Mechanisms Leading to Gut Dysbiosis in COVID-19: Current Evidence and Uncertainties Based on Adverse Outcome Pathways
Abstract
:1. Introduction
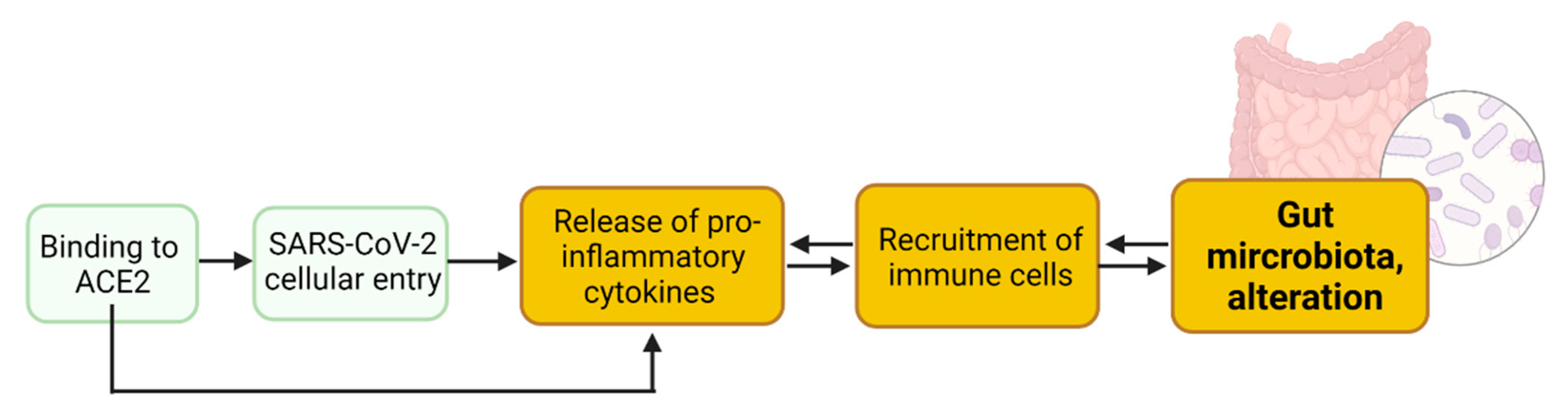
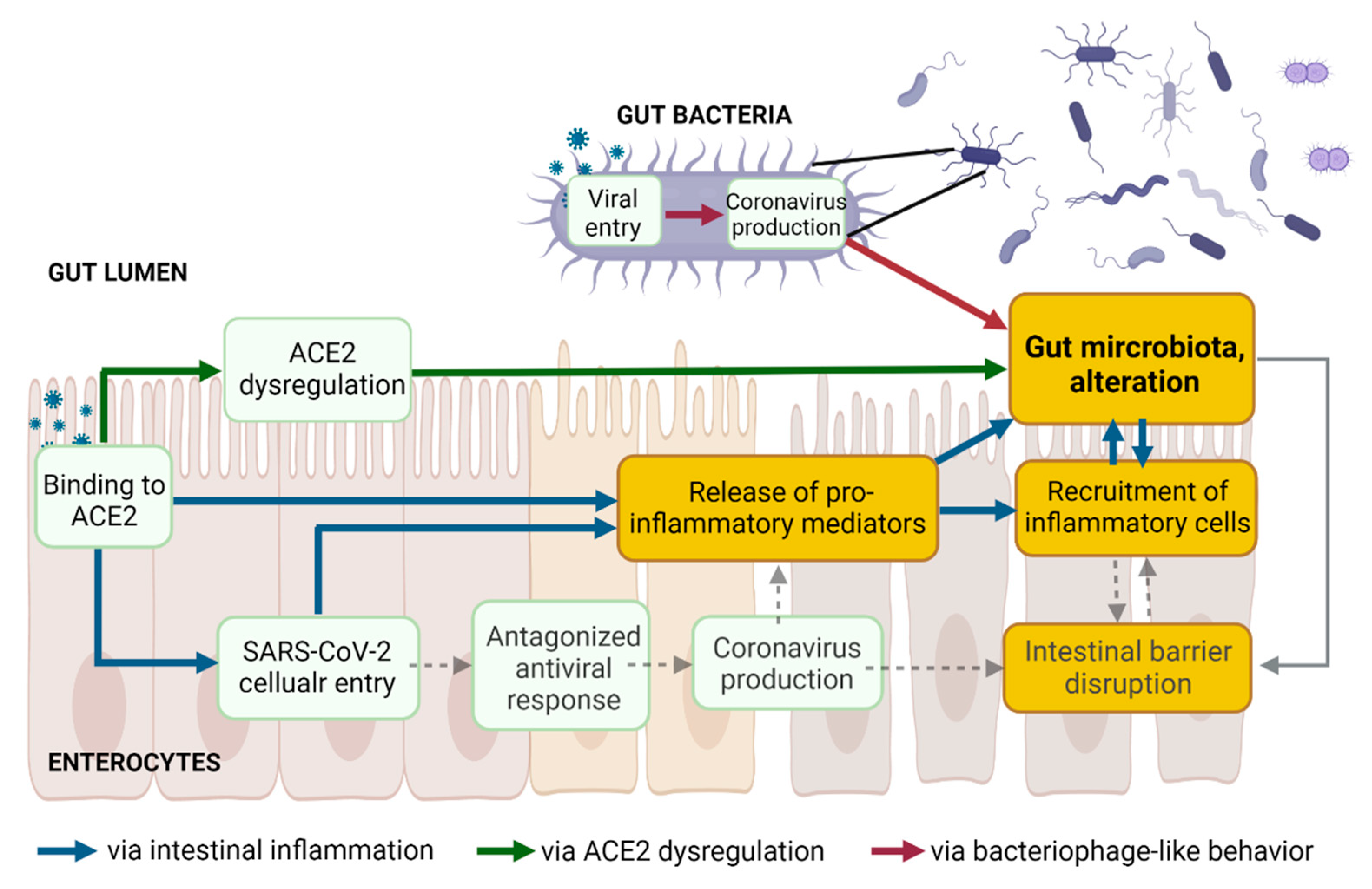
2. Enteric SARS-CoV-2 Presence Leads to Intestinal Inflammation Altering Gut Microbiota
2.1. SARS-CoV-2 Entry into Enterocytes Leads to Intestinal Inflammation
2.2. Binding to Enteric ACE2 Leads to Intestinal Inflammation
2.3. Intestinal Inflammation Leads to Alteration of Gut Microbiota
2.4. Potential Implication for Disease Management
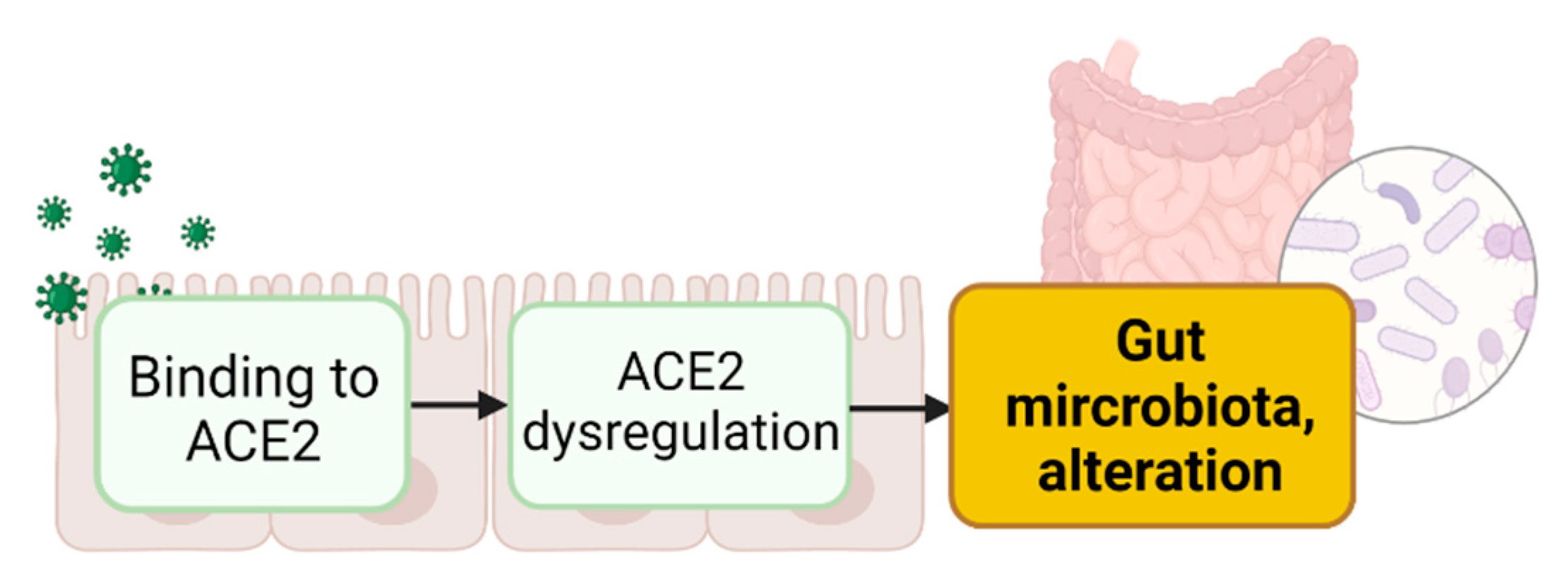
3. Intestinal ACE2 Dysregulation Inducing Gut Dysbiosis
3.1. Binding of S Proteins to Enteric ACE2 Induces Intestinal ACE2 Dysregulation
3.2. Enteric ACE2 Dysregulation Leads to Gut Microbiota Alteration
3.3. Potential Implications for Disease Management
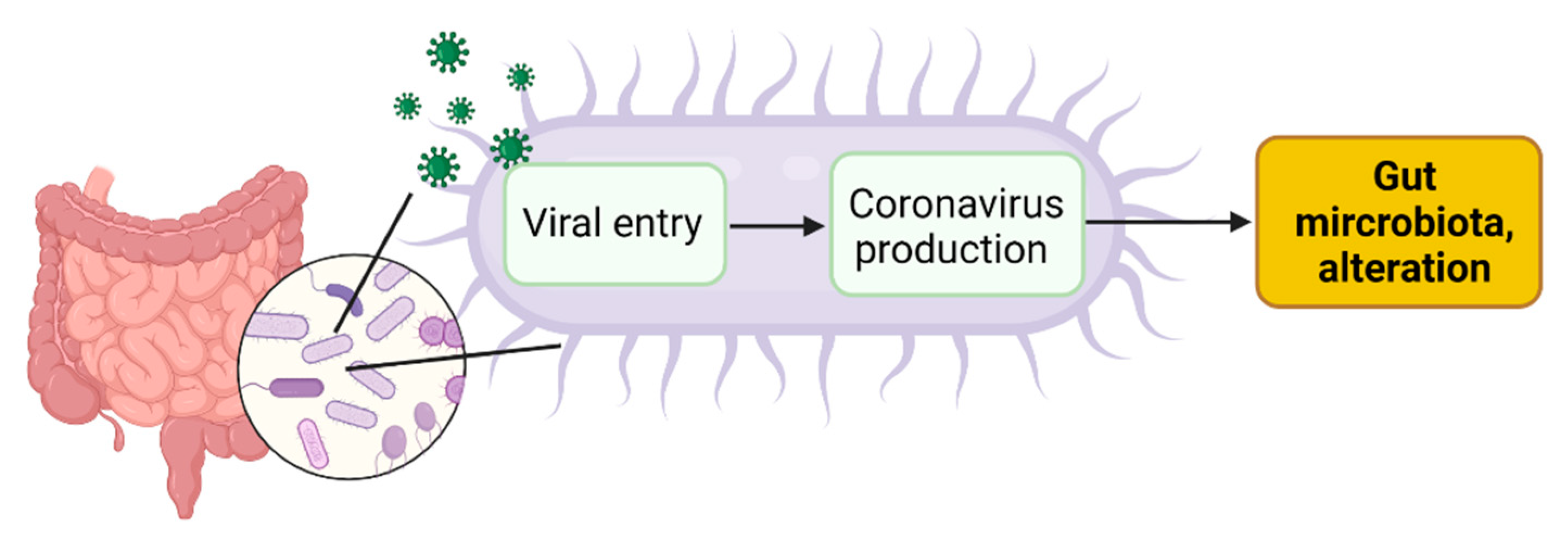
4. SARS-CoV-2 Infection of Microbial Bacteria Driving Gut Dysbiosis
4.1. Viral Entry in Gut Bacteria Leads to Coronavirus Production in Bacteria
4.2. Coronavirus Production in Gut Bacteria Leads to Alteration of Gut Microbiota
4.3. Potential Implications for Disease Management
5. Central Role of Gut Microbiota in COVID-19 and Potential Modulation
5.1. Gut Microbiota and Intestinal Barrier Integrity in COVID-19
5.2. Central Role of the Gut (Microbiota) in COVID-19 and Long COVID
5.3. Potential Modulation of Gut Microbiota to Mitigate COVID-19
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Disclaimer
References
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Chidambaram, V.; Tun, N.L.; Haque, W.Z.; Majella, M.G.; Sivakumar, R.K.; Kumar, A.; Hsu, A.T.-W.; Ishak, I.A.; Nur, A.A.; Ayeh, S.K.; et al. Factors associated with disease severity and mortality among patients with COVID-19: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0241541. [Google Scholar] [CrossRef]
- Mudatsir, M.; Fajar, J.K.; Wulandari, L.; Soegiarto, G.; Ilmawan, M.; Purnamasari, Y.; Mahdi, B.A.; Jayanto, G.D.; Suhendra, S.; Setianingsih, Y.A.; et al. Predictors of COVID-19 severity: A systematic review and meta-analysis. F1000Research 2020, 9, 1107. [Google Scholar] [CrossRef] [PubMed]
- Nishiga, M.; Wang, D.W.; Han, Y.; Lewis, D.B.; Wu, J.C. COVID-19 and cardiovascular disease: From basic mechanisms to clinical perspectives. Nat. Rev. Cardiol. 2020, 17, 543–558. [Google Scholar] [CrossRef] [PubMed]
- Clerbaux, L.-A.; Albertini, M.C.; Amigó, N.; Beronius, A.; Bezemer, G.F.G.; Coecke, S.; Daskalopoulos, E.P.; del Giudice, G.; Greco, D.; Grenga, L.; et al. Factors Modulating COVID-19: A Mechanistic Understanding Based on the Adverse Outcome Pathway Framework. J. Clin. Med. 2022, 11, 4464. [Google Scholar] [CrossRef] [PubMed]
