A Novel Preoperative Prediction Model Based on Deep Learning to Predict Neoplasm T Staging and Grading in Patients with Upper Tract Urothelial Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
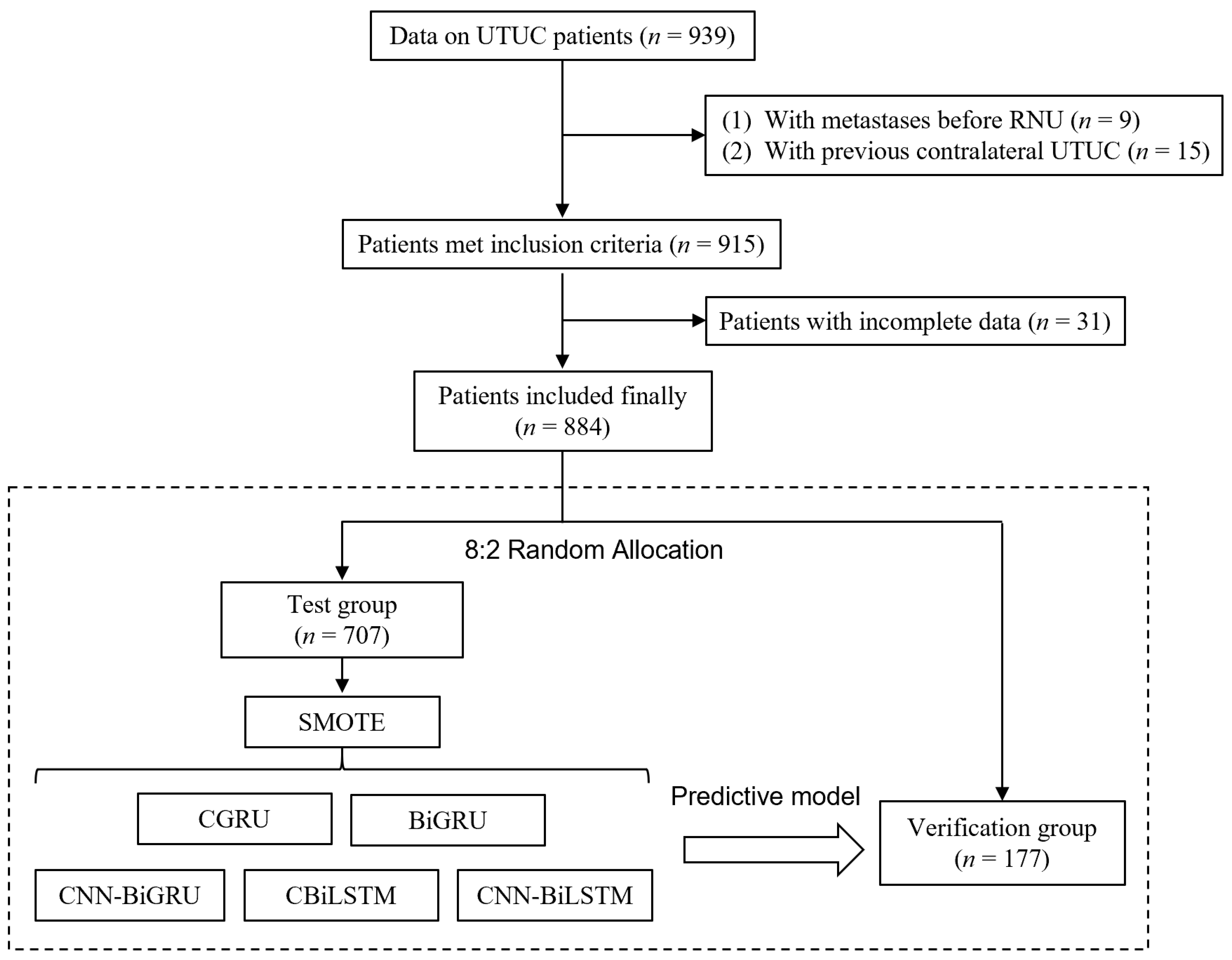
2.1. Patient Selection
2.2. Feature Selection and Model Predictive Indicators
2.3. Deep Learning and Model Construction
2.4. Performance Verification
2.5. Statistical Analysis
3. Results
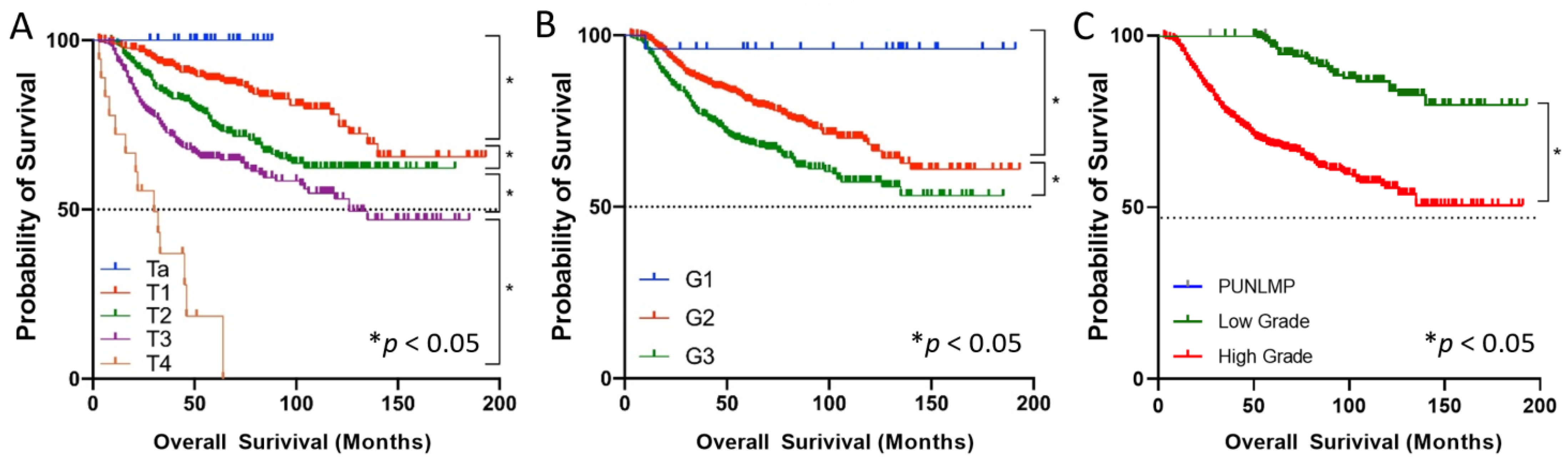
3.1. Patient Characteristics
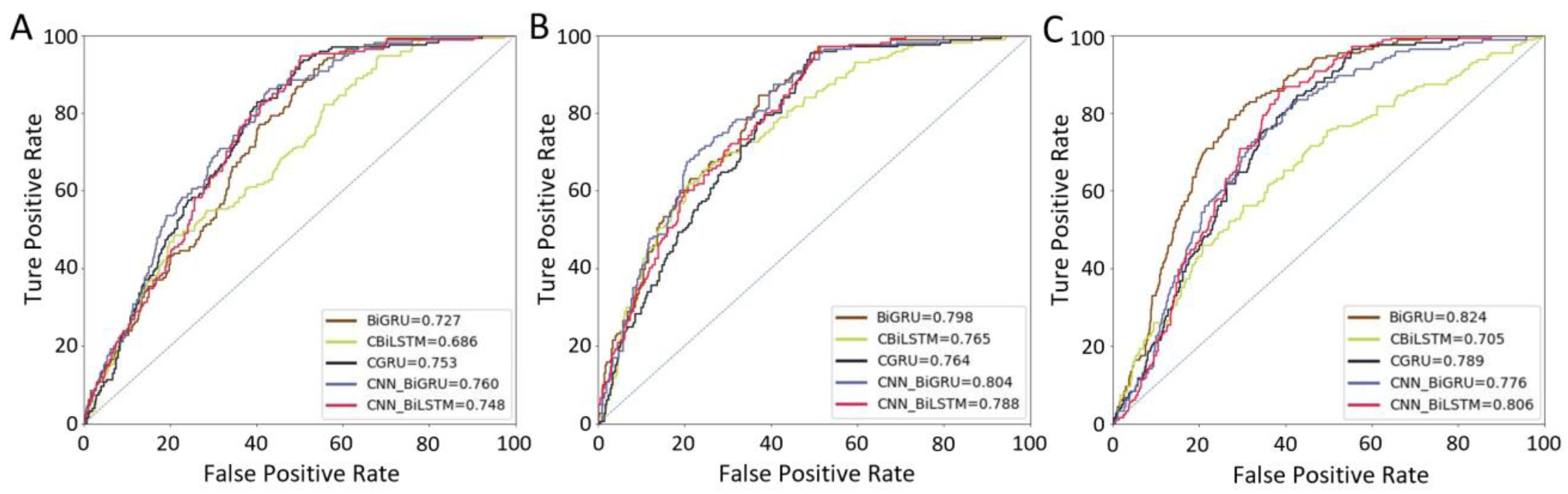
3.2. Performance of Different Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roupret, M.; Colin, P.; Yates, D.R. A new proposal to risk stratify urothelial carcinomas of the upper urinary tract (UTUCs) in a predefinitive treatment setting: Low-risk versus high-risk UTUCs. Eur. Urol. 2014, 66, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Shang, D.; Zhang, J.; Zhang, L.; Shi, R.; Fu, F.; Tian, Y. A retrospective review of patients with urothelial cancer in 3370 recipients after renal transplantation: A single-center experience. World J. Urol. 2015, 33, 713–717. [Google Scholar] [CrossRef] [PubMed]
- Seisen, T.; Peyronnet, B.; Dominguez-Escrig, J.L.; Bruins, H.M.; Yuan, C.Y.; Babjuk, M.; Bohle, A.; Burger, M.; Comperat, E.M.; Cowan, N.C.; et al. Oncologic Outcomes of Kidney-sparing Surgery Versus Radical Nephroureterectomy for Upper Tract Urothelial Carcinoma: A Systematic Review by the EAU Non-muscle Invasive Bladder Cancer Guidelines Panel. Eur. Urol. 2016, 70, 1052–1068. [Google Scholar] [CrossRef] [PubMed]
- Hosogoe, S.; Hatakeyama, S.; Kusaka, A.; Hamano, I.; Iwamura, H.; Fujita, N.; Yamamoto, H.; Tobisawa, Y.; Yoneyama, T.; Yoneyama, T.; et al. Platinum-based Neoadjuvant Chemotherapy Improves Oncological Outcomes in Patients with Locally Advanced Upper Tract Urothelial Carcinoma. Eur. Urol. Focus 2018, 4, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Baard, J.; de Bruin, D.M.; Zondervan, P.J.; Kamphuis, G.; de la Rosette, J.; Laguna, M.P. Diagnostic dilemmas in patients with upper tract urothelial carcinoma. Nat. Rev. Urol. 2017, 14, 181–191. [Google Scholar] [CrossRef]
- Ng Chieng Hin, J.; Hettiarachchilage, D.; Gravestock, P.; Rai, B.; Somani, B.K.; Veeratterapillay, R. Role of Ureteroscopy in Treatment of Upper Tract Urothelial Carcinoma. Curr. Urol. Rep. 2021, 22, 49. [Google Scholar] [CrossRef]
