A Survey of Radiomics in Precision Diagnosis and Treatment of Adult Gliomas
Abstract
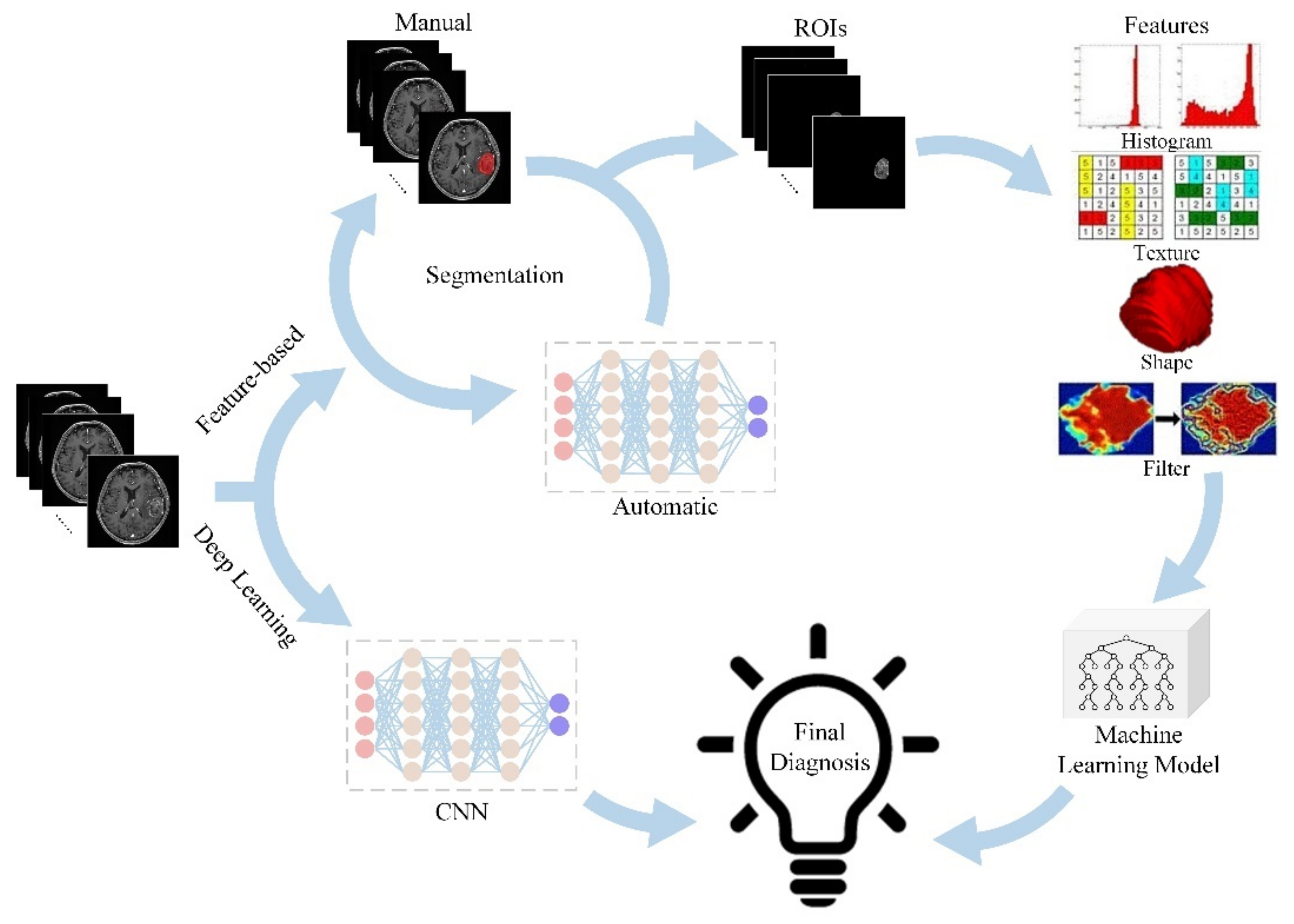
:1. Introduction
2. Radiomics in the Differential Diagnosis of Adult Gliomas
2.1. Differentiating High-Grade Gliomas (HGGs) from Solitary Brain Metastases (SBM)
2.2. Differentiating GBMs from Primary Central Nervous System Lymphomas (PCNSLs)
3. Radiomics in Preoperative Grading of Adult Gliomas
4. Radiomics in Predicting Genotyping of Adult Gliomas
4.1. IDH Mutation
4.2. MGMT Promoter Methylation
4.3. 1p/19q Co-Deletion
4.4. TERT Promoter Mutation
4.5. Combined Prediction of Multiple Genotypes
5. Radiomics in the Treatment and Efficacy Evaluation of Adult Gliomas
5.1. Identification of Tumor Infiltration Region
5.2. Discrimination of True Tumor Progression (TTP) and Pseudoprogression (PsP) after Chemotherapy and Radiotherapy
5.3. Judgment of Sensitivity to Chemotherapy and Targeted Drugs
5.4. Survival Prediction
6. Limitations and Prospects
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Aldape, K.; Brat, D.J.; Capper, D.; Ellison, D.W.; Hawkins, C.; Paulus, W.; Perry, A.; Reifenberger, G.; Figarella-Branger, D.; et al. Announcing cIMPACT-NOW: The Consortium to Inform Molecular and Practical Approaches to CNS Tumor Taxonomy. Acta Neuropathol. 2017, 133, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro. Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- D’Angelo, L.; Armocida, D.; Sampirisi, L.; Paglia, F.; Berra, L.V.; Santoro, A. Role of endoscopic surgical biopsy in diagnoses of intraventricular/periventricular tumors: Review of literature including a monocentric case series. Acta Neurol. Belg. 2020, 120, 517–530. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [Green Version]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Ou, X.; Wang, J.; Guo, W.; Ma, X. Radiomics-Based Machine Learning in Differentiation Between Glioblastoma and Metastatic Brain Tumors. Front. Oncol. 2019, 9, 806. [Google Scholar] [CrossRef] [Green Version]
- Artzi, M.; Bressler, I.; Ben Bashat, D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J. Magn. Reson. Imaging 2019, 50, 519–528. [Google Scholar] [CrossRef]
- Bae, S.; An, C.; Ahn, S.S.; Kim, H.; Han, K.; Kim, S.W.; Park, J.E.; Kim, H.S.; Lee, S.K. Robust performance of deep learning for distinguishing glioblastoma from single brain metastasis using radiomic features: Model development and validation. Sci. Rep. 2020, 10, 12110. [Google Scholar] [CrossRef]
- Ortiz-Ramón, R.; Ruiz-España, S.; Mollá-Olmos, E.; Moratal, D. Glioblastomas and brain metastases differentiation following an MRI texture analysis-based radiomics approach. Phys. Med. 2020, 76, 44–54. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, R.; Gao, J.; Tan, D.; Yang, X.; Wen, M.; Wang, J.; Xie, X.; Liao, R.; Tang, Y.; et al. An Integrated Radiomics Model Incorporating Diffusion-Weighted Imaging and 18F-FDG PET Imaging Improves the Performance of Differentiating Glioblastoma From Solitary Brain Metastases. Front. Oncol. 2021, 11, 732704. [Google Scholar] [CrossRef]