- Ragonnaud, E.; Biragyn, A. Gut microbiota as the key controllers of “healthy” aging of elderly people. Immun. Ageing 2021, 18, 2. [Google Scholar] [CrossRef] [PubMed]
- Bosco, N.; Noti, M. The aging gut microbiome and its impact on host immunity. Genes Immun. 2021, 22, 289–303. [Google Scholar] [CrossRef]
- Vodnar, D.-C.; Mitrea, L.; Teleky, B.-E.; Szabo, K.; Călinoiu, L.-F.; Nemeş, S.-A.; Martău, G.-A. Coronavirus Disease (COVID-19) Caused by (SARS-CoV-2) Infections: A Real Challenge for Human Gut Microbiota. Front. Cell. Infect. Microbiol. 2020, 10, 575559. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with Coronavirus Disease 2019 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef]
- Cheung, C.C.L.; Goh, D.; Lim, X.; Tien, T.Z.; Lim, J.C.T.; Lee, J.N.; Tan, B.; Tay, Z.E.A.; Wan, W.Y.; Chen, E.X.; et al. Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19. Gut 2022, 71, 226–229. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e948. [Google Scholar] [CrossRef]
- Petrillo, M.; Brogna, C.; Cristoni, S.; Querci, M.; Piazza, O.; Eede, G.V.D. Increase of SARS-CoV-2 RNA load in faecal samples prompts for rethinking of SARS-CoV-2 biology and COVID-19 epidemiology. F1000Research 2021, 10, 370. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Harty, S.; Moeller, A.H.; Klein, S.L.; Erdman, S.E.; Friston, K.J.; Carmody, R.N. The gut microbiome as a biomarker of differential susceptibility to SARS-CoV-2. Trends Mol. Med. 2021, 27, 1115–1134. [Google Scholar] [CrossRef]
- Schult, D.; Reitmeier, S.; Koyumdzhieva, P.; Lahmer, T.; Middelhoff, M.; Erber, J.; Schneider, J.; Kager, J.; Frolova, M.; Horstmann, J.; et al. Gut bacterial dysbiosis and instability is associated with the onset of complications and mortality in COVID-19. Gut Microbes 2022, 14, 2031840. [Google Scholar] [CrossRef]
- Spor, A.; Koren, O.; Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat. Rev. Microbiol. 2011, 9, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Li, S.; Gan, R.-Y.; Zhou, T.; Xu, D.-P.; Li, H.-B. Impacts of Gut Bacteria on Human Health and Diseases. Int. J. Mol. Sci. 2015, 16, 7493–7519. [Google Scholar] [CrossRef]
- Sartor, R.B. Mechanisms of Disease: Pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pr. Gastroenterol. Hepatol. 2006, 3, 390–407. [Google Scholar] [CrossRef]
- Dang, A.T.; Marsland, B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019, 12, 843–850. [Google Scholar] [CrossRef]
- Tripathi, A.; Debelius, J.; Brenner, D.A.; Karin, M.; Loomba, R.; Schnabl, B.; Knight, R. The gut–liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 397–411. [Google Scholar] [CrossRef]
- Manzoor, R.; Ahmed, W.; Afify, N.; Memon, M.; Yasin, M.; Memon, H.; Rustom, M.; Al Akeel, M.; Alhajri, N. Trust Your Gut: The Association of Gut Microbiota and Liver Disease. Microorganisms 2022, 10, 1045. [Google Scholar] [CrossRef] [PubMed]