- Favaretto, R.L.; Shariat, S.F.; Savage, C.; Godoy, G.; Chade, D.C.; Kaag, M.; Bochner, B.H.; Coleman, J.; Dalbagni, G. Combining imaging and ureteroscopy variables in a preoperative multivariable model for prediction of muscle-invasive and non-organ confined disease in patients with upper tract urothelial carcinoma. BJU Int. 2012, 109, 77–82. [Google Scholar] [CrossRef]
- Brien, J.C.; Shariat, S.F.; Herman, M.P.; Ng, C.K.; Scherr, D.S.; Scoll, B.; Uzzo, R.G.; Wille, M.; Eggenner, S.E.; Terrell, J.D.; et al. Preoperative hydronephrosis, ureteroscopic biopsy grade and urinary cytology can improve prediction of advanced upper tract urothelial carcinoma. J. Urol. 2010, 184, 69–73. [Google Scholar] [CrossRef]
- Margulis, V.; Youssef, R.F.; Karakiewicz, P.I.; Lotan, Y.; Wood, C.G.; Zigeuner, R.; Kikuchi, E.; Weizer, A.; Raman, J.D.; Remzi, M.; et al. Preoperative multivariable prognostic model for prediction of nonorgan confined urothelial carcinoma of the upper urinary tract. J. Urol. 2010, 184, 453–458. [Google Scholar] [CrossRef]
- Lucas, M.; Jansen, I.; van Leeuwen, T.G.; Oddens, J.R.; de Bruin, D.M.; Marquering, H.A. Deep Learning-based Recurrence Prediction in Patients with Non-muscle-invasive Bladder Cancer. Eur. Urol Focus 2022, 8, 165–172. [Google Scholar] [CrossRef]
- Churpek, M.M.; Yuen, T.C.; Winslow, C.; Meltzer, D.O.; Kattan, M.W.; Edelson, D.P. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Crit. Care Med. 2016, 44, 368–374. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.Z.; Wang, Y.; Peng, J.; Sun, M.W.; Zeng, J.; Jiang, H. Comparison between logistic regression and machine learning algorithms on survival prediction of traumatic brain injuries. J. Crit. Care 2019, 54, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.P.; Xiong, G.Y.; Li, X.S.; Matin, S.F.; Garcia, M.; Fang, D.; Wang, T.Y.; Yu, W.; Gong, K.; Song, Y.; et al. Predictive factors for worse pathological outcomes of upper tract urothelial carcinoma: Experience from a nationwide high-volume centre in China. BJU Int. 2013, 112, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Moons, K.G.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef] [PubMed]
- Rouprêt, M.; Babjuk, M.; Burger, M.; Capoun, O.; Cohen, D.; Compérat, E.M.; Cowan, N.C.; Dominguez-Escrig, J.L.; Gontero, P.; Mostafid, A.H.; et al. European Association of Urology Guidelines on Upper Urinary Tract Urothelial Carcinoma: 2020 Update. Eur. Urol. 2021, 79, 62–79. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- El Rifai, H.; Al Qadi, L.; Elnagar, A. Arabic text classification: The need for multi-labeling systems. Neural Comput. Appl. 2022, 34, 1135–1159. [Google Scholar] [CrossRef]
