Development and Multicenter Validation of a Novel Immune-Inflammation-Based Nomogram to Predict Survival in Western Resectable Gastric and Gastroesophageal Junction Adenocarcinoma (GEA): The NOMOGAST
Abstract
:1. Introduction
2. Materials and Methods
Statistical Analysis, Model Development and Validation
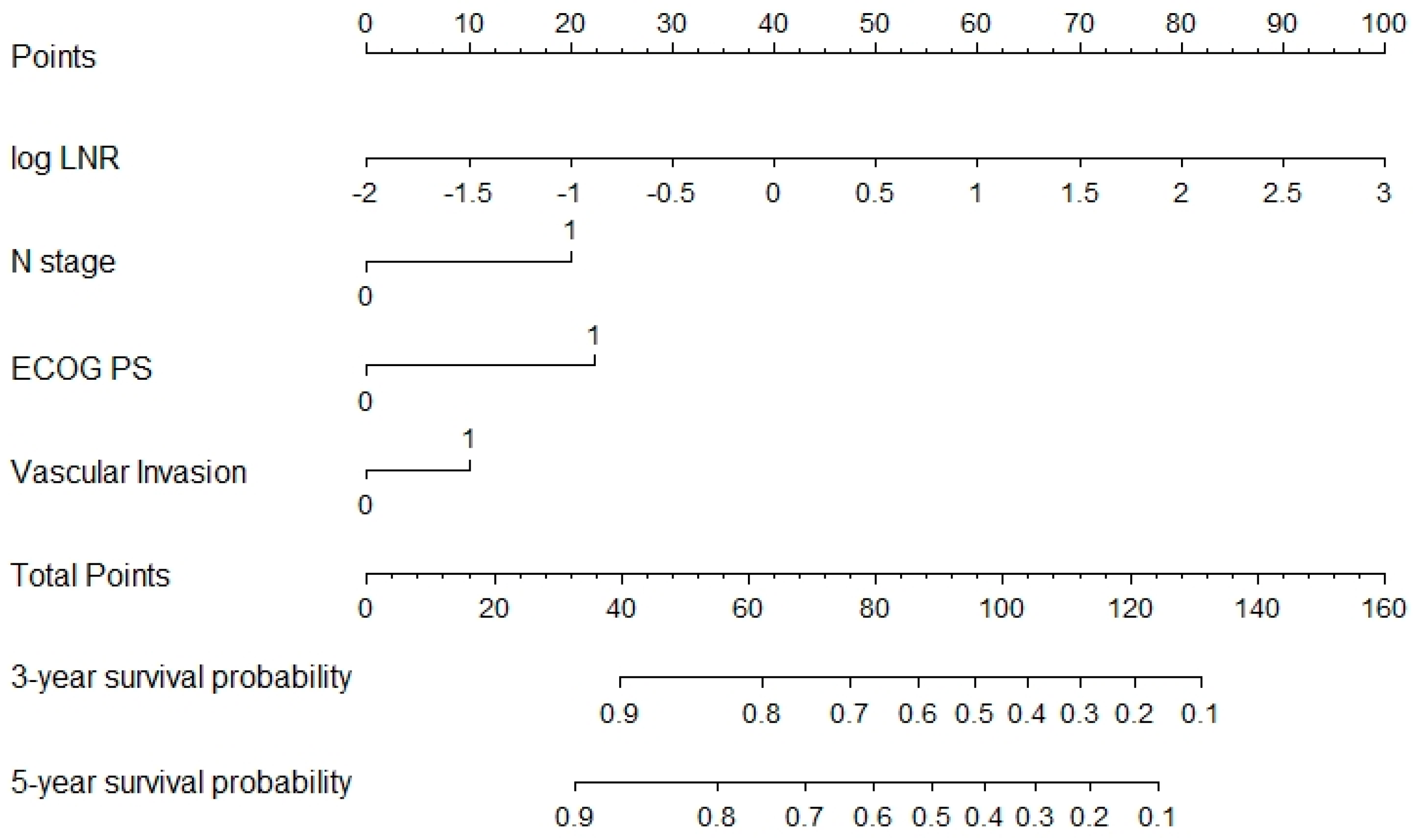
3. Results
3.1. Patient Population and Prognostic Factors
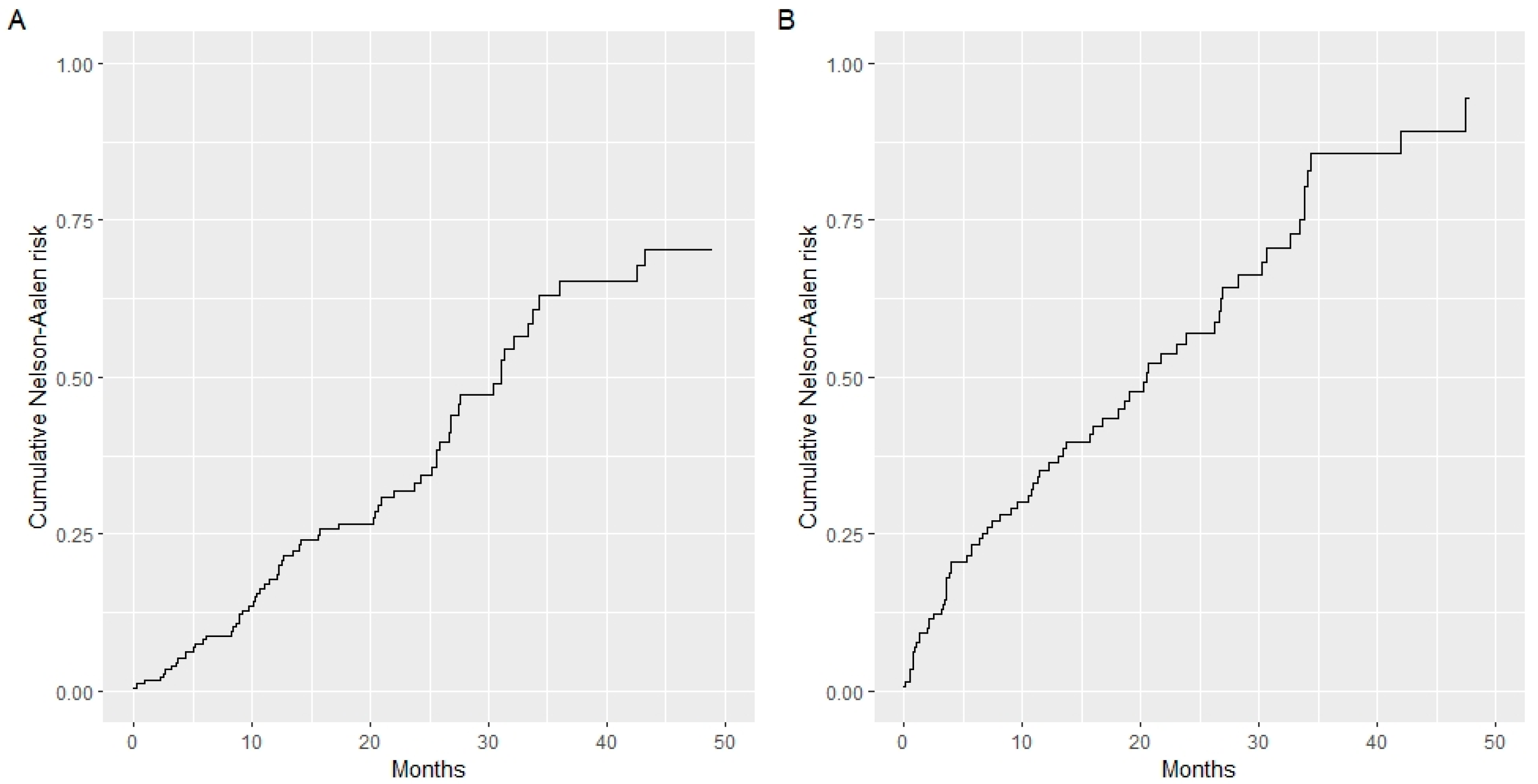
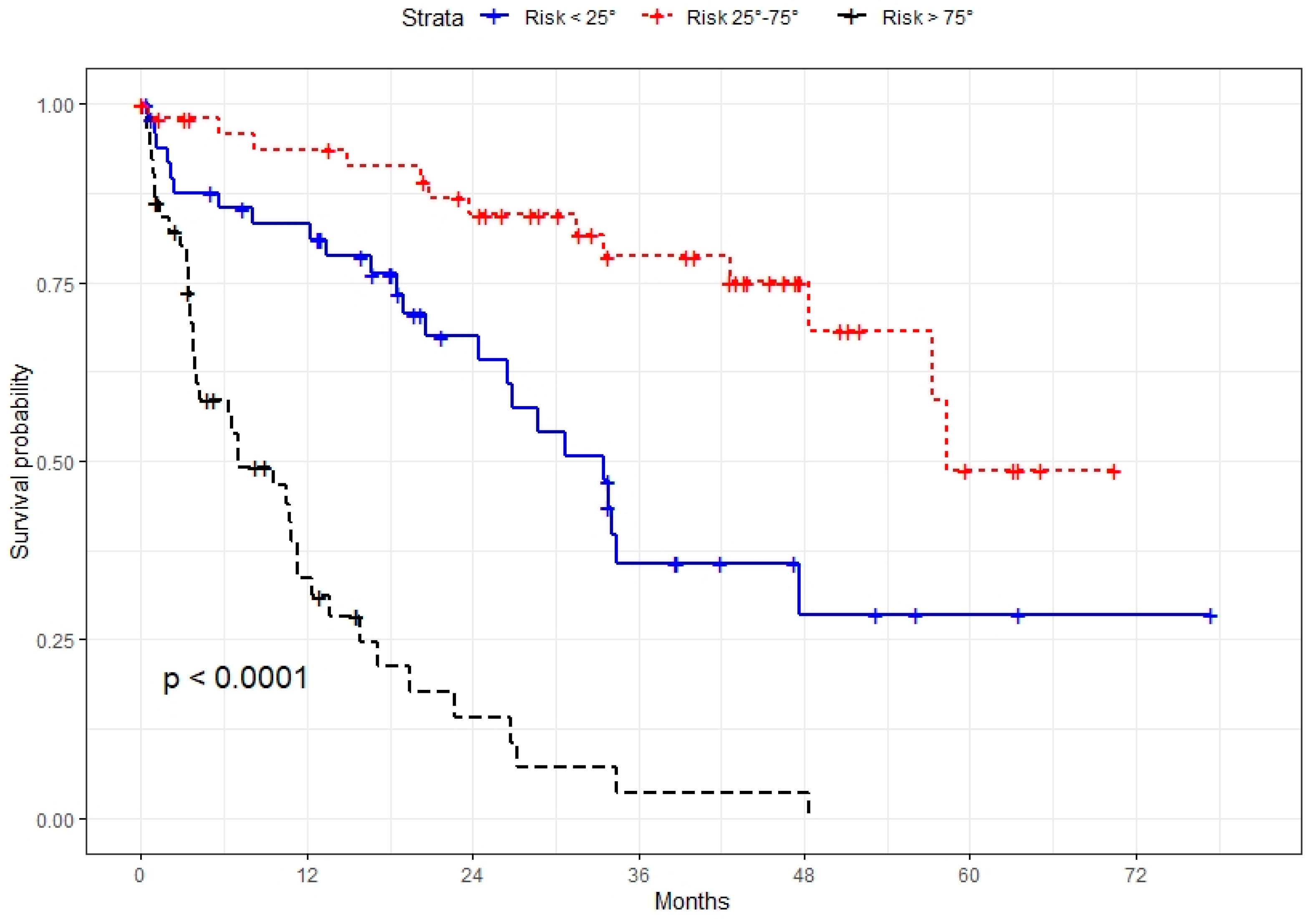
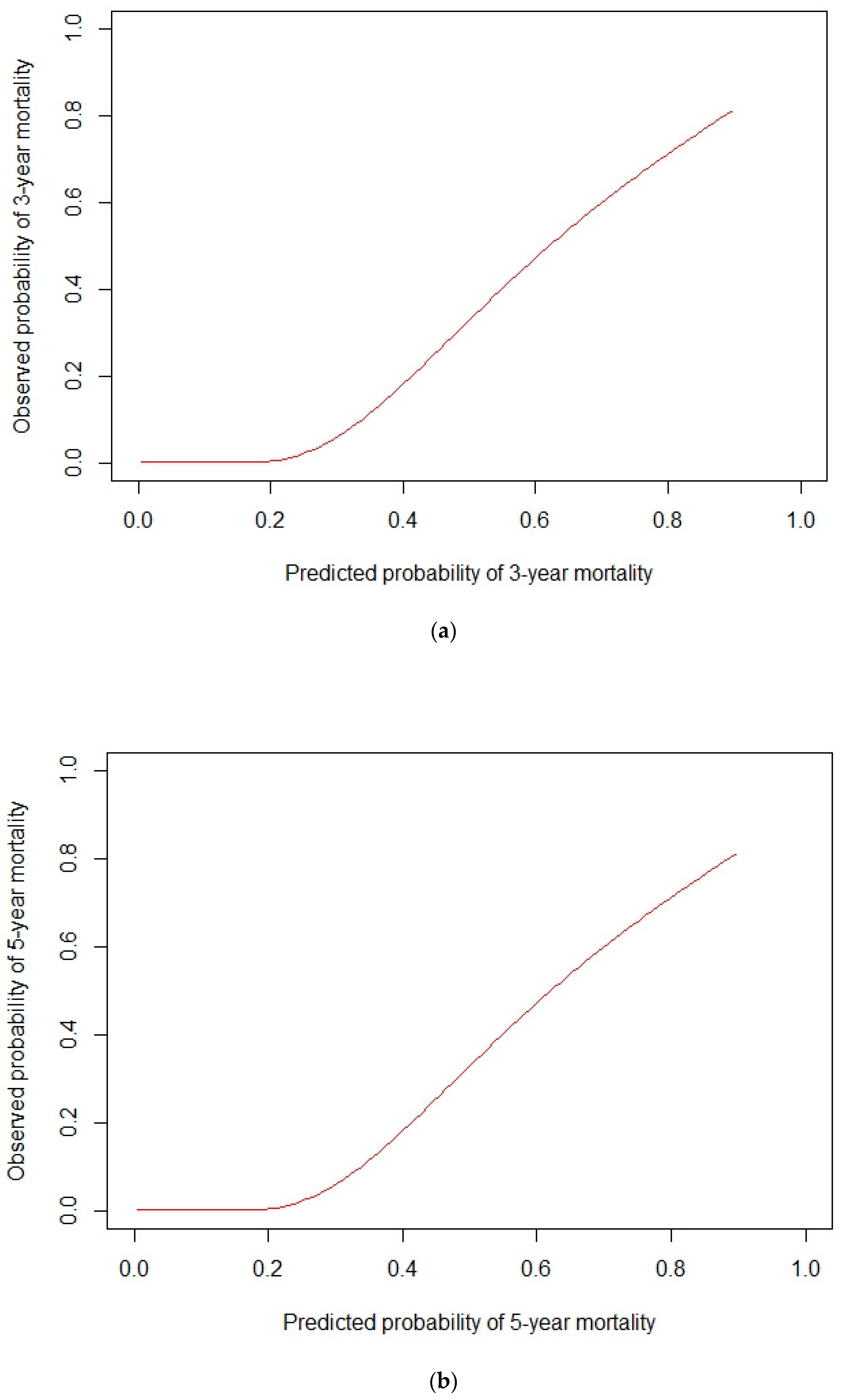
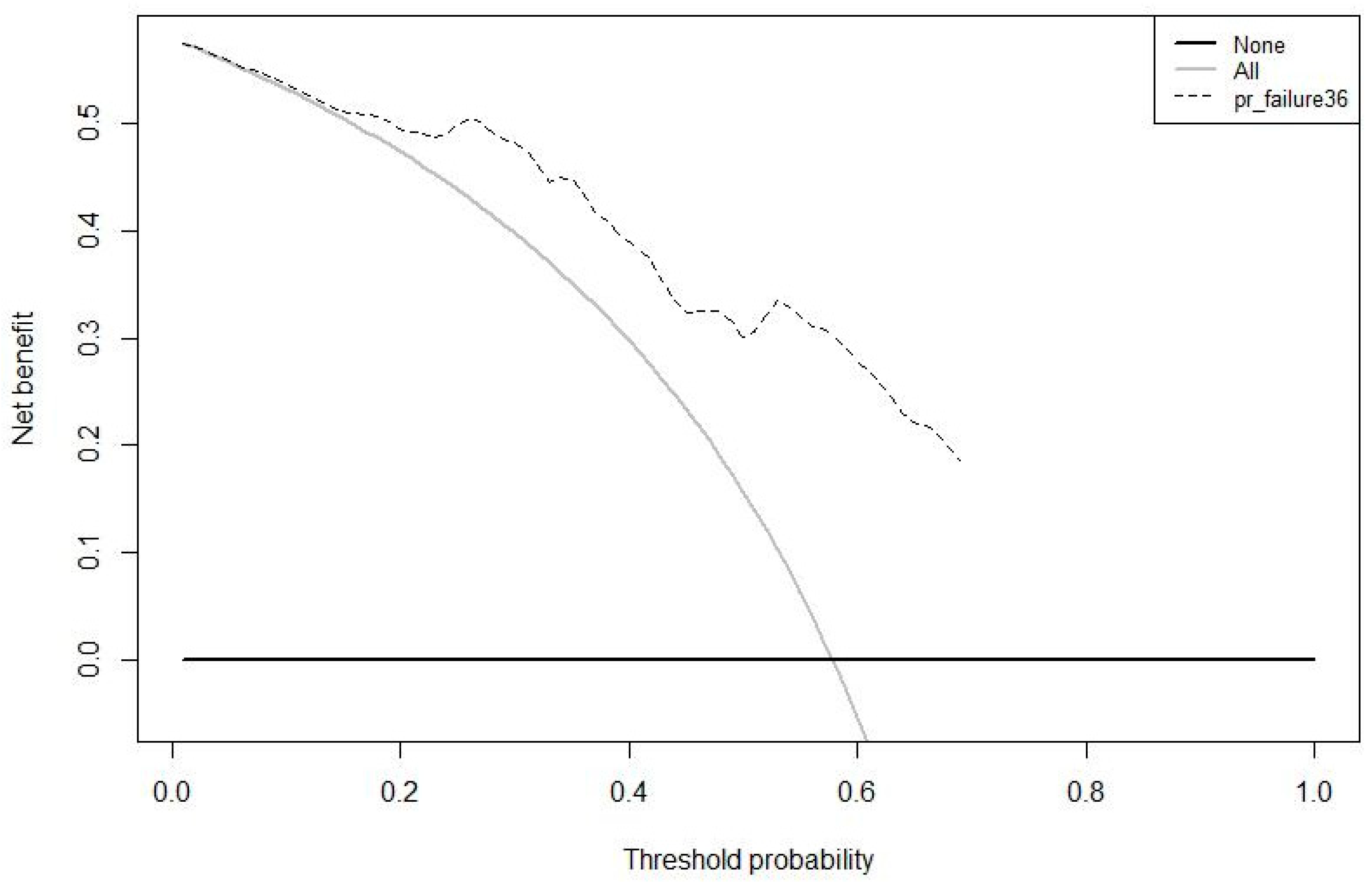
3.2. Prognostic Model Development and Validation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphate |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| CA19.9 | Carbohydrate antigen 19-9 |
| CEA | Carcinoembryonic antigen |
| GEA | Gastric and gastroesophageal junction adenocarcinoma |
| LDH | Lactate dehydrogenase |
| NLR | Neutrophil to lymphocyte ratio |
| OS | Overall Survival |
| PNI | Prognostic nutritional index |
| SII | Systemic inflammatory index |
| TC | Training cohort |
| VC | Validation cohort |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Anderson, W.F.; Camargo, M.C.; Fraumeni, J.F., Jr.; Correa, P.; Rosenberg, P.S.; Rabkin, C.S. Agespecific trends in incidence of noncardia gastric cancer in US adults. JAMA 2010, 303, 1723–1728. [Google Scholar] [CrossRef] [PubMed]