- Mitrea, L.; Nemes, S.A.; Szabo, K.; Teleky, B.E.; Vodnar, D.C. Guts Imbalance Imbalances the Brain: A Review of Gut Microbiota Association with Neurological and Psychiatric Disorders. Front. Med. 2022, 9, 813204. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.; Baindara, P.; Mandal, S.M. Molecular pathogenesis of secondary bacterial infection associated to viral infections including SARS-CoV-2. J. Infect. Public Health 2020, 13, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hall, S.; Vitetta, L. Altered gut microbial metabolites could mediate the effects of risk factors in COVID-19. Rev. Med. Virol. 2021, 31, 1–13. [Google Scholar] [CrossRef]
- Venzon, M.; Bernard-Raichon, L.; Klein, J.; Axelrad, J.; Hussey, G.; Sullivan, A.; Casanovas-Massana, A.; Noval, M.; Valero-Jimenez, A.; Gago, J.; et al. Gut microbiome dysbiosis during COVID-19 is associated with increased risk for bacteremia and microbial translocation. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Sencio, V.; Machelart, A.; Robil, C.; Benech, N.; Hoffmann, E.; Galbert, C.; Deryuter, L.; Heumel, S.; Hantute-Ghesquier, A.; Flourens, A.; et al. Alteration of the gut microbiota following SARS-CoV-2 infection correlates with disease severity in hamsters. Gut Microbes 2022, 14, 2018900. [Google Scholar] [CrossRef]
- Sokol, H.; Contreras, V.; Maisonnasse, P.; Desmons, A.; Delache, B.; Sencio, V.; Machelart, A.; Brisebarre, A.; Humbert, L.; Deryuter, L.; et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes 2021, 13, 1893113. [Google Scholar] [CrossRef]
- Nymark, P.; Sachana, M.; Leite, S.B.; Sund, J.; Krebs, C.E.; Sullivan, K.; Edwards, S.; Viviani, L.; Willett, C.; Landesmann, B.; et al. Systematic Organization of COVID-19 Data Supported by the Adverse Outcome Pathway Framework. Front. Public Health 2021, 9, 638605. [Google Scholar] [CrossRef]
- Wittwehr, C.; Amorim, M.J.; Clerbaux, L.A.; Krebs, C.; Landesmann, B.; Macmillan, D.S.; Nymark, P.; Ram, R.; Garcia-Reyero, N.; Sachana, M.; et al. Understanding COVID-19 through adverse outcome pathways–2nd CIAO AOP Design Workshop. ALTEX 2021, 38, 351–357. [Google Scholar] [CrossRef]
- Clerbaux, L.-A.; Amigó, N.; Amorim, M.J.; Bal-Price, A.; Leite, S.B.; Beronius, A.; Bezemer, G.F.G.; Bostroem, A.-C.; Carusi, A.; Coecke, S.; et al. COVID-19 through Adverse Outcome Pathways: Building networks to better understand the disease—3rd CIAO AOP Design Workshop. ALTEX 2022, 39, 322–335. [Google Scholar] [CrossRef]
- CIAO. Modelling the Pathogenesis of COVID-19 Using the Adverse Outcome Pathway Framework. Available online: www.ciao-covid.net (accessed on 29 June 2022).
