Compared the Microbiota Profiles between Samples from Bronchoalveolar Lavage and Endotracheal Aspirates in Severe Pneumonia: A Real-World Experience
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Patients, ICU Setting, and Severity Scores
2.3. Data Collection, Sample Collection, and DNA Extraction
2.4. Library Construction and 16S Ribosomal RNA Sequencing
2.5. Filtering 16S rRNA Sequencing Data for Quality
- (i)
- The command “fastq_quality_filter −Q 33 −q 20 −p 70”. “−q 20” designated 20 as the minimum quality score. “−p 70” designated the minimum percentage of the bases required for “−q” quality to be ≥70%.
- (ii)
- The command “fastq_quality_trimmer −t 20 −l 100 −Q 33”. “−t 20” designated that bases with lower qualities (Q < 20) would be trimmed (checking from the end of the sequence). “−l 100” indicated that the minimum acceptable sequence length was 100 after trimming.
- (iii)
- Sequences were retained if both forward and reverse sequencing reads passed the first and second steps.
2.6. Taxonomy Assignment Based on Bacterial 16S rRNA Sequences
2.7. Computational and Statistical Analysis
3. Results
3.1. Cohort Demographic Data and Antibiotic Prescription Pattern
3.2. Subgroup Analysis of Clinical Outcomes and Antibiotic Prescription Pattern According to BAL Culture Results
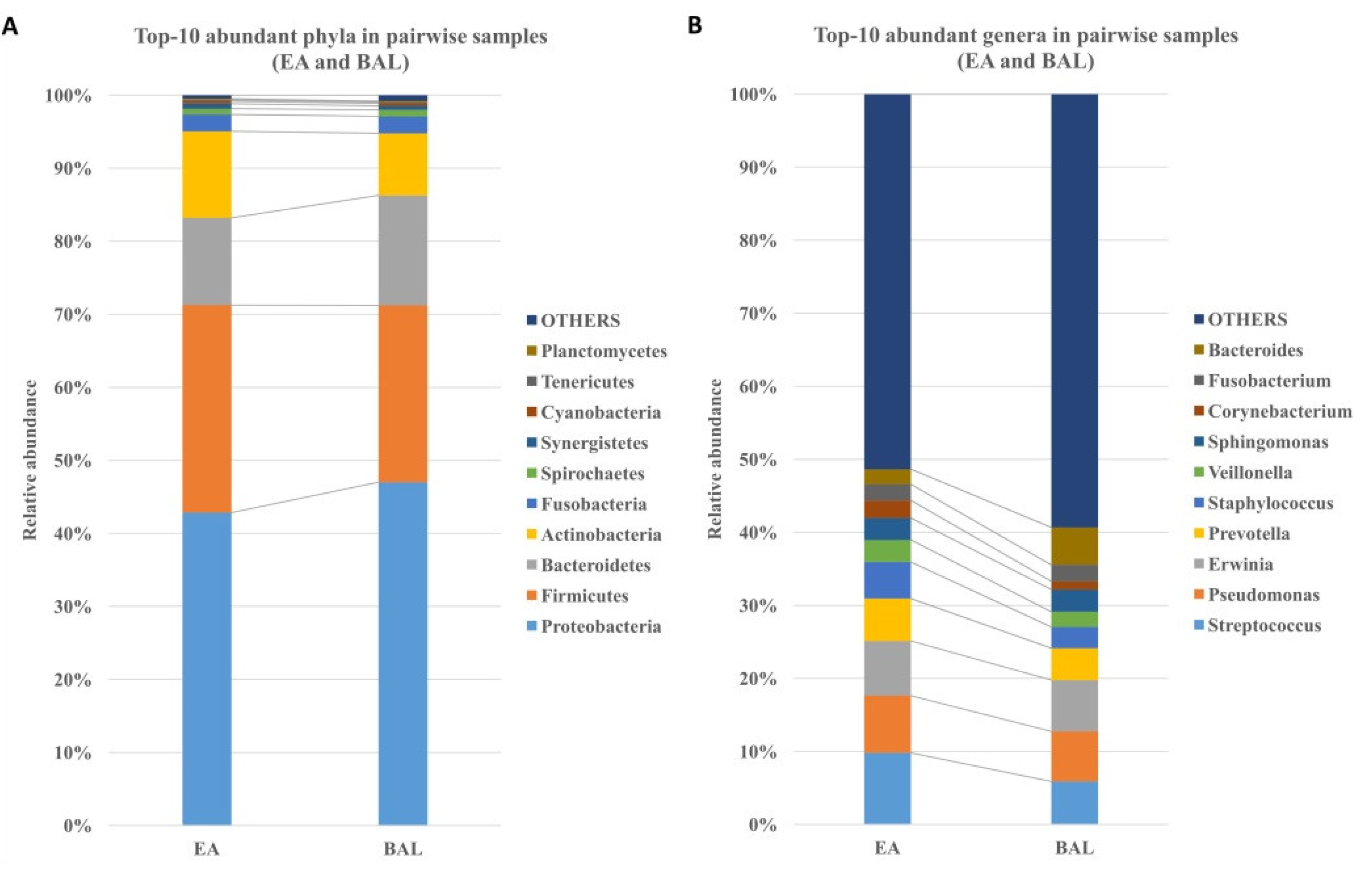
3.3. The EA Microbiota Resembles BAL Microbiota
3.4. Similar Microbial Diversity in EA and BAL Samples
3.5. Consistent Microbial Relationships between EA and BAL Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalil, A.C.; Metersky, M.L.; Klompas, M.; Muscedere, J.; Sweeney, D.A.; Palmer, L.B.; Napolitano, L.M.; O’Grady, N.P.; Bartlett, J.G.; Carratala, J.; et al. Management of Adults with Hospital-acquired and Ventilator-associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 2016, 63, e61–e111. [Google Scholar] [CrossRef]
- World Health Organization. The Top 10 Causes of Death in 2016. Available online: http://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death/ (accessed on 19 November 2021).
- Murdoch, D.R.; O’Brien, K.L.; Scott, J.A.; Karron, R.A.; Bhat, N.; Driscoll, A.J.; Knoll, M.D.; Levine, O.S. Breathing new life into pneumonia diagnostics. J. Clin. Microbiol. 2009, 47, 3405–3408. [Google Scholar] [CrossRef] [Green Version]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Kim, T.H.; Hong, S.B.; Lim, C.M.; Koh, Y.; Jang, E.Y.; Huh, J.W. The Role of Exosomes in Bronchoalveloar Lavage from Patients with Acute Respiratory Distress Syndrome. J. Clin. Med. 2019, 8, 1148. [Google Scholar] [CrossRef] [Green Version]