- Camargo, M.C.; Anderson, W.F.; King, J.B.; Correa, P.; Thomas, C.C.; Rosenberg, P.; Eheman, C.R.; Rabkin, C.S. Divergent trends for gastric cancer incidence by anatomical subsite in US adults. Gut 2011, 60, 1644–1649. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Verheij, M.; Allum, W.; Cunningham, D.; Cervantes, A.; Arnold, D. Gastric cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27 (Suppl. S5), v38–v49. [Google Scholar] [CrossRef]
- Ajani, J.A.; Lee, J.; Sano, T.; Janjigian, Y.Y.; Fan, D.; Song, S. Gastric adenocarcinoma. Nat. Rev. Dis. Primers 2017, 3, 17036. [Google Scholar] [CrossRef] [PubMed]
- Al-Batran, S.E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.-G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastrooesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Salati, M.; Spallanzani, A. New Horizons for Personalised Treatment in Gastroesophageal Cancer. J. Clin. Med. 2022, 11, 311. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.; Fassan, M.; Cunningham, D.; Allum, W.; Okines, A.; Lampis, A.; Hahne, J.C.; Rugge, M.; Peckitt, C.; Nankivell, M.; et al. Effect of pathologic tumor response and nodal status on survival in the medical research council adjuvant gastric infusional chemotherapy trial. J. Clin. Oncol. 2016, 34, 2721–2727. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.H.; Yang, H.K.; Kim, H.; Kim, W.H.; Kim, Y.W.; Kook, M.C.; Park, Y.