- Wu, H.; Xing, Y.; Ge, W.; Liu, X.; Zou, J.; Zhou, C. Drug-drug interaction extraction via hybrid neural networks on biomedical literature. J. Biomed. Inform. 2020, 106, 103432. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Lu, B.; Nie, B.; Peng, Z.; Liu, H.; Pi, X. Hybrid Network with Attention Mechanism for Detection and Location of Myocardial Infarction Based on 12-Lead Electrocardiogram Signals. Sensors 2020, 20, 1020. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Huang, X.; Li, Y.; Yousaf Iqbal, M. A Double-Channel Hybrid Deep Neural Network Based on CNN and BiLSTM for Remaining Useful Life Prediction. Sensors 2020, 20, 7109. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, G.; Huang, Y.; Joshi, R.; O’Connor, T.; Javidi, B. Spatio-temporal continuous gesture recognition under degraded environments: Performance comparison between 3D integral imaging (InIm) and RGB-D sensors. Opt. Express 2021, 29, 30937–30951. [Google Scholar] [CrossRef] [PubMed]
- Acharya, J.; Basu, A. Deep Neural Network for Respiratory Sound Classification in Wearable Devices Enabled by Patient Specific Model Tuning. IEEE Trans. Biomed. Circuits Syst. 2020, 14, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Chicco, D.; Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Raman, J.D.; Park, R. Endoscopic management of upper-tract urothelial carcinoma. Expert Rev. Anticancer. Ther. 2017, 17, 545–554. [Google Scholar] [CrossRef]
- Azizi, M.; Cheriyan, S.K.; Peyton, C.C.; Foerster, B.; Shariat, S.F.; Spiess, P.E. Optimal Management of Upper Tract Urothelial Carcinoma: An Unmet Need. Curr. Treat. Options Oncol. 2019, 20, 40. [Google Scholar] [CrossRef]
- Margolin, E.J.; Matulay, J.T.; Li, G.; Meng, X.; Chao, B.; Vijay, V.; Silver, H.; Clinton, T.N.; Krabbe, L.M.; Woldu, S.L.; et al. Discordance between Ureteroscopic Biopsy and Final Pathology for Upper Tract Urothelial Carcinoma. J. Urol. 2018, 199, 1440–1445. [Google Scholar] [CrossRef]
- Mori, K.; Katayama, S.; Laukhtina, E.; Schuettfort, V.M.; Pradere, B.; Quhal, F.; Motlagh, R.S.; Mostafaei, H.; Grossmann, N.C.; Rajwa, P.; et al. Discordance Between Clinical and Pathological Staging and Grading in Upper Tract Urothelial Carcinoma. Clin. Genitourin. Cancer 2022, 20, 95.e1–95.e6. [Google Scholar] [CrossRef]