- De Causans, A.; Carré, A.; Roux, A.; Tauziède-Espariat, A.; Ammari, S.; Dezamis, E.; Dhermain, F.; Reuzé, S.; Deutsch, E.; Oppenheim, C.; et al. Development of a Machine Learning Classifier Based on Radiomic Features Extracted From Post-Contrast 3D T1-Weighted MR Images to Distinguish Glioblastoma From Solitary Brain Metastasis. Front. Oncol. 2021, 11, 638262. [Google Scholar] [CrossRef]
- Su, C.Q.; Chen, X.T.; Duan, S.F.; Zhang, J.X.; You, Y.P.; Lu, S.S.; Hong, X.N. A radiomics-based model to differentiate glioblastoma from solitary brain metastases. Clin. Radiol. 2021, 76, 629.e11–629.e18. [Google Scholar] [CrossRef]
- Sartoretti, E.; Sartoretti, T.; Wyss, M.; Reischauer, C.; van Smoorenburg, L.; Binkert, C.A.; Sartoretti-Schefer, S.; Mannil, M. Amide proton transfer weighted (APTw) imaging based radiomics allows for the differentiation of gliomas from metastases. Sci. Rep. 2021, 11, 5506. [Google Scholar] [CrossRef]
- Mărginean, L.; Ștefan, P.A.; Lebovici, A.; Opincariu, I.; Csutak, C.; Lupean, R.A.; Coroian, P.A.; Suciu, B.A. CT in the Differentiation of Gliomas from Brain Metastases: The Radiomics Analysis of the Peritumoral Zone. Brain Sci. 2022, 12, 109. [Google Scholar] [CrossRef]
- Cao, X.; Tan, D.; Liu, Z.; Liao, M.; Kan, Y.; Yao, R.; Zhang, L.; Nie, L.; Liao, R.; Chen, S.; et al. Differentiating solitary brain metastases from glioblastoma by radiomics features derived from MRI and 18F-FDG-PET and the combined application of multiple models. Sci. Rep. 2022, 12, 5722. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Z.; Wu, G.; Yu, J.; Wang, Y.; Lv, X.; Ju, X.; Chen, Z. Primary central nervous system lymphoma and glioblastoma differentiation based on conventional magnetic resonance imaging by high-throughput SIFT features. Int. J. Neurosci. 2018, 128, 608–618. [Google Scholar] [CrossRef]
- Suh, H.B.; Choi, Y.S.; Bae, S.; Ahn, S.S.; Chang, J.H.; Kang, S.G.; Kim, E.H.; Kim, S.H.; Lee, S.K. Primary central nervous system lymphoma and atypical glioblastoma: Differentiation using radiomics approach. Eur. Radiol. 2018, 28, 3832–3839. [Google Scholar] [CrossRef]
- Bathla, G.; Priya, S.; Liu, Y.; Ward, C.; Le, N.H.; Soni, N.; Maheshwarappa, R.P.; Monga, V.; Zhang, H.; Sonka, M. Radiomics-based differentiation between glioblastoma and primary central nervous system lymphoma: A comparison of diagnostic performance across different MRI sequences and machine learning techniques. Eur. Radiol. 2021, 31, 8703–8713. [Google Scholar] [CrossRef]
- Xia, W.; Hu, B.; Li, H.; Geng, C.; Wu, Q.; Yang, L.; Yin, B.; Gao, X.; Li, Y.; Geng, D. Multiparametric-MRI-Based Radiomics Model for Differentiating Primary Central Nervous System Lymphoma From Glioblastoma: Development and Cross-Vendor Validation. J. Magn. Reson. Imaging 2021, 53, 242–250. [Google Scholar] [CrossRef]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. Computer-Aided Grading of Gliomas Combining Automatic Segmentation and Radiomics. Int. J. Biomed. Imaging 2018, 2018, 2512037. [Google Scholar] [CrossRef] [Green Version]