-K.; Kim, H.-H.; Lee, H.S.; Lee, K.H.; et al. Predictive test for chemotherapy response in resectable gastric cancer: A multi-cohort, retrospective analysis. Lancet Oncol. 2018, 19, 629–638. [Google Scholar] [CrossRef]
- Smyth, E.C.; Nyamundanda, G.; Cunningham, D.; Fontana, E.; Ragulan, C.; Tan, I.B.; Lin, S.; Wotherspoon, A.; Nankivell, M.; Fassan, M.; et al. A sevenGene Signature assay improves prognostic risk stratification of perioperative chemotherapy treated gastroesophageal cancer patients from the MAGIC trial. Ann. Oncol. 2018, 29, 2356–2362. [Google Scholar] [CrossRef] [PubMed]
- Pietrantonio, F.; Miceli, R.; Raimondi, A.; Kim, Y.W.; Kang, W.K.; Langley, R.E.; Choi, Y.Y.; Kim, K.-M.; Nankivell, M.G.; Morano, F.; et al. Individual Patient Data Meta-Analysis of the Value of Microsatellite Instability as a Biomarker in Gastric Cancer. J. Clin. Oncol. 2019, 37, 3392–3400. [Google Scholar] [CrossRef] [PubMed]
- Raimondi, A.; Palermo, F.; Prisciandaro, M.; Aglietta, M.; Antonuzzo, L.; Aprile, G.; Berardi, R.; Cardellino, G.G.; de Manzoni, G.; de Vita, F.; et al. TremelImumab and Durvalumab Combination for the Non-OperatIve Man-agement (NOM) of Microsatellite InstabiliTY (MSI)-High Resectable Gastric or Gastroesophageal Junction Cancer: The Multicentre, Single-Arm, Multi-Cohort, Phase II INFINITY Study. Cancers 2021, 13, 2839. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, V.P.; Gonen, M.; Smith, J.J.; DeMatteo, R.P. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015, 16, e173–e180. [Google Scholar] [CrossRef]
- Iasonos, A.; Schrag, D.; Raj, G.V.; Panageas, K.S. How To Build and Interpret a Nomogram for Cancer Prognosis. J. Clin. Oncol. 2008, 26, 1364–1370. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Elkin, E.B. Decision curve analysis: A novel method for evaluating prediction models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Bando, E.; Ji, X.; Kattan, M.; Seo, H.S.; Song, K.Y.; Park, C.H.; Bencivenga, M.; De Manzoni, G.; Terashima, M. Development and validation of a pretreatment nomogram to predict overall survival in gastric cancer. Cancer Med. 2020, 9, 5708–5728. [Google Scholar] [CrossRef] [PubMed]
- Bando, E.; Ji, X.; Kattan, M.; Bencivenga, M.; De Manzoni, G.; Terashima, M. Development and validation of a pretreatment nomogram for disease specific mortality in gastric cancer- A competing risk analysis. Cancer Med. 2021, 10, 7561–7571. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, R.; Inagawa, S.; Sano, N.; Tadano, S.; Adachi, S.; Yamamoto, M. The neutrophil-to-lymphocyte ratio (NLR) predicts short-term and long-term outcomes in gastric cancer patients. Eur. J. Surg. Oncol. 