- OECD. Users’ Handbook supplement to the Guidance Document for developing and assessing Adverse Outcome Pathways. In OECD Series on Adverse Outcome Pathways; OECD Publishing: Paris, France, 2018. [Google Scholar] [CrossRef]
- Draskau, M.K.; Spiller, C.M.; Boberg, J.; Bowles, J.; Svingen, T.; Kam, M. Developmental biology meets toxicology: Contributing reproductive mechanisms to build adverse outcome pathways. Mol. Hum. Reprod. 2020, 26, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Siwicki, A.K.; Terech-Majewska, E.; Grudniewska, J.; Malaczewska, J.; Kazun, K.; Lepa, A. Influence of deltamethrin on nonspecific cellular and humoral defense mechanisms in rainbow trout (Oncorhynchus mykiss). Environ. Toxicol. Chem. 2010, 29, 489–491. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, D.L.; Crump, D.; Garcia-Reyero, N.; Hecker, M.; Hutchinson, T.H.; Lalone, C.A.; Landesmann, B.; Lettieri, T.; Munn, S.; Nepelska, M.; et al. Adverse Outcome Pathway (AOP) Development I: Strategies and Principles. Toxicol. Sci. 2014, 142, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Villeneuve, D.L.; Crump, D.; Garcia-Reyero, N.; Hecker, M.; Hutchinson, T.H.; Lalone, C.A.; Landesmann, B.; Lettieri, T.; Munn, S.; Nepelska, M.; et al. Adverse Outcome Pathway Development II: Best Practices. Toxicol. Sci. 2014, 142, 321–330. [Google Scholar] [CrossRef]
- Svingen, T.; Villeneuve, D.L.; Knapen, D.; Panagiotou, E.M.; Draskau, M.K.; Damdimopoulou, P.; O’Brien, J.M. A Pragmatic Approach to Adverse Outcome Pathway Development and Evaluation. Toxicol. Sci. 2021, 184, 183–190. [Google Scholar] [CrossRef]
- Clerbaux, L.A.; Muñoz, A.; Soares, H.; Petrillo, M.; Albertini, M.C.; Lanthier, N.; Grenga, L.; Amorim, M.J. Gut as an alternative entry route for SARS-CoV-2: Current evidence and uncertainties of productive enteric infection in COVID-19. J. Clin. Med. 2022. submitted. [Google Scholar]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef]
- Suárez-Fariñas, M.; Tokuyama, M.; Wei, G.; Huang, R.; Livanos, A.; Jha, D.; Levescot, A.; Irizar, H.; Kosoy, R.; Cording, S.; et al. Intestinal Inflammation Modulates the Expression of ACE2 and TMPRSS2 and Potentially Overlaps with the Pathogenesis of SARS-CoV-2–related Disease. Gastroenterology 2021, 160, 287–301.e20. [Google Scholar] [CrossRef]
- Guo, M.; Tao, W.; Flavell, R.A.; Zhu, S. Potential intestinal infection and faecal–oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 269–283. [Google Scholar] [CrossRef]
- Lee, J.J.; Kopetz, S.; Vilar, E.; Shen, J.P.; Chen, K.; Maitra, A. Relative Abundance of SARS-CoV-2 Entry Genes in the Enterocytes of the Lower Gastrointestinal Tract. Genes 2020, 11, 645. [Google Scholar] [CrossRef]
- Beumer, J.; Geurts, M.H.; Lamers, M.M.; Puschhof, J.; Zhang, J.; van der Vaart, J.; Mykytyn, A.Z.; Breugem, T.I.; Riesebosch, S.; Schipper, D.; et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 2021, 12, 5498. [Google Scholar] [CrossRef] [PubMed]
- Stanifer, M.L.; Kee, C.; Cortese, M.; Zumaran, C.M.; Triana, S.; Mukenhirn, M.; Kraeusslich, H.-G.; Alexandrov, T.; Bartenschlager, R.; Boulant, S. Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells. Cell Rep. 2020, 32, 107863. [Google Scholar] [CrossRef] [PubMed]
- Merad, M.; Blish, C.A.; Sallusto, F.; Iwasaki, A. The immunology and immunopathology of COVID-19. Science 2022, 375, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.G.; van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q.; et al. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Sellon, R.K.; Tonkonogy, S.; Schultz, M.; Dieleman, L.A.; Grenther, W.; Balish, E.; Rennick, D.M.; Sartor, R.B. Resident Enteric Bacteria Are Necessary for Development of Spontaneous Colitis and Immune System Activation in Interleukin-10-Deficient Mice. Infect. Immun. 1998, 66, 5224–5231. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, L.; Lin, W.; Tang, W.; Chan, F.K.; Ng, S.C. Review article: Probiotics, prebiotics and dietary approaches during COVID-19 pandemic. Trends Food Sci. Technol. 2021, 108, 187–196. [Google Scholar] [CrossRef]
- Salvi, P.S.; Cowles, R.A. Butyrate and the Intestinal Epithelium: Modulation of Proliferation and Inflammation in Homeostasis and Disease. Cells 2021, 10, 1775. [Google Scholar] [CrossRef]
- Yang, W.; Yu, T.; Huang, X.; Bilotta, A.J.; Xu, L.; Lu, Y.; Sun, J.; Pan, F.; Zhou, J.; Zhang, W.; et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 2020, 11, 4457. [Google Scholar] [CrossRef]
- Sencio, V.; Gallerand, A.; Machado, M.G.; Deruyter, L.; Heumel, S.; Soulard, D.; Barthelemy, J.; Cuinat, C.; Vieira, A.T.; Barthelemy, A.; et al. Influenza Virus Infection Impairs the Gut’s Barrier Properties and Favors Secondary Enteric Bacterial Infection through Reduced Production of Short-Chain Fatty Acids. Infect. Immun. 2021, 89, e0073420. [Google Scholar] [CrossRef]
- Potdar, A.A.; Dube, S.; Naito, T.; Li, K.; Botwin, G.; Haritunians, T.; Li, D.; Casero, D.; Yang, S.; Bilsborough, J.; et al. Altered Intestinal ACE2 Levels Are Associated with Inflammation, Severe Disease, and Response to Anti-Cytokine Therapy in Inflammatory Bowel Disease. Gastroenterology 2021, 160, 809–822.e7. [Google Scholar] [CrossRef]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef] [PubMed]
- Ryan, F.J.; Ahern, A.M.; Fitzgerald, R.S.; Laserna-Mendieta, E.J.; Power, E.M.; Clooney, A.G.; O’Donoghue, K.W.; McMurdie, P.J.; Iwai, S.; Crits-Christoph, A.; et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 2020, 11, 1512. [Google Scholar] [CrossRef] [PubMed]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef]
- Wang, J.; Chen, W.-D.; Wang, Y.-D. The Relationship Between Gut Microbiota and Inflammatory Diseases: The Role of Macrophages. Front. Microbiol. 2020, 11, 1065. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic Syndrome and Altered Gut Microbiota in Mice Lacking Toll-Like Receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef]
- Knip, M.; Siljander, H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 2016, 12, 154–167. [Google Scholar] [CrossRef]
- Meijnikman, A.S.; Gerdes, V.E.; Nieuwdorp, M.; Herrema, H. Evaluating Causality of Gut Microbiota in Obesity and Diabetes in Humans. Endocr. Rev. 2018, 39, 133–153. [Google Scholar] [CrossRef]
- Atarashi, K.; Suda, W.; Luo, C.; Kawaguchi, T.; Motoo, I.; Narushima, S.; Kiguchi, Y.; Yasuma, K.; Watanabe, E.; Tanoue, T.; et al. Ectopic colonization of oral bacteria in the intestine drives T H 1 cell induction and inflammation. Science 2017, 358, 359–365. [Google Scholar] [CrossRef]
- Lupp, C.; Robertson, M.L.; Wickham, M.E.; Sekirov, I.; Champion, O.L.; Gaynor, E.C.; Finlay, B.B. Host-Mediated Inflammation Disrupts the Intestinal Microbiota and Promotes the Overgrowth of Enterobacteriaceae. Cell Host Microbe 2007, 2, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Barman, M.; Unold, D.; Shifley, K.; Amir, E.; Hung, K.; Bos, N.; Salzman, N. Enteric Salmonellosis Disrupts the Microbial Ecology of the Murine Gastrointestinal Tract. Infect. Immun. 2008, 76, 907–915. [Google Scholar] [CrossRef]
