A Critical Evaluation of Validation and Clinical Experience Studies in Non-Invasive Prenatal Testing for Trisomies 21, 18, and 13 and Monosomy X
Abstract
1. Introduction
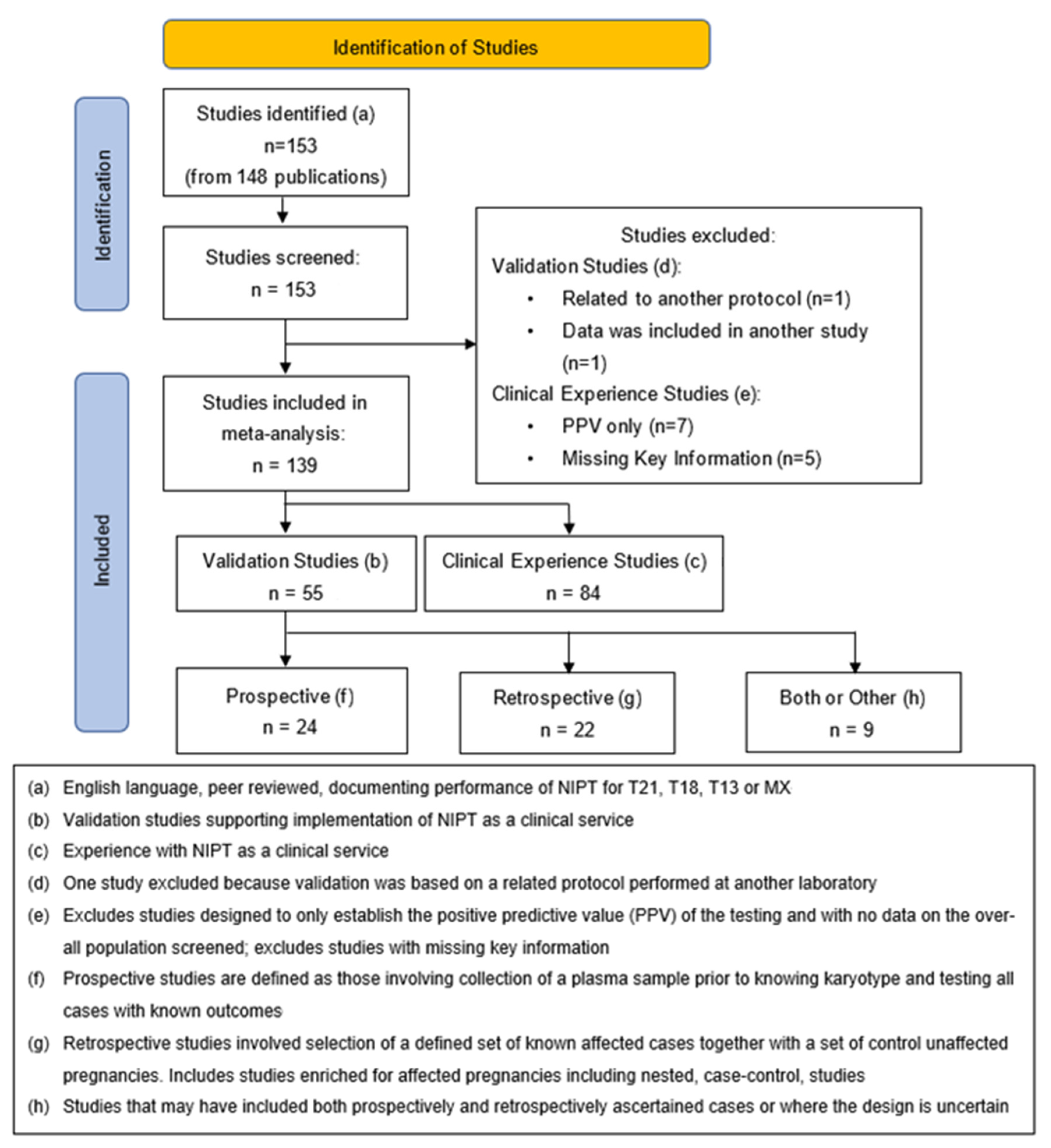
2. Methods
2.1. Validation Studies
2.2. Clinical Experience Studies
3. Results
3.1. Validation Studies
3.1.1. Overall Performance
3.1.2. Retrospective Versus Prospective
3.1.3. Method of Testing
3.1.4. Country
3.2. Clinical Experience Studies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Benn, P.; Cuckle, H.; Pergament, E. Non-invasive prenatal testing for aneuploidy: Current status and future prospects. Ultrasound Obstet. Gynecol. 2013, 42, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Palomaki, G.E.; Kloza, E.M.; Lambert-Messerlian, G.M.; Haddow, J.E.; Neveux, L.M.; Ehrich, M.; van den Boom, D.; Bombard, A.T.; Deciu, C.; Grody, W.W. DNA sequencing of maternal plasma to detect Down syndrome: An international clinical validation study. Genet. Med. 2011, 13, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Sparks, A.B.; Struble, C.A.; Wang, E.T.; Song, K.; Oliphant, A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: Evaluation for trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012, 206, 319.e311–319.e319. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W.; Platt, L.D.; Goldberg, J.D.; Abuhamad, A.Z.; Sehnert, A.J.; Rava, R.P. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet. Gynecol. 2012, 119, 890–901. [Google Scholar] [CrossRef]
- Wang, E.; Batey, A.; Struble, C.; Musci, T.; Song, K.; Oliphant, A. Gestational age and maternal weight effects on fetal cell-free DNA in maternal plasma. Prenat. Diagn. 2013, 33, 662–666. [Google Scholar] [CrossRef]
- Fan, H.C.; Blumenfeld, Y.J.; Chitkara, U.; Hudgins, L.; Quake, S.R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl. Acad. Sci. USA 2008, 105, 16266–16271. [Google Scholar] [CrossRef]
- Chiu, R.W.; Chan, K.A.; Gao, Y.; Lau, V.Y.; Zheng, W.; Leung, T.Y.; Foo, C.H.; Xie, B.; Tsui, N.B.; Lun, F.M. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc. Natl. Acad. Sci. USA 2008, 105, 20458–20463. [Google Scholar] [CrossRef]
- Zimmermann, B.; Hill, M.; Gemelos, G.; Demko, Z.; Banjevic, M.; Baner, J.; Ryan, A.; Sigurjonsson, S.; Chopra, N.; Dodd, M. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenat. Diagn. 2012, 32, 1233–1241. [Google Scholar] [CrossRef]
- Pergament, E.; Cuckle, H.; Zimmermann, B.; Banjevic, M.; Sigurjonsson, S.; Ryan, A.; Hall, M.P.; Dodd, M.; Lacroute, P.; Stosic, M. Single-nucleotide polymorphism–based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet. Gynecol. 2014, 124, 210. [Google Scholar] [CrossRef]
- Hartwig, T.S.; Ambye, L.; Sørensen, S.; Jørgensen, F.S. Discordant non-invasive prenatal testing (NIPT)–a systematic review. Prenat. Diagn. 2017, 37, 527–539. [Google Scholar] [CrossRef]
- Hui, L.; Bianchi, D.W. Fetal fraction and noninvasive prenatal testing: What clinicians need to know. Prenat. Diagn. 2020, 40, 155–163. [Google Scholar] [CrossRef] [PubMed]
- McKanna, T.; Ryan, A.; Krinshpun, S.; Kareht, S.; Marchand, K.; Grabarits, C.; Ali, M.; McElheny, A.; Gardiner, K.; LeChien, K. Fetal fraction-based risk algorithm for non-invasive prenatal testing: Screening for trisomies 13 and 18 and triploidy in women with low cell-free fetal DNA. Ultrasound Obstet. Gynecol. 2019, 53, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Gil, M.M.; Accurti, V.; Santacruz, B.; Plana, M.N.; Nicolaides, K.H. Analysis of cell-free DNA in ma-ternal blood in screening for aneuploidies: Updated meta-analysis. Ultrasound Obstet. Gyncol. 2017, 50, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Phillips, S.; Freeman, K.; Geppert, J.; Agbebiyi, A.; Uthman, O.A.; Madan, J.; Clarke, A.; Quenby, S.; Clarke, A. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: A systematic review and meta-analysis. BMJ Open 2016, 6, e010002. [Google Scholar] [CrossRef]
- Mackie, F.L.; Allen, S.; Morris, R.K.; Kilby, M.D. Cell-free fetal DNA-based noninvasive prenatal testing of aneuploidy. Obstet. Gynecol. 2017, 19, 211–218. [Google Scholar] [CrossRef]
- Iwarsson, E.; Jacobsson, B.; Dagerhamn, J.; Davidson, T.; Bernabé, E.; Heibert Arnlind, M. Analysis of cell-free fetal DNA in maternal blood for detection of trisomy 21, 18 and 13 in a general pregnant population and in a high risk population–a systematic review and meta-analysis. Acta Obstet. Et Gynecol. Scand. 2017, 96, 7–18. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef]
- Reitsma, J.B.; Glas, A.S.; Rutjes, A.W.; Scholten, R.J.; Bossuyt, P.M.; Zwinderman, A.H. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J. Clin. Epidemiol. 2005, 58, 982–990. [Google Scholar] [CrossRef]
- Benn, P.; Curnow, K.J.; Chapman, S.; Michalopoulos, S.N.; Hornberger, J.; Rabinowitz, M. An eco-nomic analysis of cell-free DNA non-invasive prenatal testing in the US general pregnancy population. PLoS ONE 2015, 10, e0132313. [Google Scholar] [CrossRef]
- DiNonno, W.; Demko, Z.; Martin, K.; Billings, P.; Egbert, M.; Zneimer, S.; Keen-Kim, D.; Benn, P. Quality assurance of non-invasive prenatal screening (NIPS) for fetal aneuploidy using positive predictive values as outcome measures. J. Clin. Med. 2019, 8, 1311. [Google Scholar] [CrossRef]
- Leeflang, M.M.; Bossuyt, P.M.; Irwig, L. Diagnostic test accuracy may vary with prevalence: Implications for evidence-based diagnosis. J. Clin. Epidemiol. 2009, 62, 5–12. [Google Scholar] [CrossRef]
- Leeflang, M.M.; Rutjes, A.W.; Reitsma, J.B.; Hooft, L.; Bossuyt, P.M. Variation of a test’s sensitivity and specificity with disease prevalence. Cmaj 2013, 185, E537–E544. [Google Scholar] [CrossRef]
- Taneja, P.A.; Snyder, H.L.; de Feo, E.; Kruglyak, K.M.; Halks-Miller, M.; Curnow, K.J.; Bhatt, S. Noninvasive prenatal testing in the general obstetric population: Clinical performance and counseling considerations in over 85 000 cases. Prenat. Diagn. 2016, 36, 237–243. [Google Scholar] [CrossRef]
- Hancock, S.; Ben-Shachar, R.; Adusei, C.; Oyolu, C.B.; Evans, E.A.; Kang, H.P.; Haverty, C.; Muzzey, D. Clinical experience across the fetal-fraction spectrum of a non-invasive prenatal screening approach with low test-failure rate. Ultrasound Obstet. Gynecol. 2020, 56, 422–430. [Google Scholar] [CrossRef]
- Alberti, A.; Salomon, L.; Le Lorc’h, M.; Couloux, A.; Bussieres, L.; Goupil, S.; Malan, V.; Pelletier, E.; Hyon, C.; Vialard, F. Non-invasive prenatal testing for trisomy 21 based on analysis of cell-free fetal DNA circulating in the maternal plasma. Prenat. Diagn. 2015, 35, 471–476. [Google Scholar] [CrossRef]
- Arigul, T.; Suwannachairob, W.; Moungklom, N.; Praphanphoj, V. Sensitivity and specificity of MGC-NIPS for trisomy 13, trisomy 18, trisomy 21, and sex chromosome aneuploidy screening in 219 Thai pregnant women. Genom. Genet. 2020, 13, 1–6. [Google Scholar]
- Ashoor, G.; Syngelaki, A.; Wagner, M.; Birdir, C.; Nicolaides, K.H. Chromosome-selective sequencing of maternal plasma cell–free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012, 206, 322.e321–322.e325. [Google Scholar] [CrossRef]
- Ashoor, G.; Syngelaki, A.; Wang, E.; Struble, C.; Oliphant, A.; Song, K.; Nicolaides, K.H. Trisomy 13 detection in the first trimester of pregnancy using a chromosome-selective cell-free DNA analysis method. Ultrasound Obstet. Gynecol. 2013, 41, 21–25. [Google Scholar] [CrossRef]