2018, 44, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Hirahara, T.; Arigami, T.; Yanagita, S.; Matsushita, D.; Uchikado, Y.; Kita, Y.; Mori, S.; Sasaki, K.; Omoto, I.; Kurahara, H.; et al. Combined neutrophil-lymphocyte ratio and platelet-lymphocyte ratio predicts chemotherapy response and prognosis in patients with advanced gastric cancer. BMC Cancer 2019, 19, 672. [Google Scholar] [CrossRef] [PubMed]

| Training (n = 185) | Validation (n = 151) | p Value | |
|---|---|---|---|
| Age Mean (SD) | 70.7 (11.3) | 73.8 (9.7) | 0.009 |
| Gender | 0.188 | ||
| Female | 83 (44.9%) | 57 (37.7%) | |
| Male | 102 (55.1%) | 94 (62.3%) | |
| ECOG PS | 0.466 | ||
| 0 | 87 (47.0%) | 65 (43.0%) | |
| ≥1 | 98 (53.0%) | 86 (57.0%) | |
| Sites | 0.008 | ||
| Anastomosis | 1 (0.5%) | 1 (0.7%) | |
| Angulus | 0 (0.0%) | 8 (5.3%) | |
| Antrum | 65 (35.1%) | 53 (35.3%) | |
| Cardias | 25 (13.5%) | 18 (12.0%) | |
| Body | 78 (42.2%) | 56 (37.3%) | |
| Fundus | 12 (6.5%) | 5 (3.3%) | |
| Linitis | 2 (1.1%) | 0 (0.0%) | |
| Stump | 1 (0.5%) | 1 (0.7%) | |
| Pylorus | 1 (0.5%) | 8 (5.3%) | |
| CEA Mean (SD) | 5.6 (19.3) | 7.1 (20.1) | 0.191 |
| CA 19-9 Mean (SD) | 46.6 (182.4) | 127.8 (784.6) | 0.270 |
| T stage | 0.825 | ||
| 0 | 0 (0.0%) | 0 (0.0%) | |
| 1 | 33 (17.8%) | 18 (11.9%) | |
| 2 | 40 (21.6%) | 20 (13.2%) | |
| 3 | 64 (34.6%) | 61 (40.4%) | |
| 4 | 44 (23.8%) | 50 (33.1%) | |