- Tian, Q.; Yan, L.F.; Zhang, X.; Zhang, X.; Hu, Y.C.; Han, Y.; Liu, Z.C.; Nan, H.Y.; Sun, Q.; Sun, Y.Z.; et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J. Magn. Reson. Imaging 2018, 48, 1518–1528. [Google Scholar] [CrossRef]
- Jeong, J.; Wang, L.; Ji, B.; Lei, Y.; Ali, A.; Liu, T.; Curran, W.J.; Mao, H.; Yang, X. Machine-learning based classification of glioblastoma using delta-radiomic features derived from dynamic susceptibility contrast enhanced magnetic resonance images: Introduction. Quant. Imaging Med. Surg. 2019, 9, 1201–1213. [Google Scholar] [CrossRef]
- Park, Y.W.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kim, S.H.; Lee, S.K. Radiomics MRI Phenotyping with Machine Learning to Predict the Grade of Lower-Grade Gliomas: A Study Focused on Nonenhancing Tumors. Korean J. Radiol. 2019, 20, 1381–1389. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, T.; Takahashi, W.; Haga, A.; Takahashi, S.; Kiryu, S.; Nawa, K.; Ohta, T.; Ozaki, S.; Nozawa, Y.; Tanaka, S.; et al. Prediction of malignant glioma grades using contrast-enhanced T1-weighted and T2-weighted magnetic resonance images based on a radiomic analysis. Sci. Rep. 2019, 9, 19411. [Google Scholar] [CrossRef] [Green Version]
- Haubold, J.; Demircioglu, A.; Gratz, M.; Glas, M.; Wrede, K.; Sure, U.; Antoch, G.; Keyvani, K.; Nittka, M.; Kannengiesser, S.; et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric 18F-FET PET-MRI and MR Fingerprinting. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1435–1445. [Google Scholar] [CrossRef]
- Zhang, Z.; Xiao, J.; Wu, S.; Lv, F.; Gong, J.; Jiang, L.; Yu, R.; Luo, T. Deep Convolutional Radiomic Features on Diffusion Tensor Images for Classification of Glioma Grades. J. Digit. Imaging 2020, 33, 826–837. [Google Scholar] [CrossRef]
- Gutta, S.; Acharya, J.; Shiroishi, M.S.; Hwang, D.; Nayak, K.S. Improved Glioma Grading Using Deep Convolutional Neural Networks. AJNR. Am. J. Neuroradiol. 2021, 42, 233–239. [Google Scholar] [CrossRef]
- Su, C.; Chen, X.; Liu, C.; Li, S.; Jiang, J.; Qin, Y.; Zhang, S. T2-FLAIR, DWI and DKI radiomics satisfactorily predicts histological grade and Ki-67 proliferation index in gliomas. Am. J. Transl. Res. 2021, 13, 9182–9194. [Google Scholar]