- Jeon, S.S.; Sung, H.H.; Jeon, H.G.; Han, D.H.; Jeong, B.C.; Seo, S.I.; Lee, H.M.; Choi, H.Y. Endoscopic management of upper tract urothelial carcinoma: Improved prediction of invasive cancer using a ureteroscopic scoring model. Surg. Oncol. 2017, 26, 252–256. [Google Scholar] [CrossRef]
- Petros, F.G.; Qiao, W.; Singla, N.; Clinton, T.N.; Robyak, H.; Raman, J.D.; Margulis, V.; Matin, S.F. Preoperative multiplex nomogram for prediction of high-risk nonorgan-confined upper-tract urothelial carcinoma. Urol. Oncol. 2019, 37, 292.e1–292.e9. [Google Scholar] [CrossRef]
- Ma, R.; Xia, H.; Qiu, M.; Tao, L.; Lu, M.; Huang, R.; Lu, J.; Ma, L. A Diagnostic Nomogram of Pathologic Grade for Preoperative Risk Stratification in Upper Tract Urothelial Carcinoma. Clin. Med. Insights Oncol. 2020, 14. [Google Scholar] [CrossRef]
- Yoshida, T.; Kobayashi, T.; Kawaura, T.; Miyake, M.; Ito, K.; Okuno, H. Development and external validation of a preoperative nomogram for predicting pathological locally advanced disease of clinically localized upper urinary tract carcinoma. Cancer Med. 2020, 9, 3733–3741. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ren, X.; Wang, G.; Sun, X.; Tang, S.; Zhang, B.; Xing, X.; Zhang, W.; Gao, G.; Du, J.; et al. Construction of a survival prediction model for high- and low-grade UTUC after tumor resection based on “SEER database”: A multicenter study. BMC Cancer 2021, 21, 999. [Google Scholar] [CrossRef] [PubMed]
- Aslan, M.F.; Unlersen, M.F.; Sabanci, K.; Durdu, A. CNN-based transfer learning-BiLSTM network: A novel approach for COVID-19 infection detection. Appl. Soft Comput. 2021, 98, 106912. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.; Xiang, W.; Atkinson, I. Continuous and automatic mortality risk prediction using vital signs in the intensive care unit: A hybrid neural network approach. Sci. Rep. 2020, 10, 21282. [Google Scholar] [CrossRef]
- Hsu, F.S.; Huang, S.R.; Huang, C.W.; Huang, C.J.; Cheng, Y.R.; Chen, C.C.; Hsiao, J.; Chen, C.W.; Chen, L.C.; Lai, Y.C.; et al. Benchmarking of eight recurrent neural network variants for breath phase and adventitious sound detection on a self-developed open-access lung sound database-HF Lung V1. PLoS ONE 2021, 16, e0254134. [Google Scholar] [CrossRef]
- Margulis, V.; Puligandla, M.; Trabulsi, E.J.; Plimack, E.R.; Kessler, E.R.; Matin, S.F.; Godoy, G.; Alva, A.; Hahn, N.M.; Carducci, M.A.; et al. Phase II Trial of Neoadjuvant Systemic Chemotherapy Followed by Extirpative Surgery in Patients with High Grade Upper Tract Urothelial Carcinoma. J. Urol. 2020, 203, 690–698. [Google Scholar] [CrossRef]