| IS | 4 (2.2%) | 2 (1.3%) | |
| N stage | 0.030 | ||
| 0 | 67 (36.2%) | 38 (25.2%) | |
| 1 | 118 (63.8%) | 113 (74.8%) | |
| log NLR | 0.044 | ||
| Mean (SD) | 0.9 (0.6) | 1.1 (0.8) | |
| Vascular Invasion | 0.022 | ||
| 0 | 101 (54.6%) | 101 (66.9%) | |
| 1 | 84 (45.4%) | 50 (33.1%) |
| Variable | All | HR (Univariable) | HR (Multivariable) | |
|---|---|---|---|---|
| Age | Mean (SD) | 72.2 (10.6) | 1.03 (1.01–1.04, p < 0.001) | 1.01 (0.99–1.04, p = 0.163) |
| Gender | Female | 196 (42.5) | - | - |
| Male | 265 (57.5) | 0.97 (0.74–1.28, p = 0.847) | - | |
| BMI | Mean (SD) | 25.6 (5.0) | 1.00 (0.96–1.04, p = 0.978) | - |
| ECOG PS | 0 | 195 (43.5) | - | - |
| ≥1 | 253 (56.5) | 2.21 (1.63–3.01, p < 0.001) | 1.97 (1.24–3.11, p = 0.004) | |
| Sites | anastomosis | 3 (0.7) | - | - |
| angulus | 16 (3.5) | 0.50 (0.21–1.23, p = 0.133) | - | |
| Antrum | 163 (35.4) | 0.80 (0.60–1.07, p = 0.134) | - | |
| Cardias | 55 (12.0) | 1.21 (0.82–1.78, p = 0.339) | - | |
| Body | 180 (39.1) | 0.76 (0.57–1.01, p = 0.057) | - | |
| Fundus | 22 (4.8) | 0.66 (0.35–1.25, p = 0.203) | - | |
| Linitis | 2 (0.4) | 0.00 (0.00–Inf, p = 0.993) | - | |
| Stump | 6 (1.3) | 0.00 (0.00–Inf, p = 0.989) | - | |
| Multifocal | 1 (0.2) | 34.57 (4.42–270.60, p = 0.001) | - | |
| Pylorus | 12 (2.6) | 0.97 (0.43–2.19, p = 0.944) | - | |
| CEA | Mean (SD) | 7.1 (25.7) | 1.01 (1.00–1.01, p = 0.055) | 1.00 (0.97–1.02, p = 0.783) |
