Diagnosing Hemophagocytic Lymphohistiocytosis with Machine Learning: A Proof of Concept
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Data Classification
2.3. Predictive Model Building
3. Results
3.1. Descriptive Statistics
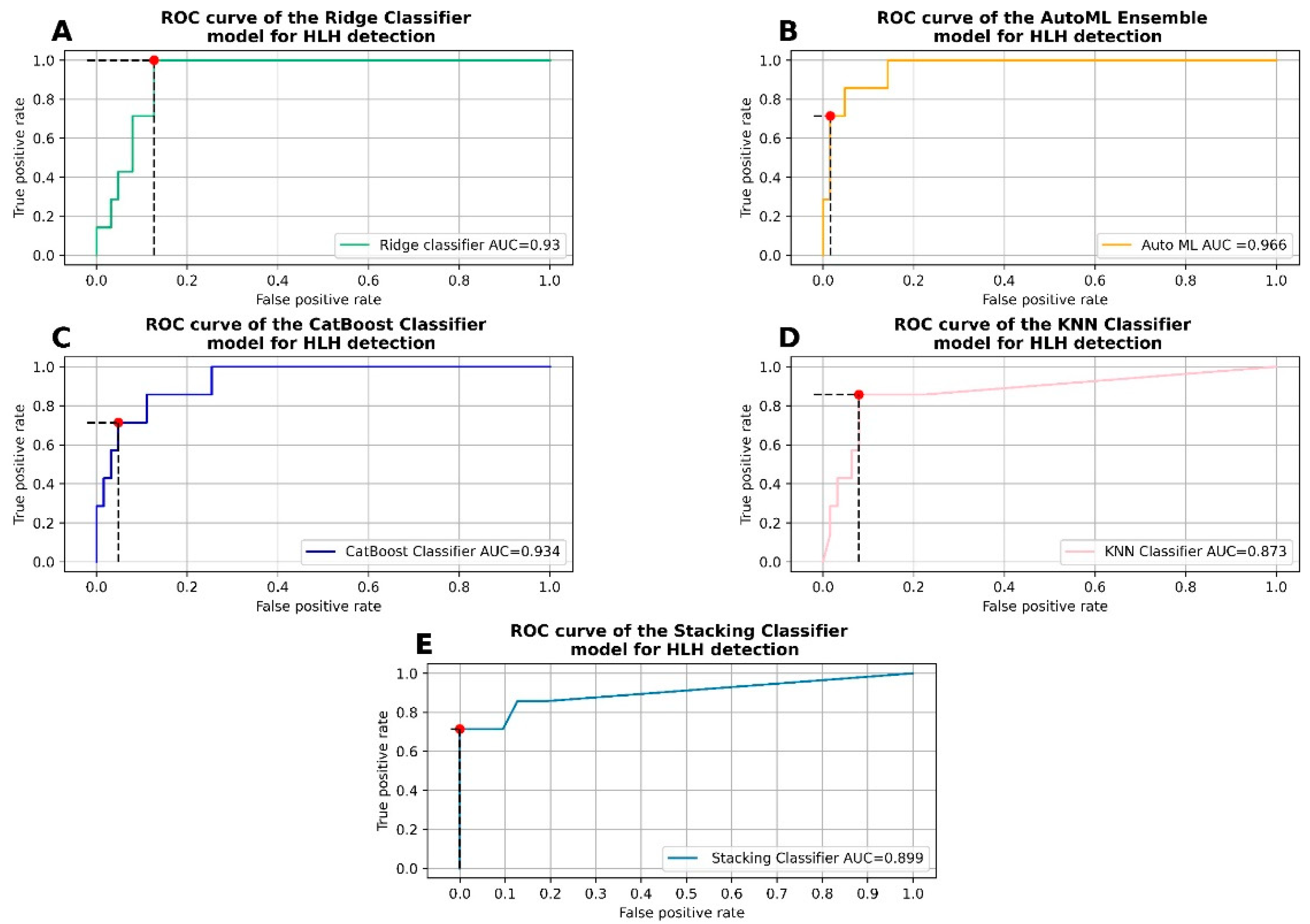
3.2. Running the Automated Machine Learning Procedure
3.3. Hyperparameter Tuning and Fitting of the Ridge Classifier
3.4. Hyperparameter Tuning and Fitting of the CatBoost Classifier
3.5. Hyperparameter Tuning and Fitting of the K Nearest Neighbors Classifier
3.6. Stacking Efficient Classifiers
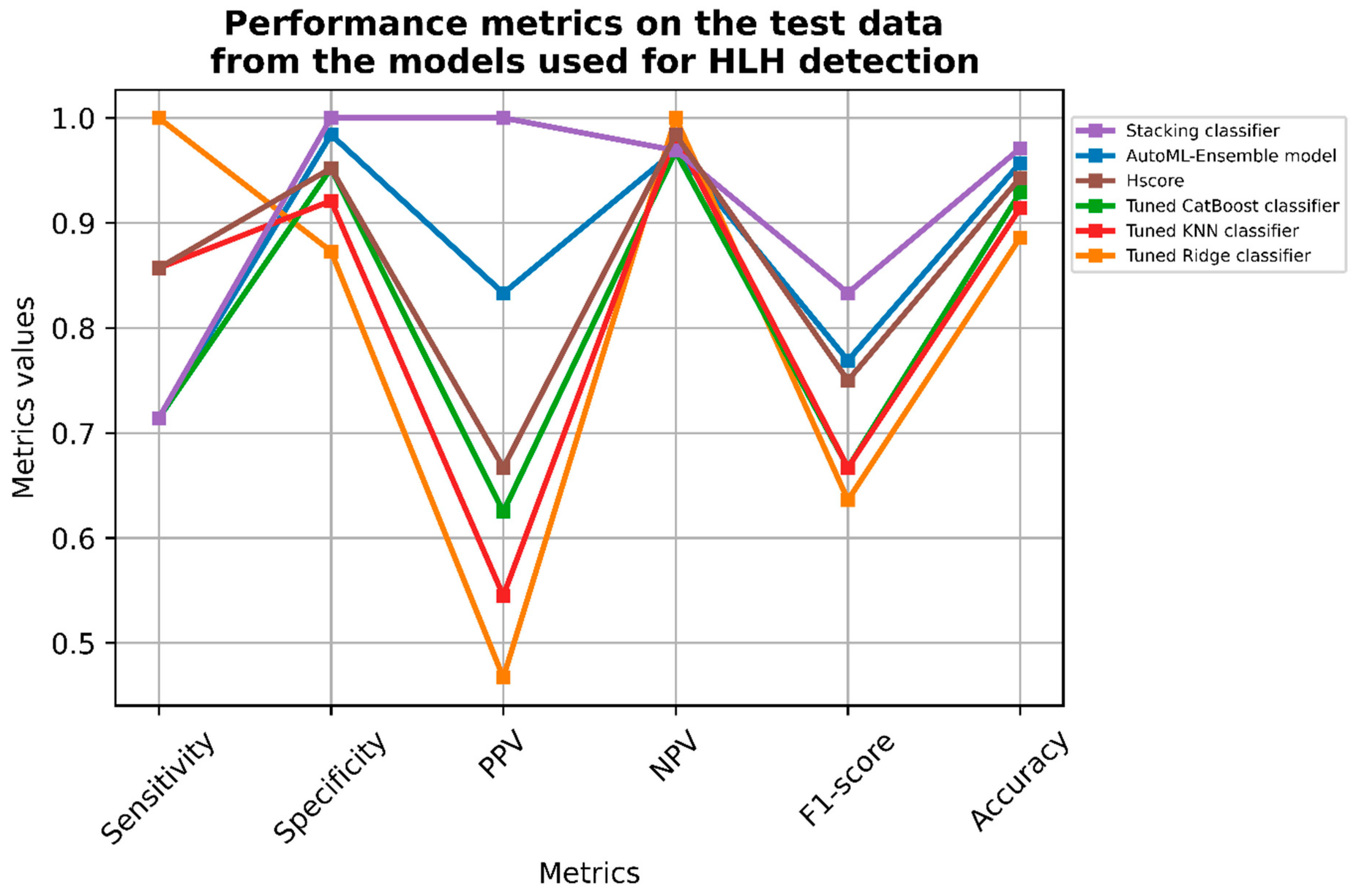
3.6.1. Classifier Metrics
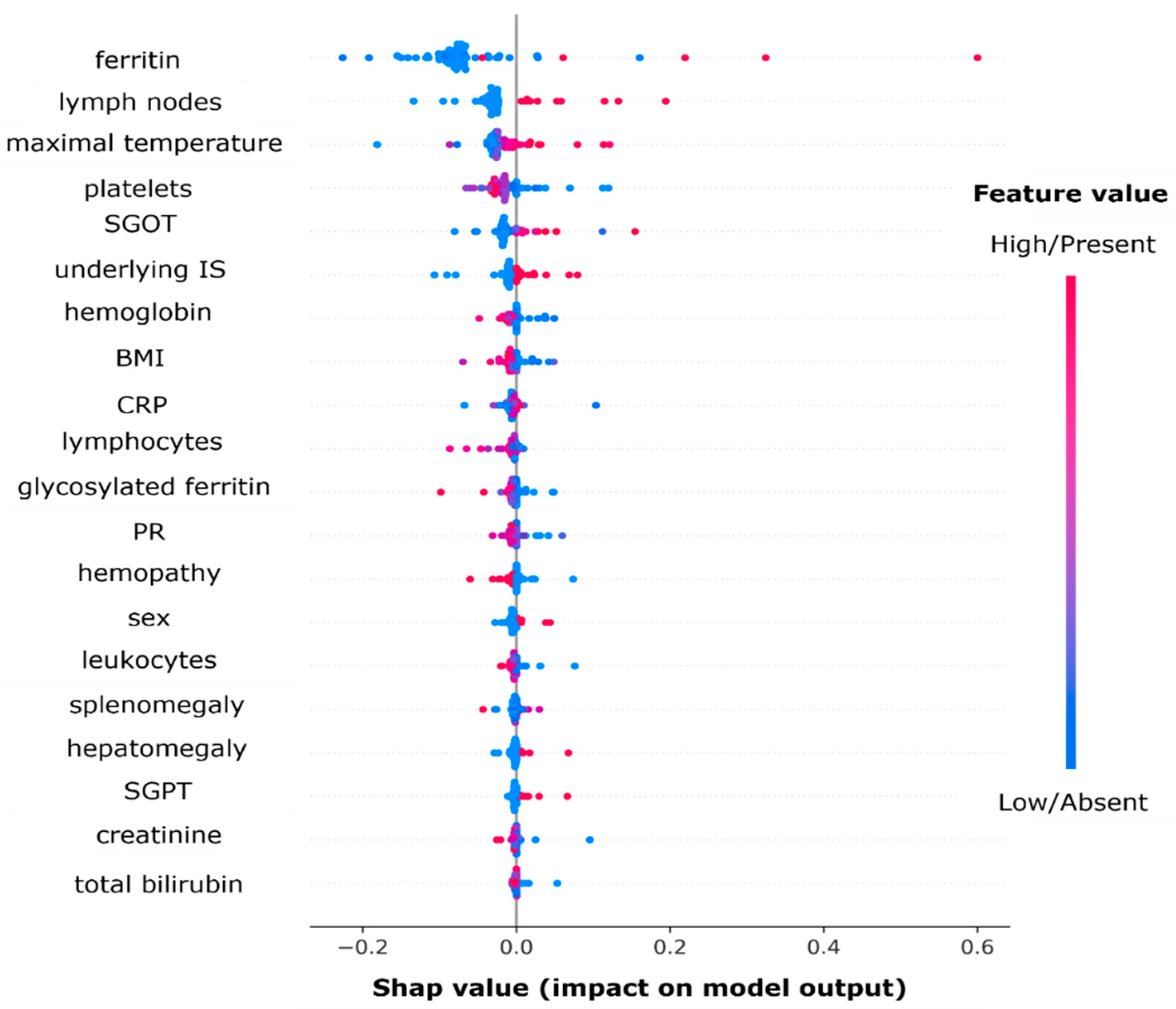
3.6.2. Variable Importance Using Shapley Values
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Al-Samkari, H.; Berliner, N. Hemophagocytic Lymphohistiocytosis. Annu. Rev. Pathol. Mech. Dis. 2018, 13, 27–49. [Google Scholar] [CrossRef] [PubMed]
- Crayne, C.B.; Albeituni, S.; Nichols, K.E.; Cron, R.Q. The Immunology of Macrophage Activation Syndrome. Front. Immunol. 2019, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Emile, J.-F.; Abla, O.; Fraitag, S.; Horne, A.; Haroche, J.; Donadieu, J.; Requena-Caballero, L.; Jordan, M.B.; Abdel-Wahab, O.; Allen, C.E.; et al. Revised classification of histiocytoses and neoplasms of the macrophage-dendritic cell lineages. Blood 2016, 127, 2672–2681. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Li, S.; Zheng, S.; Ding, Y.; Zhu, D.; Sun, C.; Hu, Y.; Qiao, J.; Fang, H. Understanding of cytokines and targeted therapy in macrophage activation syndrome. Semin. Arthritis Rheum. 2021, 51, 198–210. [Google Scholar] [CrossRef]