- Canadian Critical Care Trials Group. A randomized trial of diagnostic techniques for ventilator-associated pneumonia. N. Engl. J. Med. 2006, 355, 2619–2630. [Google Scholar] [CrossRef]
- Fagon, J.Y.; Chastre, J.; Wolff, M.; Gervais, C.; Parer-Aubas, S.; Stephan, F.; Similowski, T.; Mercat, A.; Diehl, J.L.; Sollet, J.P.; et al. Invasive and noninvasive strategies for management of suspected ventilator-associated pneumonia. A randomized trial. Ann. Intern. Med. 2000, 132, 621–630. [Google Scholar] [CrossRef]
- Sole Violan, J.; Fernandez, J.A.; Benitez, A.B.; Cardenosa Cendrero, J.A.; Rodriguez de Castro, F. Impact of quantitative invasive diagnostic techniques in the management and outcome of mechanically ventilated patients with suspected pneumonia. Crit. Care Med. 2000, 28, 2737–2741. [Google Scholar] [CrossRef]
- Raman, K.; Nailor, M.D.; Nicolau, D.P.; Aslanzadeh, J.; Nadeau, M.; Kuti, J.L. Early antibiotic discontinuation in patients with clinically suspected ventilator-associated pneumonia and negative quantitative bronchoscopy cultures. Crit. Care Med. 2013, 41, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Bregeon, F.; Papazian, L.; Thomas, P.; Carret, V.; Garbe, L.; Saux, P.; Drancourt, M.; Auffray, J.P. Diagnostic accuracy of protected catheter sampling in ventilator-associated bacterial pneumonia. Eur. Respir. J. 2000, 16, 969–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berton, D.C.; Kalil, A.C.; Teixeira, P.J. Quantitative versus qualitative cultures of respiratory secretions for clinical outcomes in patients with ventilator-associated pneumonia. Cochrane Database Syst. Rev. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, M.; Torres, A.; Ewig, S.; Marcos, M.A.; Alcon, A.; Lledo, R.; Asenjo, M.A.; Maldonaldo, A. Noninvasive versus invasive microbial investigation in ventilator-associated pneumonia: Evaluation of outcome. Am. J. Respir. Crit. Care Med. 2000, 162, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, Y.; Park, J.S.; Cho, Y.J.; Yoon, H.I.; Lee, C.T.; Lee, J.H. Characterization of Microbiota in Bronchiectasis Patients with Different Disease Severities. J. Clin. Med. 2018, 7, 429. [Google Scholar] [CrossRef] [Green Version]
- McTaggart, L.R.; Copeland, J.K.; Surendra, A.; Wang, P.W.; Husain, S.; Coburn, B.; Guttman, D.S.; Kus, J.V. Mycobiome Sequencing and Analysis Applied to Fungal Community Profiling of the Lower Respiratory Tract During Fungal Pathogenesis. Front. Microbiol. 2019, 10, 512. [Google Scholar] [CrossRef] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. The role of the bacterial microbiome in lung disease. Expert Rev. Respir Med. 2013, 7, 245–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayhew, D.; Devos, N.; Lambert, C.; Brown, J.R.; Clarke, S.C.; Kim, V.L.; Magid-Slav, M.; Miller, B.E.; Ostridge, K.K.; Patel, R.; et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018, 73, 422–430. [Google Scholar] [CrossRef] [Green Version]