- Benachi, A.; Letourneau, A.; Kleinfinger, P.; Senat, M.-V.; Gautier, E.; Favre, R.; Bidat, L.; Houf-flin-Debarge, V.; Bouyer, J.; Costa, J.-M. Cell-free DNA analysis in maternal plasma in cases of fetal abnormalities detected on ultrasound examination. Obstet. Gynecol. 2015, 125, 1330–1337. [Google Scholar] [CrossRef]
- Bianchi, D.W.; Parker, R.L.; Wentworth, J.; Madankumar, R.; Saffer, C.; Das, A.F.; Craig, J.A.; Chudova, D.I.; Devers, P.L.; Jones, K.W. DNA sequencing versus standard prenatal aneuploidy screening. N. Engl. J. Med. 2014, 370, 799–808. [Google Scholar] [CrossRef]
- Chen, E.Z.; Chiu, R.W.; Sun, H.; Akolekar, R.; Chan, K.A.; Leung, T.Y.; Jiang, P.; Zheng, Y.W.; Lun, F.M.; Chan, L.Y. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS ONE 2011, 6, e21791. [Google Scholar] [CrossRef]
- Chiu, R.W.; Akolekar, R.; Zheng, Y.W.; Leung, T.Y.; Sun, H.; Chan, K.A.; Lun, F.M.; Go, A.T.; Lau, E.T.; To, W.W. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ 2011, 342, c7401. [Google Scholar] [CrossRef]
- Dahl, F.; Ericsson, O.; Karlberg, O.; Karlsson, F.; Howell, M.; Persson, F.; Roos, F.; Stenberg, J.; Ahola, T.; Alftrén, I. Imaging single DNA molecules for high precision NIPT. Sci. Rep. 2018, 8, 4549. [Google Scholar] [CrossRef]
- Dar, P.; Jacobson, B.; MacPherson, C.; Egbert, M.; Malone, F.; Wapner, R.J.; Roman, A.S.; Khalil, A.; Faro, R.; Madankumar, R.; et al. Cell-free DNA screening for trisomies 21, 18 and 13 in pregnancies at low and high risk for aneuploidy with genetic confirmation. Am. J. Obstet. Gynecol. 2022, 227, 259.e1–259.e14. [Google Scholar] [CrossRef] [PubMed]
- Ehrich, M.; Deciu, C.; Zwiefelhofer, T.; Tynan, J.A.; Cagasan, L.; Tim, R.; Lu, V.; McCullough, R.; McCarthy, E.; Nygren, A.O. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in ma-ternal blood: A study in a clinical setting. Am. J. Obstet. Gynecol. 2011, 204, 205.e201–205.e211. [Google Scholar] [CrossRef] [PubMed]
- El Khattabi, L.A.; Brun, S.; Gueguen, P.; Chatron, N.; Guichoux, E.; Schutz, S.; Nectoux, J.; Sorlin, A.; Quere, M.; Boudjarane, J. Performance of semiconductor sequencing platform for non-invasive prenatal genetic screening for fetal aneuploidy: Results from a multicenter prospective cohort study in a clinical setting. Ultrasound Obstet. Gynecol. 2019, 54, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, O.; Ahola, T.; Dahl, F.; Karlsson, F.; Persson, F.; Karlberg, O.; Roos, F.; Alftrén, I.; Andersson, B.; Barkenäs, E. Clinical validation of a novel automated cell-free DNA screening assay for trisomies 21, 13, and 18 in maternal plasma. Prenat. Diagn. 2019, 39, 1011–1015. [Google Scholar] [CrossRef] [PubMed]
- Flöck, A.; Tu, N.-C.; Rüland, A.; Holzgreve, W.; Gembruch, U.; Geipel, A. Non-invasive prenatal testing (NIPT): Europe’s first multicenter post-market clinical follow-up study validating the quality in clinical routine. Arch. Gynecol. Obstet. 2017, 296, 923–928. [Google Scholar] [CrossRef]
- Guex, N.; Iseli, C.; Syngelaki, A.; Deluen, C.; Pescia, G.; Nicolaides, K.; Xenarios, I.; Conrad, B.; Conrad, B. A robust 2nd generation genome-wide test for fetal aneuploidy based on shotgun se-quencing cell-free DNA in maternal blood. Prenat. Diagn. 2013, 33, 707–710. [Google Scholar] [CrossRef]
- Gormus, U.; Chaubey, A.; Shenoy, S.; Wong, Y.W.; Chan, L.Y.; Choo, B.P.; Oraha, L.; Gousseva, A.; Persson, F.; Prensky, L. Assessment and Clinical Utility of a Non-Next-Generation Sequencing-Based Non-Invasive Prenatal Testing Technology. Curr. Issues Mol. Biol. 2021, 43, 958–964. [Google Scholar] [CrossRef]
- Hall, M.P.; Hill, M.; Zimmermann, B.; Sigurjonsson, S.; Westemeyer, M.; Saucier, J.; Demko, Z.; Rabinowitz, M. Non-invasive prenatal detection of trisomy 13 using a single nucleotide polymorphism-and informatics-based approach. PLoS ONE 2014, 9, e96677. [Google Scholar] [CrossRef]
- Hu, H.-J.; Lee, M.-Y.; Cho, D.-Y.; Oh, M.; Kwon, Y.-J.; Han, Y.-J.; Ryu, H.M.; Kim, Y.N.; Won, H.-S. Prospective clinical evaluation of Momguard non-invasive prenatal test in 1011 Korean high-risk pregnant women. J. Obstet. Gynaecol. 2020, 40, 1090–1095. [Google Scholar] [CrossRef]
- Jeon, Y.J.; Zhou, Y.; Li, Y.; Guo, Q.; Chen, J.; Quan, S.; Zhang, A.; Zheng, H.; Zhu, X.; Lin, J. The feasibility study of non-invasive fetal trisomy 18 and 21 detection with semiconductor sequencing platform. PLoS ONE 2014, 9, e110240. [Google Scholar] [CrossRef]
- Jiang, F.; Ren, J.; Chen, F.; Zhou, Y.; Xie, J.; Dan, S.; Su, Y.; Xie, J.; Yin, B.; Su, W. Noninvasive Fetal Trisomy (NIFTY) test: An advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Med. Genom. 2012, 5, 57. [Google Scholar] [CrossRef]
- Ke, W.-L.; Zhao, W.-H.; Wang, X.-Y. Detection of fetal cell-free DNA in maternal plasma for Down syndrome, Edward syndrome and Patau syndrome of high risk fetus. Int. J. Clin. Exp. Med. 2015, 8, 9525. [Google Scholar]
- Koumbaris, G.; Kypri, E.; Tsangaras, K.; Achilleos, A.; Mina, P.; Neofytou, M.; Velissariou, V.; Christopoulou, G.; Kallikas, I.; González-Liñán, A. Cell-free DNA analysis of targeted genomic regions in maternal plasma for non-invasive prenatal testing of trisomy 21, trisomy 18, trisomy 13, and fetal sex. Clin. Chem. 2016, 62, 848–855. [Google Scholar] [CrossRef]
- Kypri, E.; Ioannides, M.; Touvana, E.; Neophytou, I.; Mina, P.; Velissariou, V.; Vittas, S.; Santana, A.; Alexidis, F.; Tsangaras, K. Non-invasive prenatal testing of fetal chromosomal aneuploidies: Validation and clinical performance of the veracity test. Mol. Cytogenet. 2019, 12, 34. [Google Scholar] [CrossRef]
- Langlois, S.; Johnson, J.; Audibert, F.; Gekas, J.; Forest, J.C.; Caron, A.; Harrington, K.; Pastuck, M.; Meddour, H.; Tétu, A. Comparison of first-tier cell-free DNA screening for common aneuploidies with conventional publically funded screening. Prenat. Diagn. 2017, 37, 1238–1244. [Google Scholar] [CrossRef]
- Lau, T.K.; Chen, F.; Pan, X.; Pooh, R.K.; Jiang, F.; Li, Y.; Jiang, H.; Li, X.; Chen, S.; Zhang, X. Non-invasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. J. Matern. Fetal Neonatal Med. 2012, 25, 1370–1374. [Google Scholar] [CrossRef]
- Lee, D.E.; Kim, H.; Park, J.; Yun, T.; Park, D.Y.; Kim, M.; Ryu, H.M. Clinical Validation of Non-Invasive Prenatal Testing for Fetal Common Aneuploidies in 1,055 Korean Pregnant Women: A Single Center Experience. J. Korean Med. Sci. 2019, 34, e172. [Google Scholar] [CrossRef]
- Liang, D.; Lv, W.; Wang, H.; Xu, L.; Liu, J.; Li, H.; Hu, L.; Peng, Y.; Wu, L. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat. Diagn. 2013, 33, 409–415. [Google Scholar] [CrossRef]
- Mazloom, A.R.; Džakula, Ž.; Oeth, P.; Wang, H.; Jensen, T.; Tynan, J.; McCullough, R.; Saldivar, J.S.; Ehrich, M.; van den Boom, D. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma. Prenat. Diagn. 2013, 33, 591–597. [Google Scholar] [CrossRef]
- Miltoft, C.; Rode, L.; Ekelund, C.; Sundberg, K.; Kjaergaard, S.; Zingenberg, H.; Tabor, A. Contingent first-trimester screening for aneuploidies with cell-free DNA in a Danish clinical setting. Ultrasound Obstet. Gynecol. 2018, 51, 470–479. [Google Scholar] [CrossRef]