- Ramos-Casals, M.; Brito-Zerón, P.; López-Guillermo, A.; Khamashta, M.A.; Bosch, X. Adult haemophagocytic syndrome. Lancet 2014, 383, 1503–1516. [Google Scholar] [CrossRef]
- Henter, J.-I.; Horne, A.; Aricó, M.; Egeler, R.M.; Filipovich, A.H.; Imashuku, S.; Ladisch, S.; McClain, K.; Webb, D.; Winiarski, J.; et al. HLH-2004: Diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr. Blood Cancer 2007, 48, 124–131. [Google Scholar] [CrossRef]
- Bilston, L.; Croden, J.; Taparia, M.; Karkhaneh, M.; Grossman, J.; Sun, H. Validation of the HScore and the HLH—2004 diagnostic criteria for the diagnosis of hemophagocytic lymphohistiocytosis in a multicenter cohort. Eur. J. Haematol. 2022, 109, 129–137. [Google Scholar] [CrossRef]
- Suster, S.; Hilsenbeck, S.; Rywlin, A.M. Reactive histiocytic hyperplasia with hemophagocytosis in hematopoietic organs: A reevaluation of the benign hemophagocytic proliferations. Hum. Pathol. 1988, 19, 705–712. [Google Scholar] [CrossRef]
- Strauss, R.; Neureiter, D.; Westenburger, B.; Wehler, M.; Kirchner, T.; Hahn, E.G. Multifactorial risk analysis of bone marrow histiocytic hyperplasia with hemophagocytosis in critically ill medical patients—A postmortem clinicopathologic analysis. Crit. Care Med. 2004, 32, 1316–1321. [Google Scholar] [CrossRef]