- Byun, M.K.; Chang, J.; Kim, H.J.; Jeong, S.H. Differences of lung microbiome in patients with clinically stable and exacerbated bronchiectasis. PLoS ONE 2017, 12, e0183553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hogan, D.A.; Willger, S.D.; Dolben, E.L.; Hampton, T.H.; Stanton, B.A.; Morrison, H.G.; Sogin, M.L.; Czum, J.; Ashare, A. Analysis of Lung Microbiota in Bronchoalveolar Lavage, Protected Brush and Sputum Samples from Subjects with Mild-To-Moderate Cystic Fibrosis Lung Disease. PLoS ONE 2016, 11, e0149998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhodes, A.; Evans, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Intensive Care Med. 2017, 43, 304–377. [Google Scholar] [CrossRef]
- Rhodes, A.; Evans, L.E.; Alhazzani, W.; Levy, M.M.; Antonelli, M.; Ferrer, R.; Kumar, A.; Sevransky, J.E.; Sprung, C.L.; Nunnally, M.E.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Crit. Care Med. 2017, 45, 486–552. [Google Scholar] [CrossRef]
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 Update. Crit. Care Med. 2018, 46, 997–1000. [Google Scholar] [CrossRef]
- Taylor, B.E.; McClave, S.A.; Martindale, R.G.; Warren, M.M.; Johnson, D.R.; Braunschweig, C.; McCarthy, M.S.; Davanos, E.; Rice, T.W.; Cresci, G.A.; et al. Guidelines for the Provision and Assessment of Nutrition Support Therapy in the Adult Critically Ill Patient: Society of Critical Care Medicine (SCCM) and American Society for Parenteral and Enteral Nutrition (A.S.P.E.N.). Crit. Care Med. 2016, 44, 390–438. [Google Scholar] [CrossRef] [PubMed]
- Singer, P.; Blaser, A.R.; Berger, M.M.; Alhazzani, W.; Calder, P.C.; Casaer, M.P.; Hiesmayr, M.; Mayer, K.; Montejo, J.C.; Pichard, C.; et al. ESPEN guideline on clinical nutrition in the intensive care unit. Clin. Nutr. 2019, 38, 48–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.N.; Yang, M.F.; Wang, C.Y.; Hsu, C.Y.; Lee, B.J.; Fu, P.K. Optimal Time and Target for Evaluating Energy Delivery after Adjuvant Feeding with Small Bowel Enteral Nutrition in Critically Ill Patients at High Nutrition Risk. Nutrients 2019, 11, 645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoen, R.E.; Pinsky, P.F.; Weissfeld, J.L.; Yokochi, L.A.; Church, T.; Laiyemo, A.O.; Bresalier, R.; Andriole, G.L.; Buys, S.S.; Crawford, E.D.; et al. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N. Engl. J. Med. 2012, 366, 2345–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- FASTX-Toolkit. Available online: http://www.alz.org/what-is-dementia.asp (accessed on 9 October 2021).