- Jang, H.J.; Cho, K.O. Applications of deep learning for the analysis of medical data. Arch. Pharm. Res. 2019, 42, 492–504. [Google Scholar] [CrossRef]
- Krittanawong, C.; Johnson, K.W.; Rosenson, R.S.; Wang, Z.; Aydar, M.; Baber, U.; Min, J.K.; Tang, W.H.W.; Halperin, J.L.; Narayan, S.M. Deep learning for cardiovascular medicine: A practical primer. Eur. Heart J. 2019, 40, 2058–2073. [Google Scholar] [CrossRef]
- She, Y.; Jin, Z.; Wu, J.; Deng, J.; Zhang, L.; Su, H.; Jiang, G.; Liu, H.; Xie, D.; Cao, N.; et al. Development and Validation of a Deep Learning Model for Non-Small Cell Lung Cancer Survival. JAMA Netw. Open 2020, 3, e205842. [Google Scholar] [CrossRef]
- Suarez-Ibarrola, R.; Hein, S.; Reis, G.; Gratzke, C.; Miernik, A. Current and future applications of machine and deep learning in urology: A review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J. Urol. 2020, 38, 2329–2347. [Google Scholar] [CrossRef]
- Lazo, J.F.; Marzullo, A.; Moccia, S.; Catellani, M.; Rosa, B.; De Mathelin, M. Using spatial-temporal ensembles of convolutional neural networks for lumen segmentation in ureteroscopy. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 915–922. [Google Scholar] [CrossRef] [PubMed]

| Variables | No. Pts (%) |
|---|---|
| Total | 884 |
| Gender | |
| Male | 395 (44.7) |
| Female | 489 (55.3) |
| Age, median (IQR) | 69 (61, 75) |
| BMI, kg/m2, median (IQR) | 24.2 (22.0, 26.3) |
| History of UBC | |
| No | 833 (94.2) |
| Yes | 51 (5.8) |
| Smoking | |
| No | 743 (84.0) |
| Yes | 141 (16.0) |
| Hydronephrosis | |
| No | 349 (39.5) |
| Yes | 535 (60.5) |
| Tumour site | |
| Left | 450 (50.9) |
| Right | 434 (49.1) |
| Tumour location | |
| Renal pelvis | 490 (55.4) |
| Ureter | 394 (44.6) |
| Tumour diameter (cm), median (IQR) | 3.0 (2.0, 4.2) |
| Pathological T stage | |
| Ta | 24 (2.7) |
| T1 | 302 (34.2) |
| T2 | 299 (33.8) |
| T3 | 240 (27.1) |
| T4 | 19 (2.1) |
| WHO 1973 grade | |
| G1 | 25 (2.8) |
| G2 | 496 (56.1) |
| G3 | 362 (41.1) |
| WHO 2004 grade | |
| PUNLMP | 3 (0.3) |
| Low grade | 225 (25.5) |
| High grade | 656 (74.2) |
| Overall survival | |
| Number | 884 |
| Mean follow-up times | 70.3 |
| Follow-up range | [3, 193] |
| Models | T-Staging | Grading Based on the 1973 WHO Classification | Grading Based on the 2004 WHO Classification | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MMC | AUC | F1 Score | MMC | AUC | F1 Score | MMC | AUC | F1 Score | |
