Novel Approaches to Measure Spatial Release From Masking in Children with Bilateral Cochlear Implants. Ear Hear. 2022, 43, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.S.; Wang, G.; Rubinstein, J.T.; Vannier, M.W. Semiautomatic Segmentation of the Cochlea Using Real-Time Volume Rendering and Regional Adaptive Snake Modeling. J. Digit. Imaging 2001, 14, 173–181. [Google Scholar] [CrossRef]
- Xianfen, D.; Siping, C.; Changhong, L.; Yuanmei, W. 3D Semi-automatic Segmentation of the Cochlea and Inner Ear. In Proceedings of the 2005 27th Annual Conference of the IEEE Engineering in Medicine and Biology, Shanghai, China, 31 August–3 September 2005; pp. 6285–6288. [Google Scholar] [CrossRef]
- Hussain, R.; Lalande, A.; Girum, K.B.; Guigou, C.; Bozorg Grayeli, A. Automatic Segmentation of Inner Ear on CT-scan Using Auto-Context Convolutional Neural Network. Sci. Rep. 2021, 11, 4406. [Google Scholar] [CrossRef]
- Nikan, S.; Van Osch, K.; Bartling, M.; Allen, D.G.; Rohani, S.A.; Connors, B.; Agrawal, S.K.; Ladak, H.M. PWD-3DNet: A Deep Learning-Based Fully-Automated Segmentation of Multiple Structures on Temporal Bone CT Scans. IEEE Trans. Image Process. 2021, 30, 739–753. [Google Scholar] [CrossRef]
- Lv, Y.; Ke, J.; Xu, Y.; Shen, Y.; Wang, J.; Wang, J. Automatic Segmentation of Temporal Bone Structures from Clinical Conventional CT Using a CNN Approach. Int. J. Med Robot. Comput. Assist. Surg. 2021, 17, e2229. [Google Scholar] [CrossRef]
- Heutink, F.; Koch, V.; Verbist, B.; van der Woude, W.J.; Mylanus, E.; Huinck, W.; Sechopoulos, I.; Caballo, M. Multi-Scale Deep Learning Framework for Cochlea Localization, Segmentation and Analysis on Clinical Ultra-High-Resolution CT Images. Comput. Methods Programs Biomed. 2020, 191, 105387. [Google Scholar] [CrossRef]
- Greenwood, D.P. Bandwidth Specification for Adaptive Optics Systems*. JOSA 1977, 67, 390–393. [Google Scholar] [CrossRef]
- Stakhovskaya, O.; Sridhar, D.; Bonham, B.H.; Leake, P.A. Frequency Map for the Human Cochlear Spiral Ganglion: Implications for Cochlear Implants. J. Assoc. Res. Otolaryngol. 2007, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Helpard, L.; Li, H.; Rohani, S.A.; Zhu, N.; Rask-Andersen, H.; Agrawal, S.; Ladak, H.M. An Approach for Individualized Cochlear Frequency Mapping Determined From 3D Synchrotron Radiation Phase-Contrast Imaging. IEEE Trans. Biomed. Eng. 2021, 68, 3602–3611. [Google Scholar] [CrossRef] [PubMed]
- Gerber, N.; Reyes, M.; Barazzetti, L.; Kjer, H.M.; Vera, S.; Stauber, M.; Mistrik, P.; Ceresa, M.; Mangado, N.; Wimmer, W.; et al. A Multiscale Imaging and Modelling Dataset of the Human Inner Ear. Sci. Data 2017, 4, 170132. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, W.; Anschuetz, L.; Weder, S.; Wagner, F.; Delingette, H.; Caversaccio, M. Human Bony Labyrinth Dataset: Co-registered CT and Micro-CT Images, Surface Models and Anatomical Landmarks. Data Brief 2019, 27, 104782. [Google Scholar] [CrossRef] [PubMed]
- Sieber, D.; Erfurt, P.; John, S.; Santos, G.R.D.; Schurzig, D.; Sørensen, M.S.; Lenarz, T. The OpenEar Library of 3D Models of the Human Temporal Bone Based on Computed Tomography and Micro-Slicing. Sci. Data 2019, 6, 180297. [Google Scholar] [CrossRef]
- Noble, J.H.; Gifford, R.H.; Labadie, R.F.; Dawant, B.M. Statistical Shape Model Segmentation and Frequency Mapping of Cochlear Implant Stimulation Targets in CT. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI, Nice, France, 1–5 October 2012; Ayache, N., Delingette, H., Golland, P., Mori, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 421–428. [Google Scholar] [CrossRef]