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Yagmurdur, H.; Tezcan, A.H.; Karakurt, O.; Leblebici, F. The efficiency of routine endotracheal aspirate cultures compared to bronchoalveolar lavage cultures in ventilator-associated pneumonia diagnosis. Niger. J. Clin. Pract. 2016, 19, 46–51. [Google Scholar] [CrossRef]
- Shindo, Y.; Ito, R.; Kobayashi, D.; Ando, M.; Ichikawa, M.; Shiraki, A.; Goto, Y.; Fukui, Y.; Iwaki, M.; Okumura, J.; et al. Risk factors for drug-resistant pathogens in community-acquired and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 2013, 188, 985–995. [Google Scholar] [CrossRef]
- Zakharkina, T.; Martin-Loeches, I.; Matamoros, S.; Povoa, P.; Torres, A.; Kastelijn, J.B.; Hofstra, J.J.; de Wever, B.; de Jong, M.; Schultz, M.J.; et al. The dynamics of the pulmonary microbiome during mechanical ventilation in the intensive care unit and the association with occurrence of pneumonia. Thorax 2017, 72, 803–810. [Google Scholar] [CrossRef] [Green Version]
- Sze, M.A.; Dimitriu, P.A.; Hayashi, S.; Elliott, W.M.; McDonough, J.E.; Gosselink, J.V.; Cooper, J.; Sin, D.D.; Mohn, W.W.; Hogg, J.C. The lung tissue microbiome in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2012, 185, 1073–1080. [Google Scholar] [CrossRef] [Green Version]
- Cabrera-Rubio, R.; Garcia-Nunez, M.; Seto, L.; Anto, J.M.; Moya, A.; Monso, E.; Mira, A. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J. Clin. Microbiol. 2012, 50, 3562–3568. [Google Scholar] [CrossRef] [Green Version]
- Yatera, K.; Noguchi, S.; Mukae, H. The microbiome in the lower respiratory tract. Respir. Investig. 2018, 56, 432–439. [Google Scholar] [CrossRef]
- Akata, K.; Yatera, K.; Yamasaki, K.; Kawanami, T.; Naito, K.; Noguchi, S.; Fukuda, K.; Ishimoto, H.; Taniguchi, H.; Mukae, H. The significance of oral streptococci in patients with pneumonia with risk factors for aspiration: The bacterial floral analysis of 16S ribosomal RNA gene using bronchoalveolar lavage fluid. BMC Pulm. Med. 2016, 16, 79. [Google Scholar] [CrossRef] [Green Version]