- Birndt, S.; Schenk, T.; Heinevetter, B.; Brunkhorst, F.M.; Maschmeyer, G.; Rothmann, F.; Weber, T.; Müller, M.; Panse, J.; Penack, O.; et al. Hemophagocytic lymphohistiocytosis in adults: Collaborative analysis of 137 cases of a nationwide German registry. J. Cancer Res. Clin. Oncol. 2020, 146, 1065–1077. [Google Scholar] [CrossRef]
- Schram, A.M.; Berliner, N. How I treat hemophagocytic lymphohistiocytosis in the adult patient. Blood 2015, 125, 2908–2914. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, A.; Magni-Manzoni, S.; Pistorio, A.; Besana, C.; Foti, T.; Ruperto, N.; Viola, S.; Martini, A. Preliminary diagnostic guidelines for macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. J. Pediatr. 2005, 146, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Ravelli, A.; Minoia, F.; Davì, S.; Horne, A.; Bovis, F.; Pistorio, A.; Aricò, M.; Avcin, T.; Behrens, E.M.; Benedetti, F.D. 2016 Classification Criteria for Macrophage Activation Syndrome Complicating Systemic Juvenile Idiopathic Arthritis: A European League Against Rheumatism/American College of Rheumatology/Paediatric Rheumatology International Trials Organisation Collaborat. Arthritis Rheumatol. 2016, 68, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Minoia, F.; Bovis, F.; Davì, S.; Horne, A.; Fischbach, M.; Frosch, M.; Huber, A.; Jelusic, M.; Sawhney, S.; McCurdy, D.K.; et al. Development and initial validation of the MS score for diagnosis of macrophage activation syndrome in systemic juvenile idiopathic