- Cheng, J.; Liu, J.; Yue, H.; Bai, H.; Pan, Y.; Wang, J. Prediction of Glioma Grade Using Intratumoral and Peritumoral Radiomic Features From Multiparametric MRI Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Ning, Z.; Luo, J.; Xiao, Q.; Cai, L.; Chen, Y.; Yu, X.; Wang, J.; Zhang, Y. Multi-modal magnetic resonance imaging-based grading analysis for gliomas by integrating radiomics and deep features. Ann. Transl. Med. 2021, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhao, R.; Qiu, Q.; Chen, J.; Duan, J.; Cao, X.; Yin, Y. Developing and validating a deep learning and radiomic model for glioma grading using multiplanar reconstructed magnetic resonance contrast-enhanced T1-weighted imaging: A robust, multi-institutional study. Quant. Imaging Med. Surg. 2022, 12, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Cidan, W.; Qi, Y.; Wang, X. Glioma grading prediction using multiparametric magnetic resonance imaging-based radiomics combined with proton magnetic resonance spectroscopy and diffusion tensor imaging. Med. Phys. 2022. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Aldape, K.; Zadeh, G.; Mansouri, S.; Reifenberger, G.; von Deimling, A. Glioblastoma: Pathology, molecular mechanisms and markers. Acta Neuropathol. 2015, 129, 829–848. [Google Scholar] [CrossRef]
- Whitfield, B.T.; Huse, J.T. Classification of adult-type diffuse gliomas: Impact of the World Health Organization 2021 update. Brain Pathol. 2022, 32, e13062. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [Green Version]
- Lohmann, P.; Lerche, C.; Bauer, E.K.; Steger, J.; Stoffels, G.; Blau, T.; Dunkl, V.; Kocher, M.; Viswanathan, S.; Filss, C.P.; et al. Predicting IDH genotype in gliomas using FET PET radiomics. Sci. Rep. 2018, 8, 13328. [Google Scholar] [CrossRef]
- Li, Z.C.; Bai, H.; Sun, Q.; Zhao, Y.; Lv, Y.; Zhou, J.; Liang, C.; Chen, Y.; Liang, D.; Zheng, H. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018, 7, 5999–6009. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Mu, W.; Wang, Y.; Liu, Z.; Liu, Z.; Wang, Y.; Ma, W.; Kong, Z.; Wang, S.; Zhou, X.; et al. A Non-invasive Radiomic Method Using 18F-FDG PET Predicts Isocitrate Dehydrogenase Genotype and Prognosis in Patients With Glioma. Front. Oncol. 2019, 9, 1183. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Li, S.; Fan, X.; Sun, Z.; Yang, Z.; Wang, K.; Zhang, Z.; Jiang, T.; Liu, Y.; et al. IDH mutation-specific radiomic signature in lower-grade gliomas. Aging 2019, 11, 673–696. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, S.T.; Wei, J.W.; Dong, D.; Wang, X.C.; Yang, G.Q.; Tian, J.; Zhang, H. A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur. Radiol. 2019, 29, 3325–3337. [Google Scholar] [CrossRef]
- Wu, S.; Meng, J.; Yu, Q.; Li, P.; Fu, S. Radiomics-based machine learning methods for isocitrate dehydrogenase genotype prediction of diffuse gliomas. J. Cancer Res. Clin. Oncol. 2019, 145, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Park, C.J.; Choi, Y.S.; Park, Y.W.; Ahn, S.S.; Kang, S.G.; Chang, J.H.; Kim, S.H.; Lee, S.K. Diffusion tensor imaging radiomics in lower-grade glioma: Improving subtyping of isocitrate dehydrogenase mutation status. Neuroradiology 2020, 62, 319–326. [Google Scholar] [CrossRef]