- Nicolaides, K.H.; Syngelaki, A.; Ashoor, G.; Birdir, C.; Touzet, G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. Am. J. Obstet. Gynecol. 2012, 207, 374.e371–374.e376. [Google Scholar] [CrossRef]
- Nicolaides, K.; Syngelaki, A.; Gil, M.; Atanasova, V.; Markova, D. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X., and Y. Prenat. Diagn. 2013, 33, 575–579. [Google Scholar] [CrossRef]
- Nicolaides, K.H.; Syngelaki, A.; Gil, M.d.M.; Quezada, M.S.; Zinevich, Y. Prenatal detection of fetal triploidy from cell-free DNA testing in maternal blood. Fetal Diagn. Ther. 2014, 35, 212–217. [Google Scholar] [CrossRef]
- Norton, M.E.; Brar, H.; Weiss, J.; Karimi, A.; Laurent, L.C.; Caughey, A.B.; Rodriguez, M.H.; Williams III, J.; Mitchell, M.E.; Adair, C.D. Non-Invasive Chromosomal Evaluation (NICE) Study: Results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012, 207, 137.e131–137.e138. [Google Scholar] [CrossRef]
- Norton, M.E.; Jacobsson, B.; Swamy, G.K.; Laurent, L.C.; Ranzini, A.C.; Brar, H.; Tomlinson, M.W.; Pereira, L.; Spitz, J.L.; Hollemon, D. Cell-free DNA analysis for noninvasive examination of trisomy. N. Engl. J. Med. 2015, 372, 1589–1597. [Google Scholar] [CrossRef]
- Palomaki, G.E.; Deciu, C.; Kloza, E.M.; Lambert-Messerlian, G.M.; Haddow, J.E.; Neveux, L.M.; Ehrich, M.; van den Boom, D.; Bombard, A.T.; Grody, W.W. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: An international collaborative study. Genet. Med. 2012, 14, 296–305. [Google Scholar] [CrossRef]
- Papageorghiou, A.T.; Khalil, A.; Forman, M.; Hulme, R.; Mazey, R.; Mousa, H.A.; Johnstone, E.D.; McKelvey, A.; Cohen, K.E.; Risley, M. Clinical evaluation of the IONA test: A non-invasive prenatal screening test for trisomies 21, 18 and 13. Ultrasound Obstet. Gynecol. 2016, 47, 188–193. [Google Scholar] [CrossRef]
- Persico, N.; Boito, S.; Ischia, B.; Cordisco, A.; De Robertis, V.; Fabietti, I.; Periti, E.; Volpe, P.; Fedele, L.; Rembouskos, G. Cell-free DNA testing in the maternal blood in high-risk pregnancies after first-trimester combined screening. Prenat. Diagn. 2016, 36, 232–236. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Persico, N.; Boito, S.; Volpe, P.; Ischia, B.; Gentile, M.; Ronzoni, L.; De Robertis, V.; Fabietti, I.; Olivieri, C.; Periti, E. Incidence of chromosomal abnormalities in fetuses with first trimester ultrasound anomalies and a low-risk cell-free DNA test for common trisomies. Prenat. Diagn. 2020, 40, 1474–1481. [Google Scholar] [CrossRef] [PubMed]
- Pertile, M.D.; Flowers, N.; Vavrek, D.; Andrews, D.; Kalista, T.; Craig, A.; Deciu, C.; Duenwald, S.; Meier, K.; Bhatt, S. Performance of a paired-end sequencing-based noninvasive prenatal screening test in the detection of genome-wide fetal chromosomal anomalies. Clin. Chem. 2021, 67, 1210–1219. [Google Scholar] [CrossRef] [PubMed]
- Pooh, R.K.; Masuda, C.; Matsushika, R.; Machida, M.; Nakamura, T.; Takeda, M.; Ohashi, H.; Kumagai, M.; Uenishi, K.; Roos, F. Clinical Validation of Fetal cfDNA Analysis Using Roll-ing-Circle-Replication and Imaging Technology in Osaka (CRITO Study). Diagnostics 2021, 11, 1837. [Google Scholar] [CrossRef]
- Poon, L.; Dumidrascu-Diris, D.; Francisco, C.; Fantasia, I.; Nicolaides, K. IONA test for first-trimester detection of trisomies 21, 18 and 13. Ultrasound Obstet. Gynecol. 2016, 47, 184–187. [Google Scholar] [CrossRef]
- Porreco, R.P.; Garite, T.J.; Maurel, K.; Marusiak, B.; Network, O.C.R.; Ehrich, M.; van den Boom, D.; Deciu, C.; Bombard, A. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. Am. J. Obstet. Gynecol. 2014, 211, 365.e1–365.e12. [Google Scholar] [CrossRef]
- Porreco, R.P.; Sekedat, M.; Bombard, A.; Garite, T.J.; Maurel, K.; Marusiak, B.; Adair, D.; Bleich, A.; Combs, C.A.; Kramer, W. Evaluation of a novel screening method for fetal aneuploidy using cell-free DNA in maternal plasma. J. Med. Screen. 2020, 27, 1–8. [Google Scholar] [CrossRef]
- Ryan, A.; Hunkapiller, N.; Banjevic, M.; Vankayalapati, N.; Fong, N.; Jinnett, K.N.; Demko, Z.; Zimmermann, B.; Sigurjonsson, S.; Gross, S.J. Validation of an enhanced version of a single-nucleotide polymorphism-based noninvasive prenatal test for detection of fetal aneuploidies. Fetal Diagn. Ther. 2016, 40, 219–223. [Google Scholar] [CrossRef]
- Sehnert, A.J.; Rhees, B.; Comstock, D.; de Feo, E.; Heilek, G.; Burke, J.; Rava, R.P. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin. Chem. 2011, 57, 1042–1049. [Google Scholar] [CrossRef]
- Shaw, S.S.; Hsiao, C.-H.; Chen, C.-Y.; Ren, Y.; Tian, F.; Tsai, C.; Chen, M.; Cheng, P.-J. Noninvasive prenatal testing for whole fetal chromosomal aneuploidies: A multicenter prospective cohort trial in Taiwan. Fetal Diagn. Ther. 2014, 35, 13–17. [Google Scholar] [CrossRef]
- Song, Y.; Liu, C.; Qi, H.; Zhang, Y.; Bian, X.; Liu, J. Noninvasive prenatal testing of fetal aneuploidies by massively parallel sequencing in a prospective Chinese population. Prenat. Diagn. 2013, 33, 700–706. [Google Scholar] [CrossRef]
- Song, Y.; Huang, S.; Zhou, X.; Jiang, Y.; Qi, Q.; Bian, X.; Zhang, J.; Yan, Y.; Cram, D.; Liu, J. Non-invasive prenatal testing for fetal aneuploidies in the first trimester of pregnancy. Ultrasound Obstet. Gynecol. 2015, 45, 55–60. [Google Scholar] [CrossRef]
- Stokowski, R.; Wang, E.; White, K.; Batey, A.; Jacobsson, B.; Brar, H.; Balanarasimha, M.; Hollemon, D.; Sparks, A.; Nicolaides, K. Clinical performance of non-invasive prenatal testing (NIPT) using targeted cell-free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenat. Diagn. 2015, 35, 1243–1246. [Google Scholar] [CrossRef]
- Stumm, M.; Entezami, M.; Haug, K.; Blank, C.; Wüstemann, M.; Schulze, B.; Raabe-Meyer, G.; Hempel, M.; Schelling, M.; Ostermayer, E. Diagnostic accuracy of random massively parallel sequencing for non-invasive prenatal detection of common autosomal aneuploidies: A collaborative study in Europe. Prenat. Diagn. 2014, 34, 185–191. [Google Scholar] [CrossRef]
- Tsaliki, E.; Papageorgiou, E.A.; Spyrou, C.; Koumbaris, G.; Kypri, E.; Kyriakou, S.; Sotiriou, C.; Touvana, E.; Keravnou, A.; Karagrigoriou, A. MeDIP real-time qPCR of maternal peripheral blood reliably identifies trisomy 21. Prenat. Diagn. 2012, 32, 996–1001. [Google Scholar] [CrossRef]
- Tynan, J.; Kim, S.; Mazloom, A.; Zhao, C.; McLennan, G.; Tim, R.; Liu, L.; Hannum, G.; Hull, A.; Bombard, A. Application of risk score analysis to low-coverage whole genome sequencing data for the noninvasive detection of trisomy 21, trisomy 18, and trisomy 13. Prenat. Diagn. 2016, 36, 56–62. [Google Scholar] [CrossRef]
- Verweij, E.; Jacobsson, B.; Van Scheltema, P.; de Boer, M.; Hoffer, M.; Hollemon, D.; Westgren, M.; Song, K.; Oepkes, D. European non-invasive trisomy evaluation (EU-NITE) study: A multicenter prospective cohort study for non-invasive fetal trisomy 21 testing. Prenat. Diagn. 2013, 33, 996–1001. [Google Scholar] [CrossRef]
- Alyafee, Y.; Al Tuwaijri, A.; Alam, Q.; Umair, M.; Haddad, S.; Alharbi, M.; Ballow, M.; Al Drees, M.; AlAbdulrahman, A.; Al Khaldi, A.; et al. Next Generation Sequencing Based Non-invasive Prenatal Testing (NIPT): First Report From Saudi Arabia. Front. Genet. 2021, 12, 630787. [Google Scholar] [CrossRef]