arthritis. Ann. Rheum. Dis. 2019, 78, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Fardet, L.; Galicier, L.; Lambotte, O.; Marzac, C.; Aumont, C.; Chahwan, D.; Coppo, P.; Hejblum, G. Development and Validation of the HScore, a Score for the Diagnosis of Reactive Hemophagocytic Syndrome: Score for Reactive Hemophagocytic Syndrome. Arthritis Rheumatol. 2014, 66, 2613–2620. [Google Scholar] [CrossRef] [PubMed]
- Debaugnies, F.; Mahadeb, B.; Ferster, A.; Meuleman, N.; Rozen, L.; Demulder, A.; Corazza, F. Performances of the H-Score for Diagnosis of Hemophagocytic Lymphohistiocytosis in Adult and Pediatric Patients. Am. J. Clin. Pathol. 2016, 145, 862–870. [Google Scholar] [CrossRef]
- Knaak, C.; Nyvlt, P.; Schuster, F.S.; Spies, C.; Heeren, P.; Schenk, T.; Balzer, F.; La Rosée, P.; Janka, G.; Brunkhorst, F.M.; et al. Hemophagocytic lymphohistiocytosis in critically ill patients: Diagnostic reliability of HLH-2004 criteria and HScore. Crit. Care 2020, 24, 244. [Google Scholar] [CrossRef]
- Lerkvaleekul, B.; Vilaiyuk, S. Macrophage activation syndrome: Early diagnosis is key. Open Access Rheumatol. 2018, 10, 117–128. [Google Scholar] [CrossRef]
- Schram, A.M.; Campigotto, F.; Mullally, A.; Fogerty, A.; Massarotti, E.; Neuberg, D.; Berliner, N. Marked hyperferritinemia does not predict for HLH in the adult population. Blood 2015, 125, 1548–1552. [Google Scholar] [CrossRef]
- Topol, E.J. High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 2019, 25, 44–56. [Google Scholar] [CrossRef]