- Peng, H.; Huo, J.; Li, B.; Cui, Y.; Zhang, H.; Zhang, L.; Ma, L. Predicting Isocitrate Dehydrogenase (IDH) Mutation Status in Gliomas Using Multiparameter MRI Radiomics Features. J. Magn. Reson. Imaging 2021, 53, 1399–1407. [Google Scholar] [CrossRef]
- Niu, L.; Feng, W.H.; Duan, C.F.; Liu, Y.C.; Liu, J.H.; Liu, X.J. The Value of Enhanced MR Radiomics in Estimating the IDH1 Genotype in High-Grade Gliomas. Biomed. Res. Int. 2020, 2020, 4630218. [Google Scholar] [CrossRef]
- Tan, Y.; Mu, W.; Wang, X.C.; Yang, G.Q.; Gillies, R.J.; Zhang, H. Whole-tumor radiomics analysis of DKI and DTI may improve the prediction of genotypes for astrocytomas: A preliminary study. Eur. J. Radiol. 2020, 124, 108785. [Google Scholar] [CrossRef]
- Manikis, G.C.; Ioannidis, G.S.; Siakallis, L.; Nikiforaki, K.; Iv, M.; Vozlic, D.; Surlan-Popovic, K.; Wintermark, M.; Bisdas, S.; Marias, K. Multicenter DSC-MRI-Based Radiomics Predict IDH Mutation in Gliomas. Cancers 2021, 13, 3965. [Google Scholar] [CrossRef]
- Choi, Y.S.; Bae, S.; Chang, J.H.; Kang, S.G.; Kim, S.H.; Kim, J.; Rim, T.H.; Choi, S.H.; Jain, R.; Lee, S.K. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro. Oncol. 2021, 23, 304–313. [Google Scholar] [CrossRef]
- Zaragori, T.; Oster, J.; Roch, V.; Hossu, G.; Chawki, M.B.; Grignon, R.; Pouget, C.; Gauchotte, G.; Rech, F.; Blonski, M.; et al. 18F-FDOPA PET for the Noninvasive Prediction of Glioma Molecular Parameters: A Radiomics Study. J. Nucl. Med. 2022, 63, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Haque, W.; Teh, C.; Butler, E.B.; Teh, B.S. Prognostic and predictive impact of MGMT promoter methylation status in high risk grade II glioma. J. Neurooncol. 2022, 157, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.B.; Guo, F.; Xu, Z.L.; Li, C.; Wei, W.; Tian, P.; Liu, T.T.; Liu, L.; Chen, G.; Ye, J.; et al. Radiomics signature: A potential biomarker for the prediction of MGMT promoter methylation in glioblastoma. J. Magn. Reson. Imaging 2018, 47, 1380–1387. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.C.; Bai, H.; Sun, Q.; Li, Q.; Liu, L.; Zou, Y.; Chen, Y.; Liang, C.; Zheng, H. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: A multicentre study. Eur. Radiol. 2018, 28, 3640–3650. [Google Scholar] [CrossRef]
- Jiang, C.; Kong, Z.; Liu, S.; Feng, S.; Zhang, Y.; Zhu, R.; Chen, W.; Wang, Y.; Lyu, Y.; You, H.; et al. Fusion Radiomics Features from Conventional MRI Predict MGMT Promoter Methylation Status in Lower Grade Gliomas. Eur. J. Radiol. 2019, 121, 108714. [Google Scholar] [CrossRef]
- Wei, J.; Yang, G.; Hao, X.; Gu, D.; Tan, Y.; Wang, X.; Dong, D.; Zhang, S.; Wang, L.; Zhang, H.; et al. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur. Radiol. 2019, 29, 877–888. [Google Scholar] [CrossRef] [Green Version]
- Kong, Z.; Lin, Y.; Jiang, C.; Li, L.; Liu, Z.; Wang, Y.; Dai, C.; Liu, D.; Qin, X.; Wang, Y.; et al. 18F-FDG-PET-based Radiomics signature predicts MGMT promoter methylation status in primary diffuse glioma. Cancer Imaging 2019, 19, 58. [Google Scholar] [CrossRef] [Green Version]