- Bajka, A.; Bajka, M.; Chablais, F.; Burkhardt, T. Audit of the first > 7500 noninvasive prenatal aneuploidy tests in a Swiss genetics center. Arch. Gynecol. Obs. 2022, 305, 1185–1192. [Google Scholar] [CrossRef]
- Beamon, C.J.; Hardisty, E.E.; Harris, S.C.; Vora, N.L. A single center’s experience with noninvasive prenatal testing. Genet. Med. 2014, 16, 681–687. [Google Scholar] [CrossRef]
- Bianchi, D.W.; Parsa, S.; Bhatt, S.; Halks-Miller, M.; Kurtzman, K.; Sehnert, A.J.; Swanson, A. Fetal sex chromosome testing by maternal plasma DNA sequencing: Clinical laboratory experience and biology. Obstet. Gynecol. 2015, 125, 375–382. [Google Scholar] [CrossRef]
- Borth, H.; Teubert, A.; Glaubitz, R.; Knippenberg, S.; Kutur, N.; Winkler, T.; Eiben, B. Analysis of cell-free DNA in a consecutive series of 13,607 routine cases for the detection of fetal chromosomal aneuploidies in a single center in Germany. Arch. Gynecol. Obstet. 2021, 303, 1407–1414. [Google Scholar] [CrossRef]
- Bu, J.; Jiang, P.; Cui, X.; Zhou, H.; Han, F. Application values of prenatal screening and non-invasive gene sequencing in fetal birth defects. Pak. J. Med. Sci. 2020, 36, 1545–1549. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, Q.; Mao, X.; Lei, W.; He, M.; Lu, W. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 42,910 single pregnancies with different clinical features. Hum. Genom. 2019, 13, 60. [Google Scholar] [CrossRef]
- Comas, C.; Echevarria, M.; Rodriguez, M.A.; Prats, P.; Rodriguez, I.; Serra, B. Initial experience with non-invasive prenatal testing of cell-free DNA for major chromosomal anomalies in a clinical setting. J. Matern. Fetal Neonatal Med. 2015, 28, 1196–1201. [Google Scholar] [CrossRef]
- Dar, P.; Curnow, K.J.; Gross, S.J.; Hall, M.P.; Stosic, M.; Demko, Z.; Zimmermann, B.; Hill, M.; Sigurjonsson, S.; Ryan, A.; et al. Clinical experience and follow-up with large scale single-nucleotide polymorphism-based noninvasive prenatal aneuploidy testing. Am. J. Obstet. Gynecol. 2014, 211, 527.e1–527.e17. [Google Scholar] [CrossRef]
- Dai, R.; Yu, Y.; Zhang, H.; Li, L.; Jiang, Y.; Liu, R.; Zhang, H. Analysis of 17,428 pregnant women undergoing non-invasive prenatal testing for fetal chromosome in Northeast China. Medicine 2021, 100, e24740. [Google Scholar] [CrossRef]
- Dan, S.; Wang, W.; Ren, J.; Li, Y.; Hu, H.; Xu, Z.; Lau, T.K.; Xie, J.; Zhao, W.; Huang, H.; et al. Clinical application of massively parallel sequencing-based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenat. Diagn. 2012, 32, 1225–1232. [Google Scholar] [CrossRef]
- Du, Y.; Lin, J.; Lan, L.; Dong, Y.; Zhu, J.; Jiang, W.; Pan, X.; Lu, Y.; Li, D.; Wang, L. Detection of chromosome abnormalities using current noninvasive prenatal testing: A multi-center comparative study. Biosci. Trends 2018, 12, 317–324. [Google Scholar] [CrossRef]
- Eiben, B.; Krapp, M.; Borth, H.; Kutur, N.; Kreiselmaier, P.; Glaubitz, R.; Deutinger, J.; Merz, E. Single Nucleotide Polymorphism-Based Analysis of Cell-Free Fetal DNA in 3000 Cases from Germany and Austria. Ultrasound Int. Open 2015, 1, E8–E11. [Google Scholar] [CrossRef][Green Version]
- Fairbrother, G.; Johnson, S.; Musci, T.J.; Song, K. Clinical experience of noninvasive prenatal testing with cell-free DNA for fetal trisomies 21, 18, and 13, in a general screening population. Prenat. Diagn. 2013, 33, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Fiorentino, F.; Bono, S.; Pizzuti, F.; Duca, S.; Polverari, A.; Faieta, M.; Baldi, M.; Diano, L.; Spinella, F. The clinical utility of genome-wide non invasive prenatal screening. Prenat. Diagn. 2017, 37, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Futch, T.; Spinosa, J.; Bhatt, S.; de Feo, E.; Rava, R.P.; Sehnert, A.J. Initial clinical laboratory expe-rience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenat. Diagn. 2013, 33, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Garshasbi, M.; Wang, Y.; Hantoosh Zadeh, S.; Giti, S.; Piri, S.; Reza Hekmat, M. Clinical Application of Cell-Free DNA Sequencing-Based Noninvasive Prenatal Testing for Trisomies 21, 18, 13 and Sex Chromosome Aneuploidy in a Mixed-Risk Population in Iran. Fetal Diagn. Ther. 2020, 47, 220–227. [Google Scholar] [CrossRef]
- Ge, Y.; Li, J.; Zhuang, J.; Zhang, J.; Huang, Y.; Tan, M.; Li, W.; Chen, J.; Zhou, Y. Expanded noninvasive prenatal testing for fetal aneuploidy and copy number variations and parental willingness for invasive diagnosis in a cohort of 18,516 cases. BMC Med. Genom. 2021, 14, 106. [Google Scholar] [CrossRef]
- Gil, M.M.; Quezada, M.S.; Bregant, B.; Ferraro, M.; Nicolaides, K.H. Implementation of maternal blood cell-free DNA testing in early screening for aneuploidies. Ultrasound Obstet. Gynecol. 2013, 42, 34–40. [Google Scholar] [CrossRef]
- Gil, M.M.; Brik, M.; Casanova, C.; Martin-Alonso, R.; Verdejo, M.; Ramirez, E.; Santacruz, B. Screening for trisomies 21 and 18 in a Spanish public hospital: From the combined test to the cell-free DNA test. J. Matern. Fetal Neonatal Med. 2017, 30, 2476–2482. [Google Scholar] [CrossRef]
- Guy, C.; Haji-Sheikhi, F.; Rowland, C.M.; Anderson, B.; Owen, R.; Lacbawan, F.L.; Alagia, D.P. Prenatal cell-free DNA screening for fetal aneuploidy in pregnant women at average or high risk: Results from a large US clinical laboratory. Mol. Genet. Genomic. Med. 2019, 7, e545. [Google Scholar] [CrossRef]
- Guy, G.P.; Hargrave, J.; Dunn, R.; Price, K.; Short, J.; Thilaganathan, B.; The SAFE test collaborative. Secondary non-invasive prenatal screening for fetal trisomy: An effectiveness study in a public health setting. BJOG 2021, 128, 440–446. [Google Scholar] [CrossRef]
- Harasim, T.; Neuhann, T.; Behnecke, A.; Stampfer, M.; Holinski-Feder, E.; Abicht, A. Initial Clinical Experience with NIPT for Rare Autosomal Aneuploidies and Large Copy Number Variations. J. Clin. Med. 2022, 11, 372. [Google Scholar] [CrossRef]
- Hong, S.Y.; Shim, S.H.; Park, H.J.; Shim, S.S.; Kim, J.Y.; Cho, Y.K.; Kim, S.H.; Cha, D.H. Experiences and efficacy of noninvasive prenatal test using maternal plasma in single center: 1,591 cases. J. Genet. Med. 2020, 17, 11–15. [Google Scholar] [CrossRef]
- Hu, H.; Wang, L.; Wu, J.; Zhou, P.; Fu, J.; Sun, J.; Cai, W.; Liu, H.; Yang, Y. Noninvasive prenatal testing for chromosome aneuploidies and subchromosomal microdeletions/microduplications in a cohort of 8141 single pregnancies. Hum. Genom. 2019, 13, 14. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.; Hamar, B.; Lazar, E.; Lim, K.; Rodriguez, D.; Stock, K.; Wolfberg, A.J.; Dunk, R. Nuchal translucency measurement plus non-invasive prenatal testing to screen for aneuploidy in a community-based average-risk population. Ultrasound Obstet. Gynecol. 2014, 44, 491. [Google Scholar] [CrossRef] [PubMed]
- Kagan, K.O.; Hoopmann, M.; Pfaff, T.; Prodan, N.; Wagner, P.; Schmid, M.; Dufke, A.; Mau-Holzmann, U.; Brucker, S.; Marcato, L.; et al. First Trimester Screening for Common Trisomies and Microdeletion 22q11.2 Syndrome Using Cell-Free DNA: A Prospective Clinical Study. Fetal Diagn. Ther 2020, 47, 841–852. [Google Scholar] [CrossRef]
- Koc, A.; Ozer Kaya, O.; Ozyilmaz, B.; Kutbay, Y.B.; Kirbiyik, O.; Ozdemir, T.R.; Erdogan, K.M.; Saka Guvenc, M.; Oztekin, D.C.; Ozeren, M.; et al. Targeted fetal cell-free DNA screening for aneuploidies in 4,594 pregnancies: Single center study. Mol. Genet. Genom. Med. 2019, 7, e00678. [Google Scholar] [CrossRef]