- Guerber, A.; Garneret, E.; El Jammal, T.; Zaepfel, S.; Gerfaud-Valentin, M.; Sève, P.; Jamilloux, Y. Evaluation of Glycosylated Ferritin in Adult-Onset Still’s Disease and Differential Diagnoses. J. Clin. Med. 2022, 11, 5012. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Van Buuren, S.; Groothuis-Oudshoorn, K. mice: Multivariate Imputation by Chained Equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Lemaitre, G.; Nogueira, F.; Aridas, C.K. Imbalanced-learn: A Python Toolbox to Tackle the Curse of Imbalanced Datasets in Machine Learning. arXiv 2016, arXiv:1609.06570. [Google Scholar]
- Wolpert, D.H. The supervised learning no-free-lunch theorems. In Soft Computing and Industry: Recent Applications; Roy, R., Köppen, M., Ovaska, S., Furuhashi, T., Hoffmann, F., Eds.; Springer: London, UK, 2002; pp. 25–42. [Google Scholar] [CrossRef]
- Płońska, A.; Płoński, P. MLJAR, Version 0.10.3; State-of-the-art Automated Machine Learning Framework for Tabular Data; MLJAR. 2021. Available online: https://github.com/mljar/mljar-supervised (accessed on 1 July 2020).
- Wolpert, D.H. Stacked generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A unified approach to interpreting model predictions. In Advances in Neural Information Processing Systems 30; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; pp. 4765–4774. Available online: http://papers.nips.cc/paper/7062-a-unified-approach-to-interpreting-model-predictions.pdf (accessed on 27 September 2022).