- Yatera, K.; Noguchi, S.; Yamasaki, K.; Kawanami, T.; Fukuda, K.; Naito, K.; Akata, K.; Kido, T.; Ishimoto, H.; Sakamoto, N.; et al. Determining the Possible Etiology of Hospital-Acquired Pneumonia Using a Clone Library Analysis in Japan. Tohoku J. Exp. Med. 2017, 242, 9–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawanami, T.; Yatera, K.; Yamasaki, K.; Noguchi, S.; Fukuda, K.; Akata, K.; Naito, K.; Kido, T.; Ishimoto, H.; Taniguchi, H.; et al. Clinical impact of methicillin-resistant staphylococcus aureus on bacterial pneumonia: Cultivation and 16S ribosomal RNA gene analysis of bronchoalveolar lavage fluid. BMC Infect. Dis. 2016, 16, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monso, E. Microbiome in chronic obstructive pulmonary disease. Ann. Transl. Med. 2017, 5, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carr, T.F.; Alkatib, R.; Kraft, M. Microbiome in Mechanisms of Asthma. Clin. Chest Med. 2019, 40, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Characteristics (n = 50) | Median IQR (n, %) | |
|---|---|---|
| Age | 75.4 | 66–84.5 |
| Gender (n, Male %) | 33 | 66.0% |
| Body mass index | 22.5 | 19.5–25.8 |
| APACHEII | 29 | 24.8–34 |
| SOFA score | 7.5 | 5.8–10 |
| mNURTIC sore | 6 | 5–7 |
| Procalcitonin on day 1 (ng/mL) | 1.17 | 0.2–6.1 |
| Charlson comorbidity index (CCI) | 4.5 | 2–7 |
| Cardiovascular disease | 23 | (46.0%) |
| Cerebral vascular disease | 33 | (66.0%) |
| Chronic lung disease | 21 | (42.0%) |
| Chronic liver disease | 18 | (36.0%) |
| Diabetes Mellitus | 19 | (38.0%) |
| Chronic renal disease | 19 | (38.0%) |
| Malignancy | 22 | (44.0%) |
| Steroid for shock on day 1 (n, %) | 42 | 84.0% |
| Mechanical ventilator days | 9 | 6–17.3 |
| ICU stays | 12 | 8–18.5 |
| Hospital stays | 21.5 | 15.8–40 |
| ICU mortality (n,%) | 13 | 26.0% |
| Hospital mortality (n,%) | 18 | 36.0% |
| Culture results (n,%) | ||
| BAL culture (−) | 29 | 58.0% |
| BAL culture (+) | 21 | 42.0% |
| BAL (+) and EA (+) | 13 | 26.0% |
| BAL (+) but EA (−) | 8 | 16.0% |
| Antibiotics prescription on day 1 | ||
| Anti-Pseudomonas | 39 | 78.0% |
| Anti-Pseudomonas + Anti-MRSA | 3 | 6.0% |
| Non-Pseudomonas, Non-MRSA coverage | 8 | 16.0% |
| Changes of antibiotics prescription on day 3 adjusted by BAL culture results (n, %) | 9 | 18.0% |
| Rationale for changing prescription in antibiotics | ||
| De-escalation due to negative of Pseudomonas | 7 | 77.8% |
| MRSA or Pseudomonas positive from BAL | 2 | 22.2% |
| Characteristics | BAL Culture (−) (n = 29, 58%) | BAL Culture (+) (n = 21, 42%) | p |
|---|---|---|---|
| Age | 75.3 (66–84.2) | 75.8 (64.7–87.7) | 0.549 |
| Gender (n, Male %) | 15 (51.7%) | 18 (85.7%) | 0.228 * |
| Body mass index | 23.6 (20.3–26.9) | 22.3 (18.6–25.0) | 0.302 |
| APACHE II | 29.0 (24.5–34.5) | 28.0 (24.5–33.0) | 0.836 |
| SOFA score | 8.0 (6.0–12.0) | 6.0 (4.5–10.0) | 0.139 |
| mNURTIC sore | 6.0 (5.5–7.0) | 6.0 (5.0–7.0) | 0.474 |