- Korostelev, S.; Totchiev, G.; Kanivets, I.; Gnetetskaya, V. Association of non-invasive prenatal testing and chromosomal microarray analysis for prenatal diagnostics. Gynecol. Endocrinol. 2014, 30 (Suppl. S1), 13–16. [Google Scholar] [CrossRef]
- La Verde, M.; De Falco, L.; Torella, A.; Savarese, G.; Savarese, P.; Ruggiero, R.; Conte, A.; Fico, V.; Torella, M.; Fico, A. Performance of cell-free DNA sequencing-based non-invasive prenatal testing: Experience on 36,456 singleton and multiple pregnancies. BMC Med. Genom. 2021, 14, 93. [Google Scholar] [CrossRef]
- Lai, Y.; Zhu, X.; He, S.; Dong, Z.; Tang, Y.; Xu, F.; Chen, Y.; Meng, L.; Tao, Y.; Yi, S.; et al. Performance of Cell-Free DNA Screening for Fetal Common Aneuploidies and Sex Chromosomal Abnormalities: A Prospective Study from a Less Developed Autonomous Region in Mainland China. Genes 2021, 12, 478. [Google Scholar] [CrossRef]
- Lau, T.K.; Cheung, S.W.; Lo, P.S.; Pursley, A.N.; Chan, M.K.; Jiang, F.; Zhang, H.; Wang, W.; Jong, L.F.; Yuen, O.K.; et al. Non-invasive prenatal testing for fetal chromosomal abnormalities by low-coverage whole-genome sequencing of maternal plasma DNA: Review of 1982 consecutive cases in a single center. Ultrasound Obs. Gynecol. 2014, 43, 254–264. [Google Scholar] [CrossRef]
- Liang, D.; Cram, D.S.; Tan, H.; Linpeng, S.; Liu, Y.; Sun, H.; Zhang, Y.; Tian, F.; Zhu, H.; Xu, M.; et al. Clinical utility of noninvasive prenatal screening for expanded chromosome disease syndromes. Genet. Med. 2019, 21, 1998–2006. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, H.; He, Y.; Xu, W.; Ma, Q.; He, Y.; Lei, W.; Chen, G.; He, Z.; Huang, J.; et al. Clinical performance of non-invasive prenatal served as a first-tier screening test for trisomy 21, 18, 13 and sex chromosome aneuploidy in a pilot city in China. Hum. Genom. 2020, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Huang, T.; Wang, X.R.; Zhou, J.H.; Yuan, H.Z.; Yang, Y.; Huang, T.T.; Liu, D.P.; Liu, Y.Q. Next-generation sequencing: A follow-up of 36,913 singleton pregnancies with noninvasive prenatal testing in central China. J. Assist. Reprod. Genet. 2020, 37, 3143–3150. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, C.; Sun, Y.; Tang, J.; Tong, K.; Zhu, J. Noninvasive prenatal testing for assessing foetal sex chromosome aneuploidy: A retrospective study of 45,773 cases. Mol. Cytogenet. 2021, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, S.; Linpeng, S.; Ding, S.; Li, S.; Li, Y.; Shi, L.; He, J.; Liu, Y. Cell-Free DNA Screening for Sex Chromosome Abnormalities and Pregnancy Outcomes, 2018-2020: A Retrospective Analysis. J. Pers. Med. 2022, 12, 48. [Google Scholar] [CrossRef]
- Lund, I.C.B.; Petersen, O.B.; Becher, N.H.; Lildballe, D.L.; Jorgensen, F.S.; Ambye, L.; Skibsted, L.; Ernst, A.; Jensen, A.N.; Fagerberg, C.; et al. National data on the early clinical use of non-invasive prenatal testing in public and private healthcare in Denmark 2013–2017. Acta Obstet. Gynecol. Scand. 2021, 100, 884–892. [Google Scholar] [CrossRef]
- Luo, Y.; Hu, H.; Jiang, L.; Ma, Y.; Zhang, R.; Xu, J.; Pan, Y.; Long, Y.; Yao, H.; Liang, Z. A retrospective analysis the clinic data and follow-up of non-invasive prenatal test in detection of fetal chromosomal aneuploidy in more than 40,000 cases in a single prenatal diagnosis center. Eur. J. Med. Genet. 2020, 63, 104001. [Google Scholar] [CrossRef]
- Luo, Y.; Hu, H.; Zhang, R.; Ma, Y.; Pan, Y.; Long, Y.; Hu, B.; Yao, H.; Liang, Z. An assessment of the analytical performance of non-invasive prenatal testing (NIPT) in detecting sex chromosome aneuploidies: 34,717-patient sample in a single prenatal diagnosis Centre in China. J. Gene Med. 2021, 23, e3362. [Google Scholar] [CrossRef]
- Luthgens, K.; Grati, F.R.; Sinzel, M.; Habig, K.; Kagan, K.O. Confirmation rate of cell free DNA screening for sex chromosomal abnormalities according to the method of confirmatory testing. Prenat. Diagn. 2021, 41, 1258–1263. [Google Scholar] [CrossRef]
- Manotaya, S.; Xu, H.; Uerpairojkit, B.; Chen, F.; Charoenvidhya, D.; Liu, H.; Petcharaburanin, N.; Liu, Y.; Tang, S.; Wang, X.; et al. Clinical experience from Thailand: Noninvasive prenatal testing as screening tests for trisomies 21, 18 and 13 in 4736 pregnancies. Prenat. Diagn. 2016, 36, 224–231. [Google Scholar] [CrossRef]
- Margiotti, K.; Cesta, A.; Dello Russo, C.; Cima, A.; Barone, M.A.; Viola, A.; Sparacino, D.; Mesoraca, A.; Giorlandino, C. Cell-free DNA screening for sex chromosomal aneuploidies in 9985 pregnancies: Italian single experience. BMC Res. Notes 2020, 13, 167. [Google Scholar] [CrossRef]
- Mesoraca, A.; Margiotti, K.; Dello Russo, C.; Cesta, A.; Cima, A.; Longo, S.A.; Barone, M.A.; Viola, A.; Sparacino, D.; Giorlandino, C. Cell-free DNA screening for aneuploidies in 7113 pregnancies: Single Italian centre study. Genet. Res. 2020, 102, e5. [Google Scholar] [CrossRef]
- Noh, J.J.; Ryu, H.M.; Oh, S.Y.; Choi, S.J.; Roh, C.R.; Kim, J.H. A two-year experience of non-invasive prenatal testing (NIPT) at an urban tertiary medical center in South Korea. Taiwan J. Obs. Gynecol. 2019, 58, 545–551. [Google Scholar] [CrossRef]
- Oepkes, D.; Page-Christiaens, G.C.; Bax, C.J.; Bekker, M.N.; Bilardo, C.M.; Boon, E.M.; Schuring-Blom, G.H.; Coumans, A.B.; Faas, B.H.; Galjaard, R.H.; et al. Trial by Dutch laboratories for evaluation of non-invasive prenatal testing. Part I-Clin. Impact Prenat. Diagn. 2016, 36, 1083–1090. [Google Scholar] [CrossRef]
- Oneda, B.; Sirleto, P.; Baldinger, R.; Taralczak, M.; Joset, P.; Zweier, M.; Niedrist, D.; Azzarello-Burri, S.; Britschgi, C.; Breymann, C.; et al. Genome-wide non-invasive prenatal testing in single- and mul-tiple-pregnancies at any risk: Identification of maternal polymorphisms to reduce the number of un-necessary invasive confirmation testing. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 252, 19–29. [Google Scholar] [CrossRef]
- Palomaki, G.E.; Kloza, E.M.; O’Brien, B.M.; Eklund, E.E.; Lambert-Messerlian, G.M. The clinical utility of DNA-based screening for fetal aneuploidy by primary obstetrical care providers in the general pregnancy population. Genet. Med. 2017, 19, 778–786. [Google Scholar] [CrossRef]
- Panchalee, T.; Poungvarin, N.; Amornrit, W.; Pooliam, J.; Taluengjit, P.; Wataganara, T. Clinical performance of DNA-based prenatal screening using single-nucleotide polymorphisms approach in Thai women with singleton pregnancy. Mol. Genet. Genom. Med. 2020, 8, e1256. [Google Scholar] [CrossRef]
- Pang, Y.; Wang, C.; Tang, J.; Zhu, J. Clinical application of noninvasive prenatal testing in the detection of fetal chromosomal diseases. Mol. Cytogenet. 2021, 14, 31. [Google Scholar] [CrossRef]
- Pavanello, E.; Sciarrone, A.; Guaraldo, V.; Muccinelli, E.; Ciuffreda, V.P.; Sauro, P.; Bondielli, G.; Mirante, S.; Mengozzi, G.; Viora, E.; et al. Cell-free DNA screening for fetal aneuploidy using the rolling circle method: A step towards non invasive prenatal testing simplification. Prenat. Diagn. 2021, 41, 1694–1700. [Google Scholar] [CrossRef]