- Kamada, N.; Kim, Y.-G.; Sham, H.P.; Vallance, B.A.; Puente, J.L.; Martens, E.C.; Núñez, G. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science 2012, 336, 1325–1329. [Google Scholar] [CrossRef] [PubMed]
- Vijay, A.; Kouraki, A.; Gohir, S.; Turnbull, J.; Kelly, A.; Chapman, V.; Barrett, D.A.; Bulsiewicz, W.J.; Valdes, A.M. The anti-inflammatory effect of bacterial short chain fatty acids is partially mediated by endocannabinoids. Gut Microbes 2021, 13, 1997559. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Ter Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016, 167, 1125–1136.e8. [Google Scholar] [CrossRef]
- Tedelind, S.; Westberg, F.; Kjerrulf, M.; Vidal, A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: A study with relevance to inflammatory bowel disease. World J. Gastroenterol. 2007, 13, 2826–2832. [Google Scholar] [CrossRef]
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, M.; Wang, Y.; Dorfman, R.G.; Liu, H.; Yu, T.; Chen, X.; Tang, D.; Xu, L.; Yin, Y.; et al. Faecalibacterium prausnitzii Produces Butyrate to Maintain Th17/Treg Balance and to Ameliorate Colorectal Colitis by Inhibiting Histone Deacetylase 1. Inflamm. Bowel Dis. 2018, 24, 1926–1940. [Google Scholar] [CrossRef]
- Atarashi, K.; Tanoue, T.; Oshima, K.; Suda, W.; Nagano, Y.; Nishikawa, H.; Fukuda, S.; Saito, T.; Narushima, S.; Hase, K.; et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 2013, 500, 232–236. [Google Scholar] [CrossRef]
- Lührs, H.; Gerke, T.; Müller, J.G.; Melcher, R.; Schauber, J.; Boxberger, F.; Scheppach, W.; Menzel, T. Butyrate Inhibits NF-κB Activation in Lamina Propria Macrophages of Patients with Ulcerative Colitis. Scand. J. Gastroenterol. 2002, 37, 458–466. [Google Scholar] [CrossRef]
- Karin, M.; Cao, Y.; Greten, F.; Li, Z.-W. NF-κB in cancer: From innocent bystander to major culprit. Nat. Rev. Cancer 2002, 2, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Klampfer, L.; Huang, J.; Sasazuki, T.; Shirasawa, S.; Augenlicht, L. Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol. Cancer Res. 2003, 11, 855–862. [Google Scholar]
- Kinoshita, M.; Suzuki, Y.; Saito, Y. Butyrate reduces colonic paracellular permeability by enhancing PPARγ activation. Biochem. Biophys. Res. Commun. 2002, 293, 827–831. [Google Scholar] [CrossRef]
- Sears, C.L.; Geis, A.L.; Housseau, F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J. Clin. Investig. 2014, 124, 4166–4172. [Google Scholar] [CrossRef]
- Klooster, J.P.T.; Bol-Schoenmakers, M.; van Summeren, K.; van Vliet, A.L.W.; de Haan, C.A.M.; van Kuppeveld, F.J.M.; Verkoeijen, S.; Pieters, R. Enterocytes, fibroblasts and myeloid cells synergize in anti-bacterial and anti-viral pathways with IL22 as the central cytokine. Commun. Biol. 2021, 4, 631. [Google Scholar] [CrossRef]
- Vuille-Dit-Bille, R.; Camargo, S.; Emmenegger, L.; Sasse, T.; Kummer, E.; Jando, J.; Hamie, Q.M.; Meier, C.F.; Hunziker, S.; Forras-Kaufmann, Z.; et al. Human intestine luminal ACE2 and amino acid transporter expression increased by ACE-inhibitors. Amino Acids 2015, 47, 693–705. [Google Scholar] [CrossRef]
- Patra, T.; Meyer, K.; Geerling, L.; Isbell, T.S.; Hoft, D.F.; Brien, J.; Pinto, A.K.; Ray, R.B.; Ray, R. SARS-CoV-2 spike protein promotes IL-6 trans-signaling by activation of angiotensin II receptor signaling in epithelial cells. PLOS Pathog. 2020, 16, e1009128. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, J.; Schiavon, C.R.; He, M.; Chen, L.; Shen, H.; Zhang, Y.; Yin, Q.; Cho, Y.; Andrade, L.; et al. SARS-CoV-2 Spike Protein Impairs Endothelial Function via Downregulation of ACE 2. Circ. Res. 2021, 128, 1323–1326. [Google Scholar] [CrossRef]
- Glowacka, I.; Bertram, S.; Herzog, P.; Pfefferle, S.; Steffen, I.; Muench, M.O.; Simmons, G.; Hofmann, H.; Kuri, T.; Weber, F.; et al. Differential Downregulation of ACE2 by the Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus and Human Coronavirus NL63. J. Virol. 2010, 84, 1198–1205. [Google Scholar] [CrossRef]
- Penninger, J.M.; Grant, M.B.; Sung, J.J.Y. The Role of Angiotensin Converting Enzyme 2 in Modulating Gut Microbiota, Intestinal Inflammation, and Coronavirus Infection. Gastroenterology 2021, 160, 39–46. [Google Scholar] [CrossRef]
- Jaworska, K.; Koper, M.; Ufnal, M. Gut microbiota and renin-angiotensin system: A complex interplay at local and systemic levels. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 321, G355–G366. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.M.; Vuille-Dit-Bille, R.N.; Meier, C.F.; Verrey, F. ACE2 and gut amino acid transport. Clin. Sci. 2020, 134, 2823–2833. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.; Singer, D.; Makrides, V.; Huggel, K.; Pos, K.M.; Wagner, C.A.; Kuba, K.; Danilczyk, U.; Skovby, F.; Kleta, R.; et al. Tissue-Specific Amino Acid Transporter Partners ACE2 and Collectrin Differentially Interact with Hartnup Mutations. Gastroenterology 2009, 136, 872–882.e3. [Google Scholar] [CrossRef]
- Haga, S.; Nagata, N.; Okamura, T.; Yamamoto, N.; Sata, T.; Yamamoto, N.; Sasazuki, T.; Ishizaka, Y. TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antivir. Res. 2010, 85, 551–555. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Rao, S.; Gao, H.; Guo, F.; Guan, B.; Huan, Y.; Yang, P.; Zhang, Y.; Deng, W.; et al. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005, 11, 875–879. [Google Scholar] [CrossRef]
- Volbeda, M.; Jou-Valencia, D.; Heuvel, M.C.V.D.; Knoester, M.; Zwiers, P.J.; Pillay, J.; Berger, S.P.; van der Voort, P.H.J.; Zijlstra, J.G.; van Meurs, M.; et al. Comparison of renal histopathology and gene expression profiles between severe COVID-19 and bacterial sepsis in critically ill patients. Crit. Care 2021, 25, 202. [Google Scholar] [CrossRef]
- Triana, S.; Metz-Zumaran, C.; Ramirez, C.; Kee, C.; Doldan, P.; Shahraz, M.; Schraivogel, D.; Gschwind, A.R.; Sharma, A.K.; Steinmetz, L.M.; et al. Single-cell analyses reveal SARS-CoV-2 interference with intrinsic immune response in the human gut. Mol. Syst. Biol. 2021, 17, e10232. [Google Scholar] [CrossRef]
- Nataf, S.; Pays, L. Molecular Insights into SARS-CoV2-Induced Alterations of the Gut/Brain Axis. Int. J. Mol. Sci. 2021, 22, 10440. [Google Scholar] [CrossRef]
- Heuberger, J.; Trimpert, J.; Vladimirova, D.; Goosmann, C.; Lin, M.; Schmuck, R.; Mollenkopf, H.; Brinkmann, V.; Tacke, F.; Osterrieder, N.; et al. Epithelial response to IFN-γ promotes SARS-CoV-2 infection. EMBO Mol. Med. 2021, 13, e13191. [Google Scholar] [CrossRef]