- Noble, J.H.; Labadie, R.F.; Majdani, O.; Dawant, B.M. Automatic Segmentation of Intra-Cochlear Anatomy in Conventional CT. IEEE Trans. BioMed Eng. 2011, 58, 2625–2632. [Google Scholar] [CrossRef]
- Wang, Z.; Demarcy, T.; Vandersteen, C.; Gnansia, D.; Raffaelli, C.; Guevara, N.; Delingette, H. Bayesian Logistic Shape Model Inference: Application to Cochlear Image Segmentation. Med. Image Anal. 2022, 75, 102268. [Google Scholar] [CrossRef]
- Finley, C.C.; Holden, T.A.; Holden, L.K.; Whiting, B.R.; Chole, R.A.; Neely, G.J.; Hullar, T.E.; Skinner, M.W. Role of Electrode Placement as a Contributor to Variability in Cochlear Implant Outcomes. Otol. Neurotol. 2008, 29, 920–928. [Google Scholar] [CrossRef]
- Macherey, O.; Carlyon, R.P. Place-Pitch Manipulations with Cochlear Implants. J. Acoust. Soc. Am. 2012, 131, 2225–2236. [Google Scholar] [CrossRef]
- Schuman, T.A.; Noble, J.H.; Wright, C.G.; Wanna, G.B.; Dawant, B.; Labadie, R.F. Anatomic Verification of a Novel Method for Precise Intrascalar Localization of Cochlear Implant Electrodes in Adult Temporal Bones Using Clinically Available Computed Tomography. Laryngoscope 2010, 120, 2277–2283. [Google Scholar] [CrossRef]
- Reda, F.A.; McRackan, T.R.; Labadie, R.F.; Dawant, B.M.; Noble, J.H. Automatic Segmentation of Intra-Cochlear Anatomy in Post-Implantation CT of Unilateral Cochlear Implant Recipients. Med. Image Anal. 2014, 18, 605–615. [Google Scholar] [CrossRef][Green Version]
- Dillon, M.T.; Canfarotta, M.W.; Buss, E.; O’Connell, B.P. Comparison of Speech Recognition with an Organ of Corti versus Spiral Ganglion Frequency-to-Place Function in Place-Based Mapping of Cochlear Implant and Electric-Acoustic Stimulation Devices. Otol. Neurotol. Off. Publ. Am. Otol. Soc. Am. Neurotol. Soc. Eur. Acad. Otol. Neurotol. 2021, 42, 721–725. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.J.; McCormick, M.; Ibáñez, L.; Consortium, T.I.S. The ITK Software Guide, 3rd ed.; Kitware, Inc.: Clifton Park, NY, USA, 2013. [Google Scholar]
- Verbist, B.M.; Joemai, R.M.S.; Briaire, J.J.; Teeuwisse, W.M.; Veldkamp, W.J.H.; Frijns, J.H.M. Cochlear Coordinates in Regard to Cochlear Implantation: A Clinically Individually Applicable 3 Dimensional CT-Based Method. Otol. Neurotol. 2010, 31, 738–744. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Vandersteen, C.; Raffaelli, C.; Guevara, N.; Patou, F.; Delingette, H. One-Shot Learning for Landmarks Detection. In Deep Generative Models, and Data Augmentation, Labelling, and Imperfections; Engelhardt, S., Oksuz, I., Zhu, D., Yuan, Y., Mukhopadhyay, A., Heller, N., Huang, S.X., Nguyen, H., Sznitman, R., Xue, Y., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021; pp. 163–172. [Google Scholar] [CrossRef]
- Criminisi, A.; Shotton, J.; Robertson, D.; Konukoglu, E. Regression Forests for Efficient Anatomy Detection and Localization in CT Studies. In Medical Computer Vision. Recognition Techniques and Applications in Medical Imaging; Menze, B., Langs, G., Tu, Z., Criminisi, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6533, pp. 106–117. [Google Scholar] [CrossRef]
- Ghesu, F.C.; Georgescu, B.; Mansi, T.; Neumann, D.; Hornegger, J.; Comaniciu, D. An Artificial Agent for Anatomical Landmark Detection in Medical Images. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2016; Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; Volume 9902, pp. 229–237. [Google Scholar] [CrossRef]