- Crisi, G.; Filice, S. Predicting MGMT Promoter Methylation of Glioblastoma from Dynamic Susceptibility Contrast Perfusion: A Radiomic Approach. J. Neuroimaging 2020, 30, 458–462. [Google Scholar] [CrossRef]
- Qian, J.; Herman, M.G.; Brinkmann, D.H.; Laack, N.N.; Kemp, B.J.; Hunt, C.H.; Lowe, V.; Pafundi, D.H. Prediction of MGMT Status for Glioblastoma Patients Using Radiomics Feature Extraction From 18F-DOPA-PET Imaging. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, 1339–1346. [Google Scholar] [CrossRef]
- Huang, W.Y.; Wen, L.H.; Wu, G.; Pang, P.P.; Ogbuji, R.; Zhang, C.C.; Chen, F.; Zhao, J.N. Radiological model based on the standard magnetic resonance sequences for detecting methylguanine methyltransferase methylation in glioma using texture analysis. Cancer Sci. 2021, 112, 2835–2844. [Google Scholar] [CrossRef]
- Kha, Q.H.; Le, V.H.; Hung, T.; Le, N. Development and Validation of an Efficient MRI Radiomics Signature for Improving the Predictive Performance of 1p/19q Co-Deletion in Lower-Grade Gliomas. Cancers 2021, 13, 5398. [Google Scholar] [CrossRef]
- Shofty, B.; Artzi, M.; Ben Bashat, D.; Liberman, G.; Haim, O.; Kashanian, A.; Bokstein, F.; Blumenthal, D.T.; Ram, Z.; Shahar, T. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int. J. Comput. Assist. Radiol. Surg. 2018, 13, 563–571. [Google Scholar] [CrossRef]
- Han, Y.; Xie, Z.; Zang, Y.; Zhang, S.; Gu, D.; Zhou, M.; Gevaert, O.; Wei, J.; Li, C.; Chen, H.; et al. Non-invasive genotype prediction of chromosome 1p/19q co-deletion by development and validation of an MRI-based radiomics signature in lower-grade gliomas. J. Neurooncol. 2018, 140, 297–306. [Google Scholar] [CrossRef]
- Kong, Z.; Jiang, C.; Zhang, Y.; Liu, S.; Liu, D.; Liu, Z.; Chen, W.; Liu, P.; Yang, T.; Lyu, Y.; et al. Thin-Slice Magnetic Resonance Imaging-Based Radiomics Signature Predicts Chromosomal 1p/19q Co-deletion Status in Grade II and III Gliomas. Front. Neurol. 2020, 11, 551771. [Google Scholar] [CrossRef]
- Park, Y.W.; Han, K.; Ahn, S.S.; Choi, Y.S.; Chang, J.H.; Kim, S.H.; Kang, S.G.; Kim, E.H.; Lee, S.K. Whole-Tumor Histogram and Texture Analyses of DTI for Evaluation of IDH1-Mutation and 1p/19q-Codeletion Status in World Health Organization Grade II Gliomas. AJNR Am. J. Neuroradiol. 2018, 39, 693–698. [Google Scholar] [CrossRef] [Green Version]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Fan, Z.; Sun, Z.; Li, Y.; Liu, X.; Liang, Y.; Liu, Y.; Zhou, C.; Zhu, Q.; Zhang, H.; et al. Radiomics Features Predict Telomerase Reverse Transcriptase Promoter Mutations in World Health Organization Grade II Gliomas via a Machine-Learning Approach. Front. Oncol. 2021, 10, 606741. [Google Scholar] [CrossRef]