| CA 19-9 | Mean (SD) | 72.1 (485.5) | 1.00 (1.00–1.00, p = 0.018) | 1.00 (1.00–1.00, p = 0.288) |
| T stage | IS | 8 (1.7) | NA (NA–NA, p = NA) | - |
| 1 | 81 (17.6) | 2.69 (0.36–20.04, p = 0.335) | - | |
| 2 | 108 (23.4) | 4.61 (0.64–33.45, p = 0.130) | - | |
| 3 | 158 (34.3) | 6.81 (0.95–49.09, p = 0.057) | - | |
| 4 | 106 (23.0) | 8.18 (1.13–59.18, p = 0.037) | - | |
| N stage | 0 | 163 (35.7) | - | - |
| ≥1 | 293 (64.3) | 1.93 (1.39–2.66, p < 0.001) | 1.99 (1.05–3.76, p = 0.034) | |
| log NLR | Mean (SD) | 1.0 (0.7) | 1.92 (1.55–2.37, p < 0.001) | 1.84 (1.30–2.60, p = 0.001) |
| Vascular Invasion | Absent | 231 (60.5) | - | - |
| Present | 151 (39.5) | 3.04 (2.21–4.18, p < 0.001) | 2.25 (1.41–3.60, p = 0.001) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salati, M.; De Ruvo, N.; Giglio, M.C.; Sorrentino, L.; Esposito, G.; Fenocchi, S.; Cucciarrè, G.; Serra, F.; Rossi, E.G.; Vittimberga, G.; et al. Development and Multicenter Validation of a Novel Immune-Inflammation-Based Nomogram to Predict Survival in Western Resectable Gastric and Gastroesophageal Junction Adenocarcinoma (GEA): The NOMOGAST. J. Clin. Med. 2022, 11, 5439. https://doi.org/10.3390/jcm11185439