- Alansary, A.; Oktay, O.; Li, Y.; Folgoc, L.L.; Hou, B.; Vaillant, G.; Kamnitsas, K.; Vlontzos, A.; Glocker, B.; Kainz, B.; et al. Evaluating Reinforcement Learning Agents for Anatomical Landmark Detection. Med. Image Anal. 2019, 53, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Leroy, G.; Rueckert, D.; Alansary, A. Communicative Reinforcement Learning Agents for Landmark Detection in Brain Images. In Machine Learning in Clinical Neuroimaging and Radiogenomics in Neuro-Oncology; Kia, S.M., Mohy-ud-Din, H., Abdulkadir, A., Bass, C., Habes, M., Rondina, J.M., Tax, C., Wang, H., Wolfers, T., Rathore, S., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 177–186. [Google Scholar] [CrossRef]
- Alansary, A.; Folgoc, L.L.; Vaillant, G.; Oktay, O.; Li, Y.; Bai, W.; Passerat-Palmbach, J.; Guerrero, R.; Kamnitsas, K.; Hou, B.; et al. Automatic View Planning with Multi-Scale Deep Reinforcement Learning Agents. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 277–285. [Google Scholar] [CrossRef]
- López Diez, P.; Sundgaard, J.V.; Patou, F.; Margeta, J.; Paulsen, R.R. Facial and Cochlear Nerves Characterization Using Deep Reinforcement Learning for Landmark Detection. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 12904, pp. 519–528. [Google Scholar] [CrossRef]
- McCouat, J.; Voiculescu, I. Contour-Hugging Heatmaps for Landmark Detection. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 19–20 June 2022; pp. 20597–20605. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- MONAI Consortium. MONAI Consortium. MONAI: Medical Open Network for AI (1.0.0). Zenodo. 2022. Available online: https://zenodo.org/record/7086266 (accessed on 22 September 2022). [CrossRef]
- Billot, B.; Robinson, E.; Dalca, A.V.; Iglesias, J.E. Partial Volume Segmentation of Brain MRI Scans of Any Resolution and Contrast. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2020; Martel, A.L., Abolmaesumi, P., Stoyanov, D., Mateus, D., Zuluaga, M.A., Zhou, S.K., Racoceanu, D., Joskowicz, L., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; pp. 177–187. [Google Scholar] [CrossRef]
- Pérez-García, F.; Sparks, R.; Ourselin, S. TorchIO: A Python Library for Efficient Loading, Preprocessing, Augmentation and Patch-Based Sampling of Medical Images in Deep Learning. Comput. Methods Programs Biomed. 2021, 208, 106236. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2999–3007. [Google Scholar] [CrossRef]
- Schurzig, D.; Timm, M.E.; Majdani, O.; Lenarz, T.; Rau, T.S. The Use of Clinically Measurable Cochlear Parameters in Cochlear Implant Surgery as Indicators for Size, Shape, and Orientation of the Scala Tympani. Ear Hear. 2021, 42, 1034–1041. [Google Scholar] [CrossRef]
- Fauser, J.; Stenin, I.; Bauer, M.; Hsu, W.H.; Kristin, J.; Klenzner, T.; Schipper, J.; Mukhopadhyay, A. Toward an Automatic Preoperative Pipeline for Image-Guided Temporal Bone Surgery. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 967–976. [Google Scholar] [CrossRef]
- Powell, K.A.; Wiet, G.J.; Hittle, B.; Oswald, G.I.; Keith, J.P.; Stredney, D.; Andersen, S.A.W. Atlas-Based Segmentation of Cochlear Microstructures in Cone Beam CT. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 363–373. [Google Scholar] [CrossRef]
- Fan, Y.; Zhang, D.; Banalagay, R.; Wang, J.; Noble, J.H.; Dawant, B.M. Hybrid Active Shape and Deep Learning Method for the Accurate and Robust Segmentation of the Intracochlear Anatomy in Clinical Head CT and CBCT Images. J. Med. Imaging 2021, 8, 064002. [Google Scholar] [CrossRef]