- Pescia, G.; Guex, N.; Iseli, C.; Brennan, L.; Osteras, M.; Xenarios, I.; Farinelli, L.; Conrad, B. Cell-free DNA testing of an extended range of chromosomal anomalies: Clinical experience with 6,388 consecutive cases. Genet. Med. 2017, 19, 169–175. [Google Scholar] [CrossRef]
- Qi, Q.G.; Tuo, Y.; Liu, L.X.; Yu, C.X.; Wu, A.N. Amniocentesis and Next Generation Sequencing (NGS)-Based Noninvasive Prenatal DNA Testing (NIPT) for Prenatal Diagnosis of Fetal Chromosomal Disorders. Int. J. Gen. Med. 2021, 14, 1811–1817. [Google Scholar] [CrossRef]
- Quezada, M.S.; Gil, M.M.; Francisco, C.; Orosz, G.; Nicolaides, K.H. Screening for trisomies 21, 18 and 13 by cell-free DNA analysis of maternal blood at 10-11 weeks’ gestation and the combined test at 11–13 weeks. Ultrasound Obstet. Gynecol. 2015, 45, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Radoi, V.E.; Bohiltea, C.L.; Bohiltea, R.E.; Albu, D.N. Cell free fetal DNA testing in maternal blood of Romanian pregnant women. Iran. J. Reprod. Med. 2015, 13, 623–626. [Google Scholar] [PubMed]
- Sainz, J.A.; Torres, M.R.; Peral, I.; Granell, R.; Vargas, M.; Carrasco, P.; Garcia-Mejido, J.A.; Santacruz, B.; Gil, M.M. Clinical and Economic Evaluation after Adopting Contingent Cell-Free DNA Screening for Fetal Trisomies in South Spain. Fetal Diagn. Ther. 2020, 47, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Santamaria, R.; Bermejo, B.; Cigarrán, S.; Benn, P. A National Referral Laboratory’s Experience with the Implementation of SNP-Based Non-invasive Prenatal Screening for Fetal Aneuploidy and Select Microdeletion Syndromes. J. Fetal Med. 2018, 5, 7–12. [Google Scholar] [CrossRef]
- Sasaki, Y.; Yamada, T.; Tanaka, S.; Sekizawa, A.; Hirose, T.; Suzumori, N.; Kaji, T.; Kawaguchi, S.; Hasuo, Y.; Nishizawa, H.; et al. Evaluation of the clinical performance of noninvasive prenatal testing at a Japanese laboratory. J. Obstet. Gynaecol. Res. 2021, 47, 3437–3446. [Google Scholar] [CrossRef]
- Serapinas, D.; Boreikaite, E.; Bartkeviciute, A.; Norvilaite, K.; Narbekovas, A.; Bartkeviciene, D. The Level of Free Fetal DNA as Precise Noninvasive Marker for Chromosomal Aneuploidies: First Results from BALTIC Region. Medicina 2020, 56, 579. [Google Scholar] [CrossRef]
- Song, J.P.; Jiang, Y.F.; Gao, T.X.; Yao, Y.Y.; Liu, L.J.; Xu, R.H.; Yi, M.Q.; Yu, C.J.; Wang, W.P.; Li, H. Performance of non-invasive prenatal screening for sex chromosome aneuploidies and parental decision-making. Chin. Med. J. 2020, 133, 1617–1619. [Google Scholar] [CrossRef]
- Strom, C.M.; Anderson, B.; Tsao, D.; Zhang, K.; Liu, Y.; Livingston, K.; Elzinga, C.; Evans, M.; Nguyen, Q.; Wolfson, D. Improving the positive predictive value of non-invasive prenatal screening (NIPS). PLoS ONE 2017, 12, e0167130. [Google Scholar] [CrossRef]
- Togneri, F.S.; Kilby, M.D.; Young, E.; Court, S.; Williams, D.; Griffiths, M.J.; Allen, S.K. Implementation of cell-free DNA-based non-invasive prenatal testing in a National Health Service Regional Genetics Laboratory. Genet. Res. 2019, 101, e11. [Google Scholar] [CrossRef]
- van der Meij, K.R.M.; Sistermans, E.A.; Macville, M.V.E.; Stevens, S.J.C.; Bax, C.J.; Bekker, M.N.; Bilardo, C.M.; Boon, E.M.J.; Boter, M.; Diderich, K.E.M.; et al. TRIDENT-2: National Implementation of Genome-wide Non-invasive Prenatal Testing as a First-Tier Screening Test in the Netherlands. Am. J. Hum. Genet. 2019, 105, 1091–1101. [Google Scholar] [CrossRef]
- Verma, I.C.; Puri, R.; Venkataswamy, E.; Tayal, T.; Nampoorthiri, S.; Andrew, C.; Kabra, M.; Bagga, R.; Gowda, M.; Batra, M.; et al. Single Nucleotide Polymorphism-Based Noninvasive Prenatal Testing: Experience in India. J. Obstet. Gynaecol. India. 2018, 68, 462–470. [Google Scholar] [CrossRef]
- Wan, J.H.; Zhen, L.; Han, J.; Pan, M.; Yang, X.; Li, D.Z. Use of noninvasive prenatal screening with cell-free DNA in late pregnancy with sonographic soft markers. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 252, 431–433. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.; Wang, W.; Dong, Y.; Zhang, M.; Wang, X.; Yin, C. Cell-free DNA screening for sex chromosome aneuploidies by non-invasive prenatal testing in maternal plasma. Mol. Cytogenet. 2020, 13, 10. [Google Scholar] [CrossRef]
- Wang, C.; Tang, J.; Tong, K.; Huang, D.; Tu, H.; Li, Q.; Zhu, J. Expanding the application of non-invasive prenatal testing in the detection of foetal chromosomal copy number variations. BMC Med. Genom. 2021, 14, 292. [Google Scholar] [CrossRef]
- Wang, J.W.; Lyu, Y.N.; Qiao, B.; Li, Y.; Zhang, Y.; Dhanyamraju, P.K.; Bamme, Y.; Yu, M.D.; Yang, D.; Tong, Y.Q. Cell-free fetal DNA testing and its correlation with prenatal indications. BMC Pregnancy Childbirth 2021, 21, 585. [Google Scholar] [CrossRef]
- Willems, P.J.; Dierickx, H.; Vandenakker, E.; Bekedam, D.; Segers, N.; Deboulle, K.; Vereecken, A. The first 3000 Non-Invasive Prenatal Tests (NIPT) with the Harmony test in Belgium and the Neth-erlands. Facts Views Vis. Obgyn. 2014, 6, 7–12. [Google Scholar]
- Xu, L.; Huang, H.; Lin, N.; Wang, Y.; He, D.; Zhang, M.; Chen, M.; Chen, L.; Lin, Y. Non-invasive cell-free fetal DNA testing for aneuploidy: Multicenter study of 31 515 singleton pregnancies in southeastern China. Ultrasound Obstet. Gynecol. 2020, 55, 242–247. [Google Scholar] [CrossRef]
- Xu, Y.; Jin, P.; Lei, Y.; Qian, Y.; Xu, Y.; Wang, M.; Jin, J.; Yin, Y.; Dong, M. Clinical Efficiency of Non-invasive Prenatal Screening for Common Trisomies in Low-Risk and Twin Pregnancies. Front. Genet. 2021, 12, 661884. [Google Scholar] [CrossRef]
- Xue, Y.; Zhao, G.; Li, H.; Zhang, Q.; Lu, J.; Yu, B.; Wang, T. Non-invasive prenatal testing to detect chromosome aneuploidies in 57,204 pregnancies. Mol. Cytogenet. 2019, 12, 29. [Google Scholar] [CrossRef]
- Yang, J.; Hou, Y.; Guo, F.; Peng, H.; Wang, D.; Li, Y.; Oy, H.; Wang, Y.; Lu, J.; Yin, A. Noninvasive prenatal detection of fetal sex chromosome abnormalities using the semiconductor sequencing platform (SSP) in Southern China. J. Assist. Reprod. Genet. 2021, 38, 727–734. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, Y.; Jiang, F.; Fu, M.; Yuan, Y.; Guo, Y.; Zhu, Z.; Lin, M.; Liu, Q.; Tian, Z.; et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: Clinical experience from 146,958 pregnancies. Ultrasound Obstet. Gynecol. 2015, 45, 530–538. [Google Scholar] [CrossRef]
- Zheng, J.; Lu, H.; Li, M.; Guan, Y.; Yang, F.; Xu, M.; Dong, J.; Zhang, Q.; An, N.; Zhou, Y. The Clinical Utility of Non-invasive Prenatal Testing for Pregnant Women with Different Diagnostic Indications. Front. Genet. 2020, 11, 624. [Google Scholar] [CrossRef]
- Zheng, Y.; Wan, S.; Dang, Y.; Song, T.; Chen, B.; Zhang, J. Clinical experience regarding the accuracy of NIPT in the detection of sex chromosome abnormality. J. Gene Med. 2020, 22, e3199. [Google Scholar] [CrossRef]
- Zhu, H.; Jin, X.; Xu, Y.; Zhang, W.; Liu, X.; Jin, J.; Qian, Y.; Dong, M. Efficiency of non-invasive prenatal screening in pregnant women at advanced maternal age. BMC Pregnancy Childbirth 2021, 21, 86. [Google Scholar] [CrossRef]
- Dougan, S.D.; Okun, N.; Bellai-Dussault, K.; Meng, L.; Howley, H.E.; Huang, T.; Reszel, J.; Lanes, A.; Walker, M.C.; Armour, C.M. Performance of a universal prenatal screening program incorporating cell-free fetal DNA analysis in Ontario, Canada. CMAJ 2021, 193, E1156–E1163. [Google Scholar] [CrossRef]