| BiGRU | 0.532 (0.525–0.539) | 0.727 (0.722–0.732) | 0.410 (0.405–0.415) | 0.604 (0.599–0.609) | 0.798 (0.793–0.803) | 0.625 (0.620–0.630) | 0.621 (0.616–0.626) | 0.824 (0.819–0.829) | 0.617 (0.612–0.622) |
| CBiLSTM | 0.482 (0.477–0.487) | 0.686 (0.681–0.691) | 0.371 (0.366–0.376) | 0.566 (0.592–0.600) | 0.765 (0.759–0.771) | 0.576 (0.570–0.582) | 0.511 (0.507–0.515) | 0.705 (0.701–0.709) | 0.396 (0.391–0.401) |
| CGRU | 0.554 (0.549–0.559) | 0.753 (0.747–0.759) | 0.482 (0.476–0.488) | 0.565 (0.558–0.572) | 0.764 (0.758–0.770) | 0.574 (0.568–0.580) | 0.596 (0.590–0.602) | 0.789 (0.783–0.795) | 0.607 (0.601–0.613) |
| CNN-BiGRU | 0.598 (0.592–0.604) | 0.760 (0.755–0.765) | 0.484 (0.479–0.489) | 0.612 (0.609–0.615) | 0.804 (0.801–0.807) | 0.608 (0.605–0.611) | 0.578 (0.574–0.582) | 0.776 (0.772–0.780) | 0.593 (0.589–0.597) |
| CNN-BiLSTM | 0.542 (0.536–0.548) | 0.748 (0.743–0.753) | 0.451 (0.446–0.456) | 0.595 (0.588–0.602) | 0.788 (0.781–0.795) | 0.602 (0.595–0.609) | 0.615 (0.609–0.621) | 0.806 (0.800–0.812) | 0.605 (0.599–0.611) |
| Author | Publication Years | Prediction Form | Outcome | No. of Patients | Variables | Evaluation Index | Validation |
|---|---|---|---|---|---|---|---|
| Brien et al. [8] | 2010 | Preoperative risk group stratification | Nonorgan-confined disease | 172 | Hydronephrosis, ureteroscopic grade, and urinary cytology | PPV 73% NPV 100% | None |
| Brien et al. [8] | 2010 | Preoperative risk group stratification | Muscle-invasive disease | 172 | Hydronephrosis, ureteroscopic grade, and urinary cytology | PPV 89% NPV 100% | None |
| Margulis et al. [9] | 2010 | Preoperative nomogram | Nonorgan-confined disease | 659 | Grade, architecture, and location | 76.6% AUC | Internal |
| Favaretto et al. [7] | 2012 | Preoperative risk group stratification | Nonorgan-confined disease | 274 | Ureteroscopic grade, location, invasion, and hydronephrosis on imaging | 70% AUC | None |
| Favaretto et al. [7] | 2012 | Preoperative risk group stratification | Muscle-invasive disease | 274 | Ureteroscopic grade, location, invasion, and hydronephrosis on imaging | 71% AUC | None |
| Chen et al. [13] | 2013 | Preoperative nomogram | Nonorgan-confined disease | 693 | Gender, architecture, multifocality, location, and grade | 79% C-index | Internal |
| Chen et al. [13] | 2013 | Preoperative nomogram | Muscle-invasive disease | 693 | Gender, architecture, multifocality, location, and grade | 79% C-index | Internal |