- Margeta, J.; Demarcy, T.; Lopez Diez, P.; Hussain, R.; Vandersteen, C.; Guevarra, N.; Delingette, H.; Gnansia, D.; Kamaric Riis, S.; Patou, F. Nautilus: A Clinical Tool for the Segmentation of Intra-Cochlear Structures and Related Applications. In Proceedings of the Conference on Implantable Auditory Prostheses (CIAP), Lake Tahoe, CA, USA, 12–16 July 2021. [Google Scholar]
- Demarcy, T. Segmentation and Study of Anatomical Variability of the Cochlea from Medical Images. Ph.D. Thesis, Université Côte d’Azur, Nice, France, 2017. [Google Scholar]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. Self-Supervised Learning for Medical Image Analysis Using Image Context Restoration. Med. Image Anal. 2019, 58, 101539. [Google Scholar] [CrossRef]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. arXiv 2019, arXiv:1711.05101. [Google Scholar]
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Unified Focal Loss: Generalising Dice and Cross Entropy-Based Losses to Handle Class Imbalanced Medical Image Segmentation. Comput. Med. Imaging Graph. 2022, 95, 102026. [Google Scholar] [CrossRef] [PubMed]
- Schurzig, D.; Timm, M.E.; Batsoulis, C.; Salcher, R.; Sieber, D.; Jolly, C.; Lenarz, T.; Zoka-Assadi, M. A Novel Method for Clinical Cochlear Duct Length Estimation toward Patient-Specific Cochlear Implant Selection. OTO Open 2018, 2, 2473974X18800238. [Google Scholar] [CrossRef] [PubMed]
- Mertens, G.; Van Rompaey, V.; Van de Heyning, P.; Gorris, E.; Topsakal, V. Prediction of the Cochlear Implant Electrode Insertion Depth: Clinical Applicability of Two Analytical Cochlear Models. Sci. Rep. 2020, 10, 3340. [Google Scholar] [CrossRef] [PubMed]
- Shamonin, D. Fast Parallel Image Registration on CPU and GPU for Diagnostic Classification of Alzheimer’s Disease. Front. Neuroinform. 2013, 7, 50. [Google Scholar] [CrossRef]
- Klein, S.; Staring, M.; Murphy, K.; Viergever, M.; Pluim, J. Elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Trans. Med. Imaging 2010, 29, 196–205. [Google Scholar] [CrossRef]
- Mattes, D.; Haynor, D.; Vesselle, H.; Lewellen, T.; Eubank, W. PET-CT Image Registration in the Chest Using Free-Form Deformations. IEEE Trans. Med. Imaging 2003, 22, 120–128. [Google Scholar] [CrossRef]
- Eshraghi, A.A.; Van De Water, T.R. Cochlear Implantation Trauma and Noise-Induced Hearing Loss: Apoptosis and Therapeutic Strategies. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 2006, 288A, 473–481. [Google Scholar] [CrossRef]
- Ishiyama, A.; Risi, F.; Boyd, P. Potential Insertion Complications with Cochlear Implant Electrodes. Cochlear Implant. Int. 2020, 21, 206–219. [Google Scholar] [CrossRef]
- McClenaghan, F.; Nash, R. The Modified Stenver’s View for Cochlear Implants—What Do the Surgeons Want to Know? J. Belg. Soc. Radiol. 2020, 104, 37. [Google Scholar] [CrossRef]
- Bennink, E.; Peters, J.P.; Wendrich, A.W.; Vonken, E.j.; van Zanten, G.A.; Viergever, M.A. Automatic Localization of Cochlear Implant Electrode Contacts in CT. Ear Hear. 2017, 38, e376–e384. [Google Scholar] [CrossRef] [PubMed]
- Hachmann, H.; Krüger, B.; Rosenhahn, B.; Nogueira, W. Localization Of Cochlear Implant Electrodes From Cone Beam Computed Tomography Using Particle Belief Propagation. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 593–597. [Google Scholar] [CrossRef]
- Zhao, Y.; Dawant, B.M.; Labadie, R.F.; Noble, J.H. Automatic Localization of Closely Spaced Cochlear Implant Electrode Arrays in Clinical CTs. Med. Phys. 2018, 45, 5030–5040. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dawant, B.M.; Labadie, R.F.; Noble, J.H. Automatic Localization of Cochlear Implant Electrodes in CT. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2014; Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R., Eds.; Springer International Publishing: Cham, Switzerland, 2014; Volume 8673, pp. 331–338. [Google Scholar] [CrossRef]