- Gug, C.; Mozos, I.; Ratiu, A.; Tudor, A.; Gorduza, E.V.; Caba, L.; Gug, M.; Cojocariu, C.; Furau, C.; Furau, G.; et al. Genetic Counseling and Management: The First Study to Report NIPT Findings in a Romanian Population. Medicina 2022, 58, 79. [Google Scholar] [CrossRef]
- McCullough, R.M.; Almasri, E.A.; Guan, X.; Geis, J.A.; Hicks, S.C.; Mazloom, A.R.; Deciu, C.; Oeth, P.; Bombard, A.T.; Paxton, B.; et al. Non-invasive prenatal chromosomal aneuploidy testing--clinical experience: 100,000 clinical samples. PLoS ONE 2014, 9, e109173. [Google Scholar] [CrossRef]
- Meck, J.M.; Kramer Dugan, E.; Matyakhina, L.; Aviram, A.; Trunca, C.; Pineda-Alvarez, D.; Aradhya, S.; Klein, R.T.; Cherry, A.M. Noninvasive prenatal screening for aneuploidy: Positive predictive values based on cytogenetic findings. Am. J. Obstet. Gynecol. 2015, 213, 214.e1–214.e5. [Google Scholar] [CrossRef]
- Saito, M.; Tokunaka, M.; Goto, M.; Takita, H.; Arakaki, T.; Miyagami, K.; Hamada, S.; Oba, T.; Matsuoka, R.; Sekizawa, A. The role of first-trimester ultrasound screening for women with positive noninvasive prenatal testing results. J. Obstet. Gynaecol. Res. 2022, 48, 328–332. [Google Scholar] [CrossRef]
- Suzumori, N.; Sekizawa, A.; Takeda, E.; Samura, O.; Sasaki, A.; Akaishi, R.; Wada, S.; Hamanoue, H.; Hirahara, F.; Sawai, H.; et al. Retrospective details of false-positive and false-negative results in non-invasive prenatal testing for fetal trisomies 21, 18 and 13. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021, 256, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Tekesin, I. Cell-free DNA Testing in Routine Practice: Characterisation of a Cohort with Positive Results for Trisomies, Sex Chromosome Anomalies and Microdeletions. Geburtshilfe Frauenheilkd 2021, 81, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Togneri, F.S.; Allen, S.K.; Mann, K.; Holgado, E.; Morgan, S. Cytogenomic results following high-chance non-invasive prenatal testing: A UK national audit. Genet. Res. 2020, 102, e7. [Google Scholar] [CrossRef] [PubMed]
- Valderramos, S.G.; Rao, R.R.; Scibetta, E.W.; Silverman, N.S.; Han, C.S.; Platt, L.D. Cell-free DNA screening in clinical practice: Abnormal autosomal aneuploidy and microdeletion results. Am. J. Obstet. Gynecol. 2016, 215, 626.e1–626.e10. [Google Scholar] [CrossRef] [PubMed]
- Van Den Bogaert, K.; Lannoo, L.; Brison, N.; Gatinois, V.; Baetens, M.; Blaumeiser, B.; Boemer, F.; Bourlard, L.; Bours, V.; De Leener, A.; et al. Outcome of publicly funded nationwide first-tier noninvasive prenatal screening. Genet. Med. 2021, 23, 1137–1142. [Google Scholar] [CrossRef]
- Wang, J.C.; Sahoo, T.; Schonberg, S.; Kopita, K.A.; Ross, L.; Patek, K.; Strom, C.M. Discordant noninvasive prenatal testing and cytogenetic results: A study of 109 consecutive cases. Genet. Med. 2015, 17, 234–236. [Google Scholar] [CrossRef]
- Wu, X.; Li, Y.; Xie, X.; Su, L.; Cai, M.; Lin, N.; Du, S.; Xu, L.; Huang, H. Clinical Review of Noninvasive Prenatal Testing: Experience from 551 Pregnancies with Noninvasive Prenatal Testing-Positive Results in a Tertiary Referral Center. J. Mol. Diagn. 2020, 22, 1469–1475. [Google Scholar] [CrossRef]
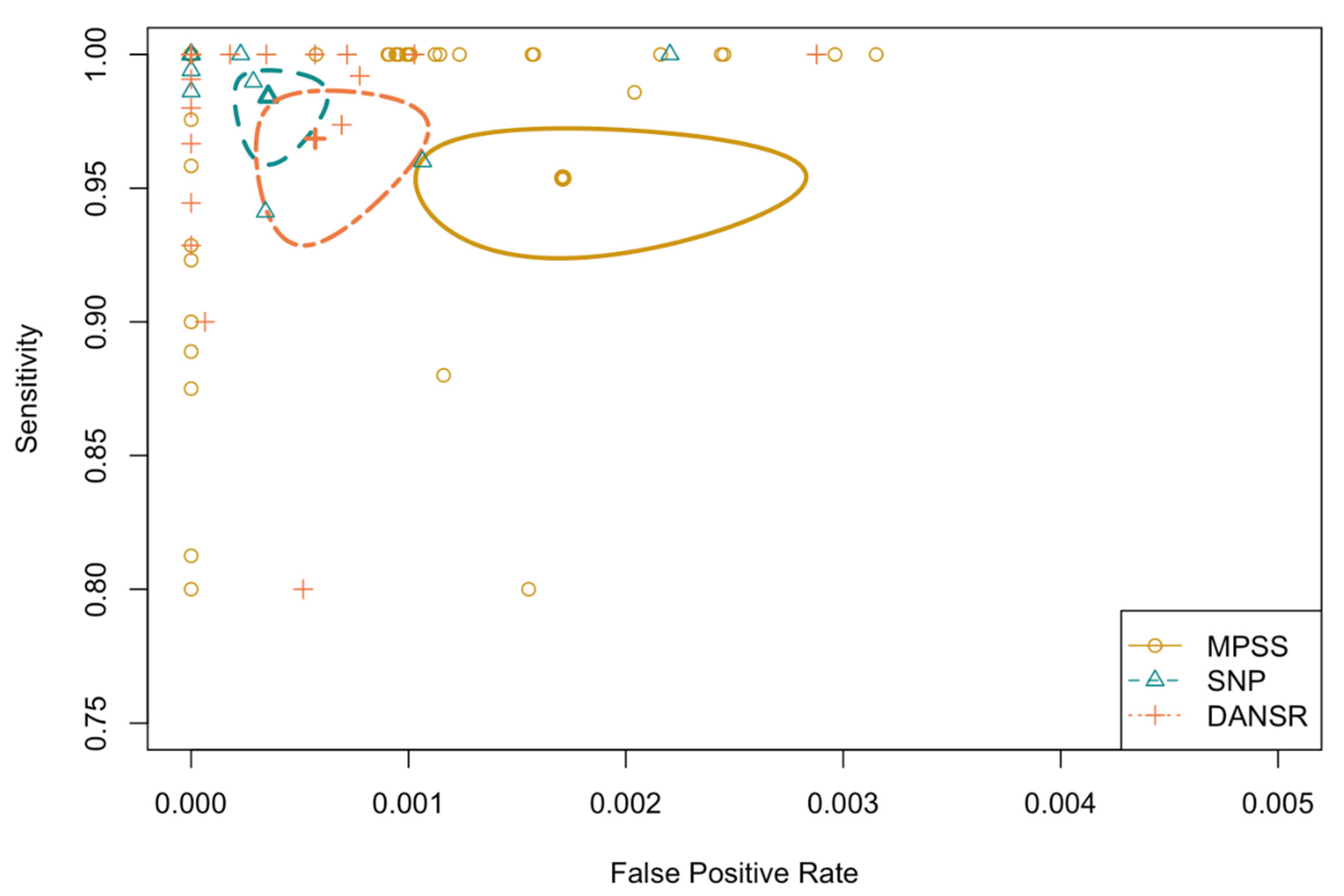
| Syndrome | Method | No. Called | Prevalence | Sensitivity | Specificity | PPV | Std PPV |
|---|---|---|---|---|---|---|---|
| T21 | All | 74697 | 3.35 (3.22, 3.48) | 99.44 (99.06, 99.67) | 99.93 (99.91, 99.95) | 98.03 (97.41, 98.50) | 79.78 (78.21, 81.34) |
| MPSS | 24308 | 5.14 (4.87, 5.42) | 99.68 (99.18, 99.88) | 99.88 (99.83, 99.92) | 97.88 (96.93, 98.54) | 70.05 (67.53, 72.57) | |
| DANSR | 26208 | 1.91 (1.75, 2.08) | 99.20 (97.96, 99.69) | 99.96 (99.92, 99.98) | 97.83 (96.16, 98.78) | 86.43 (83.45, 89.41) | |
| SNP | 19822 | 2.12 (1.93, 2.33) | 99.29 (97.92, 99.76) | 99.97 (99.94, 99.99) | 98.82 (97.26, 99.49) | 91.37 (88.69, 94.05) | |
| T18 | All | 72847 | 1.11 (1.04, 1.19) | 96.43 (94.92, 97.50) | 99.92 (99.90, 99.94) | 93.21 (91.31, 94.73) | 50.22 (46.84, 53.60) |
| MPSS | 23508 | 1.77 (1.61, 1.95) | 95.68 (93.28, 97.25) | 99.89 (99.84, 99.93) | 94.10 (91.44, 95.97) | 42.27 (37.57, 46.97) | |
| DANSR | 25619 | 0.67 (0.57, 0.77) | 97.66 (94.14, 99.09) | 99.96 (99.93, 99.98) | 94.35 (89.91, 96.90) | 67.27 (60.35, 74.18) | |
| SNP | 19823 | 0.53 (0.44, 0.65) | 98.11 (93.38, 99.48) | 99.96 (99.93, 99.98) | 93.69 (87.55, 96.91) | 69.60 (61.04, 78.16) | |
| T13 | All | 64640 | 0.50 (0.44, 0.55) | 97.19 (94.74, 98.51) | 99.94 (99.91, 99.95) | 88.35 (84.58, 91.30) | 28.94 (24.20, 33.68) |
| MPSS | 23470 | 0.72 (0.62, 0.84) | 96.47 (92.51, 98.37) | 99.88 (99.83, 99.92) | 85.42 (79.73, 89.71) | 17.66 (12.26, 23.05) | |
| DANSR | 17629 | 0.29 (0.22, 0.38) | 96.08 (86.78, 98.92) | 99.97 (99.93, 99.99) | 90.74 (80.09, 95.98) | 47.43 (34.11, 60.75) | |
| SNP | 19823 | 0.30 (0.24, 0.39) | 100 (93.98, 100) | 99.97 (99.94, 99.99) | 92.31 (83.22, 96.67) | 51.36 (39.21, 63.51) | |
| MX | All | 15079 | 1.34 (1.17, 1.54) | 96.04 (92.38, 97.98) | 99.51 (99.38, 99.61) | 72.66 (67.02, 77.66) | 13.17 (9.12, 17.23) |
| MPSS | 10552 | 1.05 (0.87, 1.27) | 94.59 (88.71, 97.50) | 99.40 (99.23, 99.53) | 62.50 (54.98, 69.46) | 10.84 (6.14, 15.54) | |
| DANSR | 2481 | 2.78 (2.20, 3.50) | 100 (94.73, 100) | 99.63 (99.29, 99.80) | 88.46 (79.50, 93.81) | 17.20 (8.83, 25.58) | |
| SNP | 2046 | 1.08 (0.71, 1.62) | 90.91 (72.19, 97.47) | 99.95 (99.72, 100) | 95.24 (77.33, 99.76) | 58.79 (37.73, 79.84) | |
| T21, T13, T18 | All | 212184 | 1.71 (1.66, 1.77) | 98.57 (98.13, 98.91) | 99.93 (99.92, 99.94) | 96.03 (95.35, 96.61) | --- |
| MPSS | 71286 | 2.58 (2.46, 2.69) | 98.47 (97.80, 98.94) | 99.88 (99.86, 99.91) | 95.76 (94.76, 96.58) | --- | |
| DANSR | 69456 | 1.04 (0.97, 1.12) | 98.61 (97.47, 99.25) | 99.96 (99.94, 99.97) | 96.48 (94.89, 97.58) | --- | |
| SNP | 59468 | 0.99 (0.91, 1.07) | 99.15 (98.02, 99.64) | 99.97 (99.95, 99.98) | 97.16 (95.49, 98.22) | --- | |
| All | All | 227263 | 1.69 (1.63, 1.74) | 98.43 (97.99, 98.78) | 99.90 (99.89, 99.91) | 94.47 (93.71, 95.13) | --- |
| MPSS | 81838 | 2.38 (2.28, 2.49) | 98.25 (97.57, 98.75) | 99.82 (99.79, 99.85) | 93.04 (91.86, 94.07) | --- | |
| DANSR | 71937 | 1.10 (1.03, 1.18) | 98.74 (97.69, 99.31) | 99.95 (99.93, 99.96) | 95.71 (94.09, 96.90) | --- | |
| SNP | 61514 | 0.99 (0.91, 1.07) | 98.85 (97.64, 99.44) | 99.97 (99.95, 99.98) | 97.09 (95.45, 98.15) | --- |
| Syndrome | Method | No. Studies | Mean Sens (95% CI) | Mean Spec (95% CI) |
|---|---|---|---|---|
| T21 | All | 48 | 98.72 (97.97, 99.19) | 99.88 (99.79, 99.93) |
| MPSS | 26 | 99.05 (97.95, 99.56) | 99.82 (99.63, 99.91) | |
| DANSR | 10 | 98.15 (94.97, 99.33) | 99.95 (99.91, 99.97) | |
| SNP | 5 | 99.07 (97.34, 99.68) | 99.97 (99.93, 99.99) | |
| T18 | All | 43 | 93.54 (90.99, 95.40) | 99.86 (99.77, 99.92) |
| MPSS | 24 | 92.28 (88.28, 94.99) | 99.83 (99.66, 99.91) | |
| DANSR | 9 | 96.76 (91.70, 98.78) | 99.93 (99.80, 99.98) | |
| SNP | 5 | 96.25 (87.26, 98.97) | 99.96 (99.92, 99.98) | |
| T13 | All | 40 | 91.87 (86.13, 95.37) | 99.90 (99.83, 99.95) |
| MPSS | 23 | 90.90 (82.39, 95.53) | 99.86 (99.69, 99.94) | |
| DANSR | 7 | 87.65 (64.99, 96.44) | 99.96 (99.91, 99.98) | |
| SNP | 6 | 98.77 (86.17, 99.90) | 99.95 (99.76, 99.99) | |