- Henter, J.-I.; Samuelsson-Horne, A.; Aricò, M.; Egeler, R.M.; Elinder, G.; Filipovich, A.H.; Gadner, H.; Imashuku, S.; Komp, D.; Ladisch, S.; et al. Treatment of hemophagocytic lymphohistiocytosis with HLH-94 immunochemotherapy and bone marrow transplantation. Blood 2002, 100, 2367–2373. [Google Scholar] [CrossRef]
- Henter, J.I.; Elinder, G.; Ost, A. Diagnostic guidelines for hemophagocytic lymphohistiocytosis. The FHL Study Group of the Histiocyte Society. Semin Oncol. 1991, 18, 29–33. [Google Scholar]
- Hejblum, G.; Lambotte, O.; Galicier, L.; Coppo, P.; Marzac, C.; Aumont, C.; Fardet, L. A Web-Based Delphi Study for Eliciting Helpful Criteria in the Positive Diagnosis of Hemophagocytic Syndrome in Adult Patients. PLoS ONE 2014, 9, e94024. [Google Scholar] [CrossRef]
- Adamson, A.S.; Welch, H.G. Machine Learning and the Cancer-Diagnosis Problem—No Gold Standard. N. Engl. J. Med. 2019, 381, 2285–2287. [Google Scholar] [CrossRef]
| Control | HLH | p. Overall | |
|---|---|---|---|
| N = 207 | N = 24 | ||
| Clinical characteristics | |||
| Sex (male) | 122 (58.9%) | 9 (37.5%) | 0.074 |
| Age at onset | 53.0 [37.0; 65.5] | 49.5 [32.8; 64.2] | 0.562 |
| Etiological subgroups | <0.001 | ||
| Hematological malignancies | 13 (6.28%) | 11 (45.8%) | |
| Inflammatory diseases | 90 (43.5%) | 8 (33.3%) | |
| Infectious diseases | 48 (23.2%) | 3 (12.5%) | |
| Liver diseases | 10 (4.83%) | 0 (0.00%) | |
| Solid neoplasia | 10 (4.83%) | 1 (4.17%) | |
| Other | 36 (17.4%) | 1 (4.17%) | |
| Chronic liver disease | 7 (3.38%) | 2 (8.33%) | 0.237 |
| CKD | 12 (5.80%) | 1 (4.17%) | 1.000 |
| Underlying IS | 69 (33.3%) | 18 (75.0%) | <0.001 |
| HIV | 2 (0.97%) | 2 (8.33%) | 0.055 |
| Solid neoplasia | 18 (8.70%) | 1 (4.17%) | 0.701 |
| Hematological malignancy | 24 (11.6%) | 9 (37.5%) | 0.002 |
| HLH-inducer treatment | 56 (27.1%) | 10 (41.7%) | 0.207 |
| Genetic predisposition | 2 (0.97%) | 1 (4.17%) | 0.282 |
| Physical exam features | |||
| Max body temperature | 37.7 [36.8; 38.9] | 39.5 [39.0; 40.0] | <0.001 |
| Hepatomegaly | 17 (8.21%) | 6 (25.0%) | 0.020 |
| Splenomegaly | 15 (7.25%) | 10 (41.7%) | <0.001 |
| Lymph nodes | 26 (12.6%) | 15 (62.5%) | <0.001 |
| Papular rash | 36 (17.4%) | 7 (29.2%) | 0.170 |
| Transient rash | 6 (2.90%) | 1 (4.17%) | 0.541 |
| Arthritis | 37 (17.9%) | 2 (8.33%) | 0.386 |
| Arthralgias | 67 (32.4%) | 5 (20.8%) | 0.357 |
| Odynophagia | 25 (12.1%) | 3 (12.5%) | 1.000 |
| BMI | 24.0 [21.0; 27.0] | 21.0 [20.0; 25.0] | 0.166 |
| Biology | |||
| Ferritin | 633 [276; 1618] | 8749 [2402; 25,713] | <0.001 |
| Glycosylated ferritin (%) | 39.0 [26.0; 53.5] | 23.5 [8.00; 34.0] | <0.001 |
| TSR | 0.16 [0.10; 0.31] | 0.12 [0.09; 0.31] | 0.628 |
| CRP | 71.0 [18.0; 155] | 77.0 [29.2; 205] | 0.203 |
| PCT | 0.50 [0.10;1.50] | 4.00 [0.20; 10.2] | 0.112 |
| AST | 30.0 [20.0; 54.0] | 112 [67.0; 172] | <0.001 |
| ALT | 32.0 [18.2; 62.2] | 79.0 [46.2; 160] | 0.001 |
| Total bilirubin | 8.00 [6.00; 13.0] | 15.5 [6.75; 33.8] | 0.009 |