- Zhao, Y.; Chakravorti, S.; Labadie, R.F.; Dawant, B.M.; Noble, J.H. Automatic Graph-Based Method for Localization of Cochlear Implant Electrode Arrays in Clinical CT with Sub-Voxel Accuracy. Med. Image Anal. 2019, 52, 1–12. [Google Scholar] [CrossRef]
- Chi, Y.; Wang, J.; Zhao, Y.; Noble, J.H.; Dawant, B.M. A Deep-Learning-Based Method for the Localization of Cochlear Implant Electrodes in CT Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; IEEE: Venice, Italy, 2019; pp. 1141–1145. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. NeuroImage 2006, 31, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.; Jia, H.; Drouillard, M.; Bensimon, J.L.; Sterkers, O.; Ferrary, E.; Nguyen, Y. An Optimized Robot-Based Technique for Cochlear Implantation to Reduce Array Insertion Trauma. Otolaryngol. Head Neck Surg. 2018, 159, 019459981879223. [Google Scholar] [CrossRef]
- Bento, R.; Danieli, F.; Magalhães, A.; Gnansia, D.; Hoen, M. Residual Hearing Preservation with the Evo® Cochlear Implant Electrode Array: Preliminary Results. Int. Arch. Otorhinolaryngol. 2016, 20, 353–358. [Google Scholar] [CrossRef]
- Escudé, B.; James, C.; Deguine, O.; Cochard, N.; Eter, E.; Fraysse, B. The Size of the Cochlea and Predictions of Insertion Depth Angles for Cochlear Implant Electrodes. Audiol. Neurotol. 2006, 11, 27–33. [Google Scholar] [CrossRef]
- Pietsch, M.; Aguirre Dávila, L.; Erfurt, P.; Avci, E.; Lenarz, T.; Kral, A. Spiral Form of the Human Cochlea Results from Spatial Constraints. Sci. Rep. 2017, 7, 7500. [Google Scholar] [CrossRef]
- Fitzgibbon, A.; Pilu, M.; Fisher, R. Direct Least Square Fitting of Ellipses. IEEE Trans. Pattern Anal. Mach. Intell. 1999, 21, 476–480. [Google Scholar] [CrossRef]
- Burger, W.; Burge, M.J. Principles of Digital Image Processing: Core Algorithms; Springer Science & Business Media: New York, NY, USA, 2010. [Google Scholar]
- Maier-Hein, L.; Reinke, A.; Christodoulou, E.; Glocker, B.; Godau, P.; Isensee, F.; Kleesiek, J.; Kozubek, M.; Reyes, M.; Riegler, M.A.; et al. Metrics Reloaded: Pitfalls and Recommendations for Image Analysis Validation. arXiv 2022, arXiv:2206.01653. [Google Scholar]
- Ruiz Pujadas, E.; Kjer, H.M.; Piella, G.; Ceresa, M.; González Ballester, M.A. Random Walks with Shape Prior for Cochlea Segmentation in Ex Vivo µCT. Int. J. Comput. Assist. Radiol. Surg. 2016, 11, 1647–1659. [Google Scholar] [CrossRef] [PubMed]
- Girum, K.B.; Lalande, A.; Hussain, R.; Créhange, G. A Deep Learning Method for Real-Time Intraoperative US Image Segmentation in Prostate Brachytherapy. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Painchaud, N.; Skandarani, Y.; Judge, T.; Bernard, O.; Lalande, A.; Jodoin, P.M. Cardiac Segmentation With Strong Anatomical Guarantees. IEEE Trans. Med. Imaging 2020, 39, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Soldati, E.; Pithioux, M.; Guenoun, D.; Bendahan, D.; Vicente, J. Assessment of Bone Microarchitecture in Fresh Cadaveric Human Femurs: What Could Be the Clinical Relevance of Ultra-High Field MRI. Diagnostics 2022, 12, 439. [Google Scholar] [CrossRef]
- Wang, Z.; Vandersteen, C.; Demarcy, T.; Gnansia, D.; Raffaelli, C.; Guevara, N.; Delingette, H. Inner-Ear Augmented Metal Artifact Reduction with Simulation-Based 3D Generative Adversarial Networks. Comput. Med. Imaging Graph. 2021, 93, 101990. [Google Scholar] [CrossRef]