| MX | All | 21 | 90.90 (83.09, 95.31) | 99.70 (99.29, 99.88) |
| MPSS | 13 | 90.83 (81.24, 95.77) | 99.70 (99.04, 99.90) | |
| DANSR | 4 | 98.92 (78.66, 99.96) | 99.45 (97.35, 99.89) | |
| SNP | 4 | 85.04 (57.03, 96.05) | 99.92 (99.56, 99.99) |
| Syndrome | Method | Mean DOR | (95% CI) |
|---|---|---|---|
| T21 | All | 35,173 | (22,313, 55,444) |
| MPSS | 28,585 | (15,104, 54,097) | |
| DANSR | 61,486 | (22,113, 170,963) | |
| SNP | 108,829 | (30,768, 384,936) | |
| T18 | All | 7,212 | (4395, 11,833) |
| MPSS | 5,195 | (2655, 10,165) | |
| DANSR | 18,403 | (6755, 50,135) | |
| SNP | 20,455 | (6053, 69125) | |
| T13 | All | 6,798 | (3959, 11,673) |
| MPSS | 4,915 | (2415, 10,004) | |
| DANSR | 9,197 | (2659, 31,807) | |
| SNP | 14,206 | (3215, 62,775) | |
| MX | All | 2,177 | (1065, 4451) |
| MPSS | 1,826 | (650, 5135) | |
| DANSR | 3,898 | (761, 19,976) | |
| SNP | 3,262 | (655, 16,256) |
| Syndrome | Study Type | No. Called | Prevalence | Sensitivity | Specificity | PPV | Std PPV |
|---|---|---|---|---|---|---|---|
| T21 | Retrospective | 11847 | 8.92 (8.42, 9.45) | 99.53 (98.9, 99.8) | 99.88 (99.79, 99.93) | 98.78 (97.92, 99.29) | 69.41 (66.65, 72.18) |
| Prospective | 57892 | 1.68 (1.58, 1.79) | 99.18 (98.38, 99.58) | 99.94 (99.92, 99.96) | 96.69 (95.38, 97.63) | 82.45 (80.09, 84.82) | |
| T18 | Retrospective | 11181 | 2.77 (2.48, 3.09) | 98.39 (96.28, 99.31) | 99.88 (99.79, 99.93) | 95.91 (93.13, 97.60) | 40.42 (35.03, 45.82) |
| Prospective | 57049 | 0.60 (0.54, 0.67) | 95.63 (92.91, 97.33) | 99.95 (99.93, 99.96) | 91.88 (88.58, 94.28) | 60.77 (55.71, 65.84) | |
| T13 | Retrospective | 10737 | 1.23 (1.04, 1.46) | 95.45 (90.44, 97.90) | 99.82 (99.72, 99.89) | 86.90 (80.44, 91.45) | 12.46 (7.08, 17.83) |
| Prospective | 49321 | 0.22 (0.18, 0.27) | 97.25 (92.22, 99.06) | 99.97 (99.94, 99.98) | 86.18 (78.98, 91.19) | 42.92 (34.17, 51.67) | |
| MX | Retrospective | 5499 | 2.38 (2.01, 2.82) | 96.18 (91.38, 98.36) | 98.99 (98.69, 99.23) | 70.00 (62.94, 76.22) | 6.90 (3.20, 10.60) |
| Prospective | 6880 | 0.39 (0.27, 0.57) | 88.89 (71.94, 96.15) | 99.78 (99.64, 99.87) | 61.54 (45.90, 75.11) | 23.94 (10.55, 37.34) |
| Syndrome | Type | N Studies | Mean Sens (95% CI) | Mean Spec (95% CI) |
|---|---|---|---|---|
| T21 | Retrospective | 16 | 99.14 (98.16, 99.60) | 99.79 (99.43, 99.92) |
| Prospective | 24 | 98.02 (96.37, 98.93) | 99.91 (99.83, 99.95) | |
| T18 | Retrospective | 13 | 96.50 (92.30, 98.45) | 99.83 (99.65, 99.92) |
| Prospective | 23 | 92.81 (88.66, 95.51) | 99.91 (99.82, 99.95) | |
| T13 | Retrospective | 12 | 88.35 (76.60, 94.61) | 99.90 (99.62, 99.97) |
| Prospective | 21 | 93.17 (84.53, 97.15) | 99.92 (99.86, 99.96) | |
| MX | Retrospective | 9 | 93.84 (86.14, 97.4) | 99.55 (97.87, 99.91) |
| Prospective | 8 | 76.11 (49.09, 91.32) | 99.67 (99.46, 99.80) |
| Syndrome | Method | FPR | Min Prevalence | PPV for Confirmed Cases | Std PPV |
|---|---|---|---|---|---|
| T21 | All | 0.050 (0.047, 0.053) | 0.68 (0.67, 0.69) | 92.48 (91.88, 93.05) | 84.34 (83.80, 84.88) |
| MPSS | 0.056 (0.052, 0.060) | 0.60 (0.59, 0.61) | 91.82 (91.10, 92.48) | 82.72 (81.91, 83.53) | |
| DANSR | 0.026 (0.012, 0.056) | 1.16 (1.03, 1.30) | 97.41 (94.08, 98.89) | 91.40 (88.11, 94.69) | |
| SNP | 0.042 (0.039, 0.047) | 0.74 (0.73, 0.76) | 94.39 (93.03, 95.50) | 86.37 (85.65, 87.10) | |
| T18 | All | 0.040 (0.038, 0.043) | 0.18 (0.18, 0.19) | 78.47 (76.80, 80.06) | 62.73 (61.42, 64.03) |
| MPSS | 0.058 (0.054, 0.062) | 0.16 (0.16, 0.17) | 75.37 (73.39, 77.24) | 49.04 (47.18, 50.90) | |
| DANSR | 0.039 (0.020, 0.074) | 0.24 (0.19, 0.31) | 79.55 (65.50, 88.85) | 66.58 (54.84, 78.32) | |
| SNP | 0.019 (0.016, 0.021) | 0.20 (0.19, 0.21) | 91.77 (88.72, 94.05) | 80.81 (79.22, 82.39) | |
| T13 | All | 0.050 (0.048, 0.053) | 0.06 (0.06, 0.07) | 48.00 (45.15, 50.86) | 30.20 (28.47, 31.94) |
| MPSS | 0.059 (0.055, 0.063) | 0.05 (0.04, 0.05) | 43.66 (40.50, 46.86) | 22.43 (20.17, 24.69) | |
| DANSR | 0.083 (0.053, 0.129) | 0.10 (0.07, 0.15) | 54.17 (35.07, 72.11) | 22.16 (8.95, 35.36) | |
| SNP | 0.040 (0.036, 0.044) | 0.08 (0.08, 0.09) | 64.13 (56.98, 70.71) | 40.23 (37.58, 42.89) | |
| MX | All | 0.095 (0.091, 0.100) | 0.10 (0.10, 0.11) | 31.05 (28.87, 33.33) | 34.84 (33.37, 36.31) |
| MPSS | 0.161 (0.153, 0.169) | 0.05 (0.05, 0.06) | 25.92 (23.70, 28.26) | 18.68 (16.90, 20.46) | |
| DANSR | 0.072 (0.055, 0.094) | 0.04 (0.03, 0.05) | 31.43 (21.76, 43.03) | 51.93 (40.91, 62.94) | |
| SNP | 0.045 (0.042, 0.050) | 0.15 (0.14, 0.16) | 74.70 (67.58, 80.70) | 50.71 (48.58, 52.83) | |
| T21, T13, T18 | All | 0.047 (0.045, 0.048) | 0.31 (0.30, 0.31) | 84.89 (84.22, 85.53) | --- |
| MPSS | 0.057 (0.055, 0.060) | 0.27 (0.27, 0.28) | 83.25 (82.46, 84.01) | --- | |
| DANSR | 0.049 (0.035, 0.068) | 0.51 (0.46, 0.56) | 90.42 (86.24, 93.43) | --- | |
| SNP | 0.034 (0.032, 0.036) | 0.34 (0.33, 0.35) | 90.95 (89.59, 92.15) | --- | |
| All | All | 0.057 (0.056, 0.059) | 0.26 (0.26, 0.27) | 78.07 (77.35, 78.77) | --- |
| MPSS | 0.076 (0.074, 0.079) | 0.23 (0.23, 0.23) | 75.39 (74.54, 76.21) | --- | |
| DANSR | 0.061 (0.049, 0.075) | 0.27 (0.24, 0.30) | 77.95 (73.17, 82.08) | --- | |
| SNP | 0.037 (0.035, 0.038) | 0.29 (0.29, 0.30) | 89.67 (88.29, 90.90) | --- |
| Syndrome | Start Year | Tech | Min Prevalence | PPV for Confirmed Cases |
|---|---|---|---|---|
| T21 | 2010–2013 | All | 0.84 (0.81, 0.86) | 90.95 (89.82, 91.96) |
| 2014–2016 | All | 0.64 (0.63, 0.66) | 92.39 (91.43, 93.25) | |
| 2017–2019 | All | 0.45 (0.42, 0.47) | 94.08 (92.61, 95.27) | |
| T18 | 2010–2013 | All | 0.21 (0.19, 0.22) | 70.00 (66.82, 73.00) |
| 2014–2016 | All | 0.18 (0.17, 0.18) | 82.39 (79.99, 84.56) | |
| 2017–2019 | All | 0.13 (0.12, 0.14) | 80.21 (75.91, 83.91) | |
| T13 | 2010–2013 | All | 0.05 (0.05, 0.06) | 40.27 (35.40, 45.34) |
| 2014–2016 | All | 0.06 (0.06, 0.07) | 44.64 (40.14, 49.22) | |
| 2017–2019 | All | 0.05 (0.04, 0.06) | 56.65 (50.23, 62.86) | |
| MX | 2010–2013 | All | 0.10 (0.09, 0.12) | 30.99 (26.72, 35.61) |
| 2014–2016 | All | 0.13 (0.12, 0.13) | 30.47 (27.19, 33.95) | |
| 2017–2019 | All | 0.04 (0.03, 0.04) | 31.01 (27.02, 35.32) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demko, Z.; Prigmore, B.; Benn, P. A Critical Evaluation of Validation and Clinical Experience Studies in Non-Invasive Prenatal Testing for Trisomies 21, 18, and 13 and Monosomy X. J. Clin. Med. 2022, 11, 4760. https://doi.org/10.3390/jcm11164760
Demko Z, Prigmore B, Benn P. A Critical Evaluation of Validation and Clinical Experience Studies in Non-Invasive Prenatal Testing for Trisomies 21, 18, and 13 and Monosomy X. Journal of Clinical Medicine. 2022; 11(16):4760. https://doi.org/10.3390/jcm11164760
Chicago/Turabian StyleDemko, Zachary, Brittany Prigmore, and Peter Benn. 2022. "A Critical Evaluation of Validation and Clinical Experience Studies in Non-Invasive Prenatal Testing for Trisomies 21, 18, and 13 and Monosomy X" Journal of Clinical Medicine 11, no. 16: 4760. https://doi.org/10.3390/jcm11164760
APA StyleDemko, Z., Prigmore, B., & Benn, P. (2022). A Critical Evaluation of Validation and Clinical Experience Studies in Non-Invasive Prenatal Testing for Trisomies 21, 18, and 13 and Monosomy X. Journal of Clinical Medicine, 11(16), 4760. https://doi.org/10.3390/jcm11164760