- Lee, S.; Yoon, G.Y.; Myoung, J.; Kim, S.-J.; Ahn, D.-G. Robust and persistent SARS-CoV-2 infection in the human intestinal brush border expressing cells. Emerg. Microbes Infect. 2020, 9, 2169–2179. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, C.G.K.; Allon, S.J.; Nyquist, S.K.; Mbano, I.M.; Miao, V.N.; Tzouanas, C.N.; Cao, Y.; Yousif, A.S.; Bals, J.; Hauser, B.M.; et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell 2020, 181, 1016–1035.e19. [Google Scholar] [CrossRef] [PubMed]
- Harnik, Y.; Buchauer, L.; Ben-Moshe, S.; Averbukh, I.; Levin, Y.; Savidor, A.; Eilam, R.; Moor, A.E.; Itzkovitz, S. Spatial discordances between mRNAs and proteins in the intestinal epithelium. Nat. Metab. 2021, 3, 1680–1693. [Google Scholar] [CrossRef]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J.; et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Jankowski, J.; Lee, H.K.; Wilflingseder, J.; Hennighausen, L. Interferon-regulated genetic programs and JAK/STAT pathway activate the intronic promoter of the short ACE2 isoform in renal proximal tubules. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bártová, E.; Legartová, S.; Krejčí, J.; Arcidiacono, O.A. Cell differentiation and aging accompanied by depletion of the ACE2 protein. Aging 2020, 12, 22495–22508. [Google Scholar] [CrossRef]
- Perlot, T.; Penninger, J.M. ACE2—From the renin–angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013, 15, 866–873. [Google Scholar] [CrossRef]
- Bröer, S. Amino Acid Transport Across Mammalian Intestinal and Renal Epithelia. Physiol. Rev. 2008, 88, 249–286. [Google Scholar] [CrossRef]
- Guo, Y.; Wang, B.; Gao, H.; Gao, L.; Hua, R.; Xu, J.-D. ACE2 in the Gut: The Center of the 2019-nCoV Infected Pathology. Front. Mol. Biosci. 2021, 8, 708336. [Google Scholar] [CrossRef]
- Moal, V.L.-L.; Servin, A.L. The Front Line of Enteric Host Defense against Unwelcome Intrusion of Harmful Microorganisms: Mucins, Antimicrobial Peptides, and Microbiota. Clin. Microbiol. Rev. 2006, 19, 315–337. [Google Scholar] [CrossRef]
- Rajput, S.; Paliwal, D.; Naithani, M.; Kothari, A.; Meena, K.; Rana, S. COVID-19 and Gut Microbiota: A Potential Connection. Indian J. Clin. Biochem. 2021, 36, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Rist, V.T.S.; Weiss, E.; Eklund, M.; Mosenthin, R. Impact of dietary protein on microbiota composition and activity in the gastrointestinal tract of piglets in relation to gut health: A review. Animal 2013, 7, 1067–1078. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Iyer, A.; Russell, W.R. Impact of protein on the composition and metabolism of the human gut microbiota and health. Proc. Nutr. Soc. 2021, 80, 173–185. [Google Scholar] [CrossRef]
- Masuoka, H.; Suda, W.; Tomitsuka, E.; Shindo, C.; Takayasu, L.; Horwood, P.; Greenhill, A.R.; Hattori, M.; Umezaki, M.; Hirayama, K. The influences of low protein diet on the intestinal microbiota of mice. Sci. Rep. 2020, 10, 17077. [Google Scholar] [CrossRef]
- Trottein, F.; Sokol, H. Potential Causes and Consequences of Gastrointestinal Disorders during a SARS-CoV-2 Infection. Cell Rep. 2020, 32, 107915. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.; Angus, P.W.; Burrell, L.M.; Herath, C.; Gibson, P.R.; Lubel, J.S. Review article: The pathophysiological roles of the renin-angiotensin system in the gastrointestinal tract. Aliment. Pharmacol. Ther. 2012, 35, 414–428. [Google Scholar] [CrossRef]
- Battagello, D.S.; Dragunas, G.; Klein, M.O.; Ayub, A.L.; Velloso, F.J.; Correa, R.G. Unpuzzling COVID-19: Tissue-related signaling pathways associated with SARS-CoV-2 infection and transmission. Clin. Sci. 2020, 134, 2137–2160. [Google Scholar] [CrossRef]
- Duan, Y.; Prasad, R.; Feng, D.; Beli, E.; Li Calzi, S.; Longhini, A.L.F.; Lamendella, R.; Floyd, J.L.; DuPont, M.; Noothi, S.K.; et al. Bone Marrow-Derived Cells Restore Functional Integrity of the Gut Epithelial and Vascular Barriers in a Model of Diabetes and ACE2 Deficiency. Circ. Res. 2019, 125, 969–988. [Google Scholar] [CrossRef]
- Danilczyk, U.; Sarao, R.; Remy, C.; Benabbas, C.; Stange, G.; Richter, A.; Arya, S.; Pospisilik, J.A.; Singer, D.; Camargo, S.; et al. Essential role for collectrin in renal amino acid transport. Nature 2006, 444, 1088–1091. [Google Scholar] [CrossRef]
- Singer, D.; Camargo, S.; Ramadan, T.; Schäfer, M.; Mariotta, L.; Herzog, B.; Huggel, K.; Wolfer, D.; Werner, S.; Penninger, J.; et al. Defective intestinal amino acid absorption in Ace2 null mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G686–G695. [Google Scholar] [CrossRef]
- Aktas, B. Gut Microbiota Dysbiosis and COVID-19: Possible Links. Ref. Modul. Food Sci. 2022, 535–544. [Google Scholar] [CrossRef]
- Spak, E.; Hallersund, P.; Edebo, A.; Casselbrant, A.; Fändriks, L. The human duodenal mucosa harbors all components for a local renin angiotensin system. Clin. Sci. 2019, 133, 971–982. [Google Scholar] [CrossRef] [PubMed]
- Shenoy, S. Gut microbiome, Vitamin D, ACE2 interactions are critical factors in immune-senescence and inflammaging: Key for vaccine response and severity of COVID-19 infection. Inflamm. Res. 2022, 71, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Muema, C.; Zhang, X.; Pan, X.; Xiong, J.; Yang, H.; Yu, J.; Wei, H. Enriched Opportunistic Pathogens Revealed by Metagenomic Sequencing Hint Potential Linkages between Pharyngeal Microbiota and COVID-19. Virol. Sin. 2021, 36, 924–933. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humaran, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Yang, T.; Chakraborty, S.; Saha, P.; Mell, B.; Cheng, X.; Yeo, J.-Y.; Mei, X.; Zhou, G.; Mandal, J.; Golonka, R.; et al. Gnotobiotic Rats Reveal That Gut Microbiota Regulates Colonic mRNA of Ace2, the Receptor for SARS-CoV-2 Infectivity. Hypertension 2020, 76, e1–e3. [Google Scholar] [CrossRef]
- Das, S.; Jayaratne, R.; Barrett, K.E. The Role of Ion Transporters in the Pathophysiology of Infectious Diarrhea. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 33–45. [Google Scholar] [CrossRef]
- Day, C.J.; Bailly, B.; Guillon, P.; Dirr, L.; Jen, F.E.-C.; Spillings, B.L.; Mak, J.; von Itzstein, M.; Haselhorst, T.; Jennings, M.P. Multidisciplinary Approaches Identify Compounds that Bind to Human ACE2 or SARS-CoV-2 Spike Protein as Candidates to Block SARS-CoV-2–ACE2 Receptor Interactions. mBio 2021, 12. [Google Scholar] [CrossRef]
- Xiong, J.; Xiang, Y.; Huang, Z.; Liu, X.; Wang, M.; Ge, G.; Chen, H.; Xu, J.; Zheng, M.; Chen, L. Structure-Based Virtual Screening and Identification of Potential Inhibitors of SARS-CoV-2 S-RBD and ACE2 Interaction. Front. Chem. 2021, 9, 740702. [Google Scholar] [CrossRef]
- Prada, J.A.H.; Ferreira, A.J.; Katovich, M.J.; Shenoy, V.; Qi, Y.; Santos, R.A.; Castellano, R.K.; Lampkins, A.J.; Gubala, V.; Ostrov, D.A.; et al. Structure-Based Identification of Small-Molecule Angiotensin-Converting Enzyme 2 Activators as Novel Antihypertensive Agents. Hypertension 2008, 51, 1312–1317. [Google Scholar] [CrossRef]