- Wang, J.; Su, D.; Fan, Y.; Chakravorti, S.; Noble, J.H.; Dawant, B.M. Atlas-Based Segmentation of Intracochlear Anatomy in Metal Artifact Affected CT Images of the Ear with Co-trained Deep Neural Networks. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2021; de Bruijne, M., Cattin, P.C., Cotin, S., Padoy, N., Speidel, S., Zheng, Y., Essert, C., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 12904, pp. 14–23. [Google Scholar] [CrossRef]
- Mertens, G.; Van de Heyning, P.; Vanderveken, O.; Topsakal, V.; Van Rompaey, V. The Smaller the Frequency-to-Place Mismatch the Better the Hearing Outcomes in Cochlear Implant Recipients? Eur. Arch. Oto-Rhino 2022, 279, 1875–1883. [Google Scholar] [CrossRef]
- Canfarotta, M.W.; Dillon, M.T.; Buss, E.; Pillsbury, H.C.; Brown, K.D.; O’Connell, B.P. Frequency-to-Place Mismatch: Characterizing Variability and the Influence on Speech Perception Outcomes in Cochlear Implant Recipients. Ear Hear. 2020, 41, 1349–1361. [Google Scholar] [CrossRef]
- López Diez, P.; Sørensen, K.; Sundgaard, J.V.; Diab, K.; Margeta, J.; Patou, F.; Paulsen, R.R. Deep Reinforcement Learning for Detection of Inner Ear Abnormal Anatomy in Computed Tomography. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2022, Singapore, 18–22 September 2022; Wang, L., Dou, Q., Fletcher, P.T., Speidel, S., Li, S., Eds.; Springer Nature Switzerland: Cham, Switzerland, 2022. Lecture Notes in Computer Science. pp. 697–706. [Google Scholar] [CrossRef]
- López Diez, P.; Juhl, K.A.; Sundgaard, J.V.; Diab, H.; Margeta, J.; Patou, F.; Paulsen, R.R. Deep Reinforcement Learning for Detection of Abnormal Anatomies. In Proceedings of the Northern Lights Deep Learning Workshop, North Pole, Norway, 10–12 January 2022; Volume 3. [Google Scholar] [CrossRef]
- Hussain, R.; Lalande, A.; Guigou, C.; Bozorg-Grayeli, A. Contribution of Augmented Reality to Minimally Invasive Computer-Assisted Cranial Base Surgery. IEEE J. Biomed. Health Inform. 2020, 24, 2093–2106. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the 33rd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–14 December 2019; Number 721. Curran Associates Inc.: Red Hook, NY, USA, 2019; pp. 8026–8037. [Google Scholar]
- Schroeder, W.; Martin, K.; Lorensen, B. The Visualization Toolkit—An Object-Oriented Approach to 3D Graphics, 4th ed.; Kitware, Inc.: Clifton Park, NY, USA, 2006. [Google Scholar]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array Programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J.; et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Ziegler, E.; Urban, T.; Brown, D.; Petts, J.; Pieper, S.D.; Lewis, R.; Hafey, C.; Harris, G.J. Open Health Imaging Foundation Viewer: An Extensible Open-Source Framework for Building Web-Based Imaging Applications to Support Cancer Research. JCO Clin. Cancer Inform. 2020, 4, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, C.B.; Kaszynski, A. PyVista: 3D Plotting and Mesh Analysis through a Streamlined Interface for the Visualization Toolkit (VTK). J. Open Source Softw. 2019, 4, 1450. [Google Scholar] [CrossRef]

| Category | Flags Implemented |
|---|---|
| Image | poor image quality (resolution) |
| Segmentation | low cochlear volume |
| low segmentation reliability | |
| irregular cochlear centerline | |
| irregular voxel intensities within segmented region | |
| Registration | low correlation between pre-op and post-op |
| large difference between registered landmarks | |
| too many electrode detected outside cochlea | |
| too many electrodes detected outside scala tympani | |
| non-basal electrodes detected outside cochlea | |
| Electrode detection | incorrect number of electrodes detected |