| Jeon et al. [28] | 2017 | Preoperative nomogram | Nonorgan-confined disease or muscle-invasive disease | 172 | Urine cytology, hydronephrosis, local invasion, lamina propria invasion, high-grade tumour, and ureteroscopic scoring | 82% AUC | None |
| Petros et al. [29] | 2019 | Preoperative nomogram | Nonorgan-confined disease | 566 | Clinical stage, biopsy tumour grade, tumour architecture, and HGB levels | 82% C-index | Internal and external |
| Ma et al. [30]. | 2020 | Preoperative nomogram | Muscle-invasive disease | 245 | Age, sessile, urine cytology, ureteroscopic, and high-grade biopsy | 78% AUC | None |
| Yoshida et al. [31] | 2020 | Preoperative nomogram | Muscle-invasive disease | 1101 | Neutrophil to lymphocyte ratio, chronic kidney disease, local invasion on imaging, tumour location, and hydronephrosis | 77% AUC | Internal and external |
| Wang et al. [32] | 2021 | Preoperative nomogram | Muscle-invasive disease | 4149 | Age, tumour size, T-stage, N-stage, M-stage, LN surgery, histology, radiation, and chemotherapy | 74% C-index | Internal and external |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Y.; Gao, W.; Ying, W.; Feng, N.; Wang, Y.; Jiang, P.; Gong, Y.; Li, X. A Novel Preoperative Prediction Model Based on Deep Learning to Predict Neoplasm T Staging and Grading in Patients with Upper Tract Urothelial Carcinoma. J. Clin. Med. 2022, 11, 5815. https://doi.org/10.3390/jcm11195815
He Y, Gao W, Ying W, Feng N, Wang Y, Jiang P, Gong Y, Li X. A Novel Preoperative Prediction Model Based on Deep Learning to Predict Neoplasm T Staging and Grading in Patients with Upper Tract Urothelial Carcinoma. Journal of Clinical Medicine. 2022; 11(19):5815. https://doi.org/10.3390/jcm11195815
Chicago/Turabian StyleHe, Yuhui, Wenzhi Gao, Wenwei Ying, Ninghan Feng, Yang Wang, Peng Jiang, Yanqing Gong, and Xuesong Li. 2022. "A Novel Preoperative Prediction Model Based on Deep Learning to Predict Neoplasm T Staging and Grading in Patients with Upper Tract Urothelial Carcinoma" Journal of Clinical Medicine 11, no. 19: 5815. https://doi.org/10.3390/jcm11195815
APA StyleHe, Y., Gao, W., Ying, W., Feng, N., Wang, Y., Jiang, P., Gong, Y., & Li, X. (2022). A Novel Preoperative Prediction Model Based on Deep Learning to Predict Neoplasm T Staging and Grading in Patients with Upper Tract Urothelial Carcinoma. Journal of Clinical Medicine, 11(19), 5815. https://doi.org/10.3390/jcm11195815