| Procalcitonin on day 1 (ng/mL) | 1.0 (0.2–5.7) | 1.2 (0.5–8.1) | 0.541 |
| Charlson comorbidity index (CCI) | 5.0 (2.0–7.0) | 3.0 (0.5–6.0) | 0.281 |
| Cardiovascular disease | 16 (55.2%) | 7 (33.3%) | 0.214 |
| Cerebral vascular disease | 18 (62.1%) | 15 (71.4%) | 0.699 |
| Chronic lung disease | 12 (41.4%) | 9 (42.9%) | 1.000 |
| Chronic liver disease | 12 (41.4%) | 6 (28.6%) | 0.527 |
| Diabetes Mellitus | 10 (34.5%) | 9 (42.9%) | 0.759 |
| Chronic renal disease | 11 (37.9%) | 8 (38.1%) | 1.000 |
| Malignancy | 15 (51.7%) | 7 (33.3%) | 0.315 |
| Steroid for shock on day 1 (n, %) | 24 (82.8%) | 18 (85.7%) | 1.000 |
| Mechanical ventilator days | 11.0 (6.5–21.0) | 8.0 (5.5–12.5) | 0.110 |
| ICU stays | 15.0 (10.5–23.0) | 10.0 (7.5–14.5) | 0.061 |
| Hospital stays | 22.0 (15.5–37.0) | 20.0 (15.5–41.5) | 0.984 |
| ICU mortality (n, %) | 10 (34.5%) | 3 (14.3%) | 0.200 |
| Hospital mortality (n, %) | 14 (48.2%) | 4 (19.0%) | 0.383 |
| Antibiotics prescription on day 1 | 0.801 | ||
| Anti-Pseudomonas | 21 (72.4%) | 17 (81.0%) | |
| Anti-Pseudomonas + Anti-MRSA | 2 (6.9%) | 1 (4.8%) | |
| Non-Pseudomonas, Non-MRSA | 5 (17.2%) | 3 (14.3%) | |
| Changes of antibiotics on day 3 adjusted by BAL (n, %) a | 6 (20.7%) | 3 (14.3%) | 0.716 |
| Rationale for changing in antibiotics | 0.276 | ||
| De-escalation due to Pseudomonas (−) | 5 (83.3%) | 2 (66.7%) | |
| MRSA or Pseudomonas (+) from BAL | 1 (16.7%) | 1 (33.3%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Y.-N.; Huang, W.-C.; Wang, C.-Y.; Fu, P.-K. Compared the Microbiota Profiles between Samples from Bronchoalveolar Lavage and Endotracheal Aspirates in Severe Pneumonia: A Real-World Experience. J. Clin. Med. 2022, 11, 327. https://doi.org/10.3390/jcm11020327
Cheng Y-N, Huang W-C, Wang C-Y, Fu P-K. Compared the Microbiota Profiles between Samples from Bronchoalveolar Lavage and Endotracheal Aspirates in Severe Pneumonia: A Real-World Experience. Journal of Clinical Medicine. 2022; 11(2):327. https://doi.org/10.3390/jcm11020327
Chicago/Turabian StyleCheng, Yeong-Nan, Wei-Chih Huang, Chen-Yu Wang, and Pin-Kuei Fu. 2022. "Compared the Microbiota Profiles between Samples from Bronchoalveolar Lavage and Endotracheal Aspirates in Severe Pneumonia: A Real-World Experience" Journal of Clinical Medicine 11, no. 2: 327. https://doi.org/10.3390/jcm11020327
APA StyleCheng, Y.-N., Huang, W.-C., Wang, C.-Y., & Fu, P.-K. (2022). Compared the Microbiota Profiles between Samples from Bronchoalveolar Lavage and Endotracheal Aspirates in Severe Pneumonia: A Real-World Experience. Journal of Clinical Medicine, 11(2), 327. https://doi.org/10.3390/jcm11020327