| irregular electrode ordering | |
| incorrect intensity at electrode locations | |
| irregular electrode pitch | |
| detected electrodes clustered together | |
| incorrect distance to modiolar axis | |
| electrodes detected near image boundaries |
| Landmark Detection | ||||||||||||
| Dataset | Apex (mm) | Center (mm) | Round Window (mm) | |||||||||
| Clinical (n = 60) | 0.71 | 0.75 | 1.30 | |||||||||
| Segmentation | ||||||||||||
| Dataset | Dice (%) | ASSD (mm) | RAVD | HD95 (mm) | ||||||||
| Structure | CO | ST | SV | CO | ST | SV | CO | ST | SV | CO | ST | SV |
| TB set 1 (n = 9) | 83 | 67 | 64 | 0.17 | 0.21 | 0.18 | −0.10 | −0.02 | −0.20 | 0.43 | 0.61 | 0.43 |
| TB set 2 (n = 9) | 77 | 64 | 58 | 0.21 | 0.23 | 0.24 | −0.10 | 0.23 | −0.38 | 0.76 | 0.77 | 0.99 |
| TB set 3 (n = 5) | 79 | 64 | 56 | 0.19 | 0.22 | 0.20 | −0.21 | −0.04 | −0.40 | 0.62 | 0.71 | 0.64 |
| Clinical (n = 58) | 86 | 0.14 | −0.13 | 0.35 | ||||||||
| Mean | 84 | 65 | 60 | 0.15 | 0.22 | 0.20 | −0.14 | 0.02 | −0.32 | 0.41 | 0.68 | 0.63 |
| Electrode Detection | ||||||||||||
| Dataset | Electrode Distance (mm) | |||||||||||
| Clinical (n = 60) | 0.09 | |||||||||||
| Registration | ||||||||||||
| Dataset | Mutual Information | Mean Registration Error (mm) | ||||||||||
| Clinical (n = 15) | 0.15 | 0.88 | ||||||||||
| Robustness Analysis | ||||||||||||
| Dataset | Reviewer 1 (%) | Reviewer 2 (%) | ||||||||||
| Pre-operative (n = 156) | 98.7 | 98.1 | ||||||||||
| Post-operative (n = 156) | 88.3 (76.2) | 85.2 (78.4) | ||||||||||
| Failure Detection | ||||||||||||
| Dataset | Sensitivity (%) | Specificity (%) | Accuracy (%) | |||||||||
| Pre-operative (n = 156) | 100 | 97.4 | 97.4 | |||||||||
| Post-operative (n = 156) | 97.3 | 57.7 | 68.6 | |||||||||
| Computational Time | ||||||||||||
| Process | Approximate Time (s) | |||||||||||
| Landmark estimation | 5.9 | |||||||||||
| Cochlear view generation | 12.5 | |||||||||||
| Segmentation and pre-operative analysis | 468.9 | |||||||||||
| Electrode detection and post-operative analysis | 148.2 | |||||||||||
| Registration | 49.8 | |||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Margeta, J.; Hussain, R.; López Diez, P.; Morgenstern, A.; Demarcy, T.; Wang, Z.; Gnansia, D.; Martinez Manzanera, O.; Vandersteen, C.; Delingette, H.; et al. A Web-Based Automated Image Processing Research Platform for Cochlear Implantation-Related Studies. J. Clin. Med. 2022, 11, 6640. https://doi.org/10.3390/jcm11226640
Margeta J, Hussain R, López Diez P, Morgenstern A, Demarcy T, Wang Z, Gnansia D, Martinez Manzanera O, Vandersteen C, Delingette H, et al. A Web-Based Automated Image Processing Research Platform for Cochlear Implantation-Related Studies. Journal of Clinical Medicine. 2022; 11(22):6640. https://doi.org/10.3390/jcm11226640
Chicago/Turabian StyleMargeta, Jan, Raabid Hussain, Paula López Diez, Anika Morgenstern, Thomas Demarcy, Zihao Wang, Dan Gnansia, Octavio Martinez Manzanera, Clair Vandersteen, Hervé Delingette, and et al. 2022. "A Web-Based Automated Image Processing Research Platform for Cochlear Implantation-Related Studies" Journal of Clinical Medicine 11, no. 22: 6640. https://doi.org/10.3390/jcm11226640
APA StyleMargeta, J., Hussain, R., López Diez, P., Morgenstern, A., Demarcy, T., Wang, Z., Gnansia, D., Martinez Manzanera, O., Vandersteen, C., Delingette, H., Buechner, A., Lenarz, T., Patou, F., & Guevara, N. (2022). A Web-Based Automated Image Processing Research Platform for Cochlear Implantation-Related Studies. Journal of Clinical Medicine, 11(22), 6640. https://doi.org/10.3390/jcm11226640