- Sutton, T.D.S.; Hill, C. Gut Bacteriophage: Current Understanding and Challenges. Front. Endocrinol. 2019, 10, 784. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, K.H. SARS-CoV-2 spike glycoprotein-binding proteins expressed by upper respiratory tract bacteria may prevent severe viral infection. FEBS Lett. 2020, 594, 1651–1660. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, M.; Querci, M.; Brogna, C.; Ponti, J.; Cristoni, S.; Markov, P.V.; Valsesia, A.; Leoni, G.; Benedetti, A.; Wiss, T.; et al. Evidence of SARS-CoV-2 bacteriophage potential in human gut microbiota. F1000Research 2022, 11, 292. [Google Scholar] [CrossRef]
- Gottlieb, P.; Alimova, A. RNA Packaging in the Cystovirus Bacteriophages: Dynamic Interactions during Capsid Maturation. Int. J. Mol. Sci. 2022, 23, 2677. [Google Scholar] [CrossRef]
- Grenga, L.; Gallais, F.; Pible, O.; Gaillard, J.-C.; Gouveia, D.; Batina, H.; Bazaline, N.; Ruat, S.; Culotta, K.; Miotello, G.; et al. Shotgun proteomics analysis of SARS-CoV-2-infected cells and how it can optimize whole viral particle antigen production for vaccines. Emerg. Microbes Infect. 2020, 9, 1712–1721. [Google Scholar] [CrossRef]
- Townsend, E.M.; Kelly, L.; Muscatt, G.; Box, J.D.; Hargraves, N.; Lilley, D.; Jameson, E. The Human Gut Phageome: Origins and Roles in the Human Gut Microbiome. Front. Cell. Infect. Microbiol. 2021, 11, 643214. [Google Scholar] [CrossRef]
- Guerin, E.; Hill, C. Shining Light on Human Gut Bacteriophages. Front. Cell. Infect. Microbiol. 2020, 10, 481. [Google Scholar] [CrossRef]
- Aggarwala, V.; Liang, G.; Bushman, F.D. Viral communities of the human gut: Metagenomic analysis of composition and dynamics. Mob. DNA 2017, 8, 12. [Google Scholar] [CrossRef]
- Zajac, V. Elimination of the new coronavirus and prevention of the second wave of infection. World J. Adv. Res. Rev. 2020, 8, 148–150. [Google Scholar] [CrossRef]
- Brogna, C.; Cristoni, S.; Petrillo, M.; Querci, M.; Piazza, O.; Eede, G.V.D. Toxin-like peptides in plasma, urine and faecal samples from COVID-19 patients. F1000Research 2021, 10, 550. [Google Scholar] [CrossRef]
- Grenga, L.; Pible, O.; Miotello, G.; Culotta, K.; Ruat, S.; Roncato, M.; Gas, F.; Bellanger, L.; Claret, P.; Dunyach-Remy, C.; et al. Taxonomical and functional changes in COVID -19 faecal microbiome could be related to SARS-CoV-2 faecal load. Environ. Microbiol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Dean, P.; Kenny, B. Intestinal barrier dysfunction by enteropathogenic Escherichia coli is mediated by two effector molecules and a bacterial surface protein. Mol. Microbiol. 2004, 54, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, V.K.; Koutsouris, A.; Lukic, S.; Pilkinton, M.; Simonovic, I.; Simonovic, M.; Hecht, G. Comparative Analysis of EspF from Enteropathogenic and Enterohemorrhagic Escherichia coli in Alteration of Epithelial Barrier Function. Infect. Immun. 2004, 72, 3218–3227. [Google Scholar] [CrossRef]
- Tafazoli, F.; Magnusson, K.-E.; Zheng, L. Disruption of Epithelial Barrier Integrity by Salmonella enterica Serovar Typhimurium Requires Geranylgeranylated Proteins. Infect. Immun. 2003, 71, 872–881. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response to Prevotellabacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Rodríguez-Piñeiro, A.M.; Schütte, A.; Ermund, A.; Boysen, P.; Bemark, M.; Sommer, F.; Bäckhed, F.; Hansson, G.C.; Johansson, M.E.V. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep. 2015, 16, 164–177. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Martinez-Silgado, A.; Akkerman, N.; Saftien, A.; Boot, C.; de Waal, A.; Beumer, J.; Dutta, D.; Heo, I.; et al. Intestinal organoid cocultures with microbes. Nat. Protoc. 2021, 16, 4633–4649. [Google Scholar] [CrossRef]
- Dicksved, J.; Schreiber, O.; Willing, B.; Petersson, J.; Rang, S.; Phillipson, M.; Holm, L.; Roos, S. Lactobacillus reuteri Maintains a Functional Mucosal Barrier during DSS Treatment Despite Mucus Layer Dysfunction. PLoS ONE 2012, 7, e46399. [Google Scholar] [CrossRef]
- Kim, H.S. Do an Altered Gut Microbiota and an Associated Leaky Gut Affect COVID-19 Severity? mBio 2021, 12. [Google Scholar] [CrossRef]
- Vignesh, R.; Swathirajan, C.R.; Tun, Z.H.; Rameshkumar, M.R.; Solomon, S.S.; Balakrishnan, P. Could Perturbation of Gut Microbiota Possibly Exacerbate the Severity of COVID-19 via Cytokine Storm? Front. Immunol. 2020, 11, 607734. [Google Scholar] [CrossRef]
- Openshaw, P.J. Crossing barriers: Infections of the lung and the gut. Mucosal Immunol. 2009, 2, 100–102. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, V.; Capurso, G.; Ianiro, G.; Gasbarrini, A.; Arcidiacono, P.G.; Alvaro, D. Intestinal permeability changes with bacterial translocation as key events modulating systemic host immune response to SARS-CoV-2: A working hypothesis. Dig. Liver Dis. 2020, 52, 1383–1389. [Google Scholar] [CrossRef] [PubMed]
- Giron, L.B.; Dweep, H.; Yin, X.; Wang, H.; Damra, M.; Goldman, A.R.; Gorman, N.; Palmer, C.S.; Tang, H.-Y.; Shaikh, M.W.; et al. Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front. Immunol. 2021, 12, 686240. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Patton, M.J.; Floyd, J.L.; Fortmann, S.; DuPont, M.; Harbour, A.; Wright, J.; Lamendella, R.; Stevens, B.R.; Oudit, G.Y.; et al. Plasma Microbiome in COVID-19 Subjects: An Indicator of Gut Barrier Defects and Dysbiosis. Int. J. Mol. Sci. 2022, 23, 9141. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Song, Z.-G.; Liu, C.; Tan, S.; Lin, S.; Zhu, J.; Dai, F.-H.; Gao, J.; She, J.-L.; Mei, Z.; et al. Gut microbiome alterations and gut barrier dysfunction are associated with host immune homeostasis in COVID-19 patients. BMC Med. 2022, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Maguire, M.; Maguire, G. Gut dysbiosis, leaky gut, and intestinal epithelial proliferation in neurological disorders: Towards the development of a new therapeutic using amino acids, prebiotics, probiotics, and postbiotics. Rev. Neurosci. 2019, 30, 179–201. [Google Scholar] [CrossRef] [PubMed]
- Effenberger, M.; Grabherr, F.; Mayr, L.; Schwaerzler, J.; Nairz, M.; Seifert, M.; Hilbe, R.; Seiwald, S.; Scholl-Buergi, S.; Fritsche, G.; et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut 2020, 69, 1543–1544. [Google Scholar] [CrossRef]
- Hayashi, Y.; Wagatsuma, K.; Nojima, M.; Yamakawa, T.; Ichimiya, T.; Yokoyama, Y.; Kazama, T.; Hirayama, D.; Nakase, H. The characteristics of gastrointestinal symptoms in patients with severe COVID-19: A systematic review and meta-analysis. J. Gastroenterol. 2021, 56, 409–420. [Google Scholar] [CrossRef]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef]
- Nobel, Y.R.; Phipps, M.; Zucker, J.; Lebwohl, B.; Wang, T.C.; Sobieszczyk, M.E.; Freedberg, D.E. Gastrointestinal Symptoms and Coronavirus Disease 2019: A Case-Control Study from the United States. Gastroenterology 2020, 159, 373–375.e2. [Google Scholar] [CrossRef]