Salati M, De Ruvo N, Giglio MC, Sorrentino L, Esposito G, Fenocchi S, Cucciarrè G, Serra F, Rossi EG, Vittimberga G, et al. Development and Multicenter Validation of a Novel Immune-Inflammation-Based Nomogram to Predict Survival in Western Resectable Gastric and Gastroesophageal Junction Adenocarcinoma (GEA): The NOMOGAST. Journal of Clinical Medicine. 2022; 11(18):5439. https://doi.org/10.3390/jcm11185439
Chicago/Turabian StyleSalati, Massimiliano, Nicola De Ruvo, Mariano Cesare Giglio, Lorena Sorrentino, Giuseppe Esposito, Sara Fenocchi, Giovanni Cucciarrè, Francesco Serra, Elena Giulia Rossi, Giovanni Vittimberga, and et al. 2022. "Development and Multicenter Validation of a Novel Immune-Inflammation-Based Nomogram to Predict Survival in Western Resectable Gastric and Gastroesophageal Junction Adenocarcinoma (GEA): The NOMOGAST" Journal of Clinical Medicine 11, no. 18: 5439. https://doi.org/10.3390/jcm11185439
APA StyleSalati, M., De Ruvo, N., Giglio, M. C., Sorrentino, L., Esposito, G., Fenocchi, S., Cucciarrè, G., Serra, F., Rossi, E. G., Vittimberga, G., Radi, G., Solaini, L., Morgagni, P., Grizzi, G., Ratti, M., Gelsomino, F., Spallanzani, A., Ghidini, M., Ercolani, G., ... Gelmini, R. (2022). Development and Multicenter Validation of a Novel Immune-Inflammation-Based Nomogram to Predict Survival in Western Resectable Gastric and Gastroesophageal Junction Adenocarcinoma (GEA): The NOMOGAST. Journal of Clinical Medicine, 11(18), 5439. https://doi.org/10.3390/jcm11185439