| LDH | 295 [220; 404] | 590 [394; 946] | <0.001 |
| CPK | 43.0 [21.8; 87.5] | 106 [31.0; 173] | 0.138 |
| Creatinine | 68.0 [57.0; 81.0] | 55.0 [45.5; 71.0] | 0.017 |
| Triglycerids | 1.40 [1.00; 1.90] | 2.85 [2.15; 3.97] | <0.001 |
| PR | 78.0 [67.0; 90.0] | 74.5 [55.2; 90.2] | 0.225 |
| Fibrinogen | 5.40 [3.27; 8.40] | 2.75 [1.75; 4.75] | 0.002 |
| Hemoglobin | 114 [93.0; 135] | 90.0 [78.2; 100] | <0.001 |
| Platelets | 255 [184; 334] | 62.0 [32.8; 138] | <0.001 |
| Leukocytes | 8.40 [5.60; 11.6] | 2.30 [1.25; 4.93] | <0.001 |
| Neutrophils | 5.10 [3.25; 8.40] | 1.55 [0.75; 3.20] | <0.001 |
| Neutrophils (%) | 0.67 [0.57; 0.77] | 0.72 [0.54; 0.81] | 0.730 |
| Lymphocytes | 1.60 [1.00; 2.10] | 0.70 [0.32; 1.00] | <0.001 |
| Evolution | |||
| ICU | 36 (17.4%) | 10 (41.7%) | 0.012 |
| Death | 17 (8.21%) | 4 (16.7%) | 0.248 |
| Model Type | AUC | Train Time (s) |
|---|---|---|
| Ensemble | 0.997475 | 0.47 |
| CatBoost | 0.994949 | 1.57 |
| Linear | 0.989899 | 1.88 |
| Nearest neighbors | 0.988636 | 1.66 |
| Neural network | 0.987374 | 1.22 |
| LightGBM | 0.984848 | 3.37 |
| Xgboost | 0.982323 | 3.71 |
| Random forest | 0.967172 | 3.73 |
| Extra trees | 0.959596 | 2.09 |
| Decision tree | 0.767677 | 2.5 |
| Baseline | 0.5 | 1.59 |
| AutoML-Ensemble Model | Tuned Ridge Classifier | Tuned CatBoost Classifier | Tuned KNN Classifier | Stacking Classifier | Hscore | |
|---|---|---|---|---|---|---|
| Sensitivity | 0.714 | 1.000 | 0.714 | 0.857 | 0.714 | 0.857 |
| Specificity | 0.984 | 0.873 | 0.952 | 0.921 | 1.000 | 0.952 |
| Positive predictive value | 0.833 | 0.467 | 0.625 | 0.545 | 1.000 | 0.667 |
| Negative predictive value | 0.969 | 1.000 | 0.968 | 0.983 | 0.969 | 0.984 |
| F1-score | 0.769 | 0.636 | 0.667 | 0.667 | 0.8333 | 0.750 |
| Accuracy | 0.957 | 0.886 | 0.929 | 0.914 | 0.971 | 0.943 |
| Threshold for probability | 0.50 | 0.36 | 0.49 | 0.50 | 0.50 | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El Jammal, T.; Guerber, A.; Prodel, M.; Fauter, M.; Sève, P.; Jamilloux, Y. Diagnosing Hemophagocytic Lymphohistiocytosis with Machine Learning: A Proof of Concept. J. Clin. Med. 2022, 11, 6219. https://doi.org/10.3390/jcm11206219
El Jammal T, Guerber A, Prodel M, Fauter M, Sève P, Jamilloux Y. Diagnosing Hemophagocytic Lymphohistiocytosis with Machine Learning: A Proof of Concept. Journal of Clinical Medicine. 2022; 11(20):6219. https://doi.org/10.3390/jcm11206219
Chicago/Turabian StyleEl Jammal, Thomas, Arthur Guerber, Martin Prodel, Maxime Fauter, Pascal Sève, and Yvan Jamilloux. 2022. "Diagnosing Hemophagocytic Lymphohistiocytosis with Machine Learning: A Proof of Concept" Journal of Clinical Medicine 11, no. 20: 6219. https://doi.org/10.3390/jcm11206219
APA StyleEl Jammal, T., Guerber, A., Prodel, M., Fauter, M., Sève, P., & Jamilloux, Y. (2022). Diagnosing Hemophagocytic Lymphohistiocytosis with Machine Learning: A Proof of Concept. Journal of Clinical Medicine, 11(20), 6219. https://doi.org/10.3390/jcm11206219