- Redd, W.D.; Zhou, J.C.; Hathorn, K.E.; Mccarty, T.R.; Bazarbashi, A.N.; Thompson, C.C.; Shen, L.; Chan, W.W. Prevalence and Characteristics of Gastrointestinal Symptoms in Patients with Severe Acute Respiratory Syndrome Coronavirus 2 Infection in the United States: A Multicenter Cohort Study. Gastroenterology 2020, 159, 765–767.e2. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, A.; Zlitni, S.; Brooks, E.F.; Vance, S.E.; Dahlen, A.; Hedlin, H.; Park, R.M.; Han, A.; Schmidtke, D.T.; Verma, R.; et al. Gastrointestinal symptoms and fecal shedding of SARS-CoV-2 RNA suggest prolonged gastrointestinal infection. Med 2022, 3, 371–387.e9. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Brooks, E.F.; Bhatt, A.S. The gut microbiome: A missing link in understanding the gastrointestinal manifestations of COVID-19? Cold Spring Harb. Mol. Case Stud. 2021, 7, a006031. [Google Scholar] [CrossRef]
- Liu, Q.; Mak, J.W.Y.; Su, Q.; Yeoh, Y.K.; Lui, G.C.-Y.; Ng, S.S.S.; Zhang, F.; Li, A.Y.L.; Lu, W.; Hui, D.S.-C.; et al. Gut microbiota dynamics in a prospective cohort of patients with post-acute COVID-19 syndrome. Gut 2022, 71, 544–552. [Google Scholar] [CrossRef]
- Davis, H.E.; Assaf, G.S.; McCorkell, L.; Wei, H.; Low, R.J.; Re’Em, Y.; Redfield, S.; Austin, J.P.; Akrami, A. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. eClinicalMedicine 2021, 38, 101019. [Google Scholar] [CrossRef]
- Arostegui, D.; Castro, K.; Schwarz, S.; Vaidy, K.; Rabinowitz, S.; Wallach, T. Persistent SARS-CoV-2 Nucleocapsid Protein Presence in the Intestinal Epithelium of a Pediatric Patient 3 Months after Acute Infection. JPGN Rep. 2021, 3, e152. [Google Scholar] [CrossRef]
- Kim, K.O.; Gluck, M. Fecal Microbiota Transplantation: An Update on Clinical Practice. Clin. Endosc. 2019, 52, 137–143. [Google Scholar] [CrossRef]
- Kazemian, N.; Kao, D.; Pakpour, S. Fecal Microbiota Transplantation during and Post-COVID-19 Pandemic. Int. J. Mol. Sci. 2021, 22, 3004. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, P.; Zhang, X. Probiotics Regulate Gut Microbiota: An Effective Method to Improve Immunity. Molecules 2021, 26, 6076. [Google Scholar] [CrossRef]
- Baindara, P.; Chakraborty, R.; Holliday, Z.M.; Mandal, S.M.; Schrum, A.G. Oral probiotics in coronavirus disease 2019: Connecting the gut–lung axis to viral pathogenesis, inflammation, secondary infection and clinical trials. New Microbes New Infect. 2021, 40, 100837. [Google Scholar] [CrossRef] [PubMed]
- Kurian, S.J.; Unnikrishnan, M.K.; Miraj, S.S.; Bagchi, D.; Banerjee, M.; Reddy, B.S.; Rodrigues, G.S.; Manu, M.K.; Saravu, K.; Mukhopadhyay, C.; et al. Probiotics in Prevention and Treatment of COVID-19: Current Perspective and Future Prospects. Arch. Med. Res. 2021, 52, 582–594. [Google Scholar] [CrossRef] [PubMed]
- Mak, J.W.Y.; Chan, F.K.L.; Ng, S.C. Probiotics and COVID-19: One size does not fit all. Lancet Gastroenterol. Hepatol. 2020, 5, 644–645. [Google Scholar] [CrossRef]
- Guarino, M.P.L.; Altomare, A.; Emerenziani, S.; Di Rosa, C.; Ribolsi, M.; Balestrieri, P.; Iovino, P.; Rocchi, G.; Cicala, M. Mechanisms of Action of Prebiotics and Their Effects on Gastro-Intestinal Disorders in Adults. Nutrients 2020, 12, 1037. [Google Scholar] [CrossRef]
- Peters, V.; Van De Steeg, E.; van Bilsen, J.; Meijerink, M. Mechanisms and immunomodulatory properties of pre- and probiotics. Benef. Microbes 2019, 10, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Shokryazdan, P.; Jahromi, M.F.; Navidshad, B.; Liang, J.B. Effects of prebiotics on immune system and cytokine expression. Med. Microbiol. Immunol. 2016, 206, 1–9. [Google Scholar] [CrossRef]
- Khaled, J.M. Probiotics, prebiotics, and COVID-19 infection: A review article. Saudi J. Biol. Sci. 2021, 28, 865–869. [Google Scholar] [CrossRef]
- Kalantar-Zadeh, K.; Ward, S.A.; Kalantar-Zadeh, K.; El-Omar, E. Considering the Effects of Microbiome and Diet on SARS-CoV-2 Infection: Nanotechnology Roles. ACS Nano 2020, 14, 5179–5182. [Google Scholar] [CrossRef]
- Infusino, F.; Marazzato, M.; Mancone, M.; Fedele, F.; Mastroianni, C.M.; Severino, P.; Ceccarelli, G.; Santinelli, L.; Cavarretta, E.; Marullo, A.G.M.; et al. Diet Supplementation, Probiotics, and Nutraceuticals in SARS-CoV-2 Infection: A Scoping Review. Nutrients 2020, 12, 1718. [Google Scholar] [CrossRef]
- Deschasaux-Tanguy, M.; Srour, B.; Bourhis, L.; Arnault, N.; Druesne-Pecollo, N.; Esseddik, Y.; de Edelenyi, F.S.; Allègre, J.; Allès, B.; Andreeva, V.A.; et al. Nutritional risk factors for SARS-CoV-2 infection: A prospective study within the NutriNet-Santé cohort. BMC Med. 2021, 19, 290. [Google Scholar] [CrossRef]
- Salazar-Robles, E.; Kalantar-Zadeh, K.; Badillo, H.; Calderón-Juárez, M.; García-Bárcenas, C.A.; Ledesma-Pérez, P.D.; Lerma, A.; Lerma, C. Association between severity of COVID-19 symptoms and habitual food intake in adult outpatients. BMJ Nutr. Prev. Health 2021, 4, 469–478. [Google Scholar] [CrossRef] [PubMed]
| KER1739 | https://aopwiki.org/events/1739 | accessed on 29 June 2022 |
| KER1738 | https://aopwiki.org/events/1738 | accessed on 29 June 2022 |
| KER1847 | https://aopwiki.org/events/1847 | accessed on 29 June 2022 |
| KER1901 | https://aopwiki.org/events/1901 | accessed on 29 June 2022 |
| KER1493 | https://aopwiki.org/events/1493 | accessed on 29 June 2022 |
| KER1497 | https://aopwiki.org/events/1497 | accessed on 29 June 2022 |
| KER1954 | https://aopwiki.org/events/1954 | accessed on 29 June 2022 |
| KER2311 | https://aopwiki.org/events/2311 | accessed on 29 June 2022 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clerbaux, L.-A.; Filipovska, J.; Muñoz, A.; Petrillo, M.; Coecke, S.; Amorim, M.-J.; Grenga, L. Mechanisms Leading to Gut Dysbiosis in COVID-19: Current Evidence and Uncertainties Based on Adverse Outcome Pathways. J. Clin. Med. 2022, 11, 5400. https://doi.org/10.3390/jcm11185400
Clerbaux L-A, Filipovska J, Muñoz A, Petrillo M, Coecke S, Amorim M-J, Grenga L. Mechanisms Leading to Gut Dysbiosis in COVID-19: Current Evidence and Uncertainties Based on Adverse Outcome Pathways. Journal of Clinical Medicine. 2022; 11(18):5400. https://doi.org/10.3390/jcm11185400
Chicago/Turabian StyleClerbaux, Laure-Alix, Julija Filipovska, Amalia Muñoz, Mauro Petrillo, Sandra Coecke, Maria-Joao Amorim, and Lucia Grenga. 2022. "Mechanisms Leading to Gut Dysbiosis in COVID-19: Current Evidence and Uncertainties Based on Adverse Outcome Pathways" Journal of Clinical Medicine 11, no. 18: 5400. https://doi.org/10.3390/jcm11185400
APA StyleClerbaux, L.-A., Filipovska, J., Muñoz, A., Petrillo, M., Coecke, S., Amorim, M.-J., & Grenga, L. (2022). Mechanisms Leading to Gut Dysbiosis in COVID-19: Current Evidence and Uncertainties Based on Adverse Outcome Pathways. Journal of Clinical Medicine, 11(18), 5400. https://doi.org/10.3390/jcm11185400