- Jiang, C.; Kong, Z.; Zhang, Y.; Liu, S.; Liu, Z.; Chen, W.; Liu, P.; Liu, D.; Wang, Y.; Lyu, Y.; et al. Conventional magnetic resonance imaging-based radiomic signature predicts telomerase reverse transcriptase promoter mutation status in grade II and III gliomas. Neuroradiology 2020, 62, 803–813. [Google Scholar] [CrossRef]
- Tian, H.; Wu, H.; Wu, G.; Xu, G. Noninvasive Prediction of TERT Promoter Mutations in High-Grade Glioma by Radiomics Analysis Based on Multiparameter MRI. Biomed. Res. Int. 2020, 2020, 3872314. [Google Scholar] [CrossRef]
- Lu, C.F.; Hsu, F.T.; Hsieh, K.L.; Kao, Y.J.; Cheng, S.J.; Hsu, J.B.; Tsai, P.H.; Chen, R.J.; Huang, C.C.; Yen, Y.; et al. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin. Cancer Res. 2018, 24, 4429–4436. [Google Scholar] [CrossRef] [Green Version]
- Arita, H.; Kinoshita, M.; Kawaguchi, A.; Takahashi, M.; Narita, Y.; Terakawa, Y.; Tsuyuguchi, N.; Okita, Y.; Nonaka, M.; Moriuchi, S.; et al. Lesion location implemented magnetic resonance imaging radiomics for predicting IDH and TERT promoter mutations in grade II/III gliomas. Sci. Rep. 2018, 8, 11773. [Google Scholar] [CrossRef] [Green Version]
- Tanboon, J.; Williams, E.A.; Louis, D.N. The Diagnostic Use of Immunohistochemical Surrogates for Signature Molecular Genetic Alterations in Gliomas. J. Neuropathol. Exp. Neurol. 2016, 75, 4–18. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, B.W.; Priesterbach-Ackley, L.P.; Petersen, J.K.; Wesseling, P. Molecular pathology of tumors of the central nervous system. Ann. Oncol. 2019, 30, 1265–1278. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, X.; Rui, W.; Sheng, Y.; Yu, Y.; Zhang, Y.; Yao, Z.; Qiu, T.; Ren, Y. A nomogram strategy for identifying the subclassification of IDH mutation and ATRX expression loss in lower-grade gliomas. Eur. Radiol. 2022, 32, 3187–3198. [Google Scholar] [CrossRef]
- Sohn, B.; An, C.; Kim, D.; Ahn, S.S.; Han, K.; Kim, S.H.; Kang, S.G.; Chang, J.H.; Lee, S.K. Radiomics-based prediction of multiple gene alteration incorporating mutual genetic information in glioblastoma and grade 4 astrocytoma, IDH-mutant. J. Neurooncol. 2021, 155, 267–276. [Google Scholar] [CrossRef]
- Zhang, X.; Tian, Q.; Wang, L.; Liu, Y.; Li, B.; Liang, Z.; Gao, P.; Zheng, K.; Zhao, B.; Lu, H. Radiomics Strategy for Molecular Subtype Stratification of Lower-Grade Glioma: Detecting IDH and TP53 Mutations Based on Multimodal MRI. J. Magn. Reson. Imaging 2018, 48, 916–926. [Google Scholar] [CrossRef]
- Park, C.J.; Han, K.; Kim, H.; Ahn, S.S.; Choi, D.; Park, Y.W.; Chang, J.H.; Kim, S.H.; Cha, S.; Lee, S.K. MRI Features May Predict Molecular Features of Glioblastoma in Isocitrate Dehydrogenase Wild-Type Lower-Grade Gliomas. AJNR Am. J. Neuroradiol. 2021, 42, 448–456. [Google Scholar] [CrossRef]
- Muscas, G.; Orlandini, S.; Becattini, E.; Battista, F.; Staartjes, V.E.; Serra, C.; Della Puppa, A. Radiomic Features Associated with Extent of Resection in Glioma Surgery. Acta Neurochir. Suppl. 2022, 134, 341–347. [Google Scholar]
- Akbari, H.; Macyszyn, L.; Da, X.; Bilello, M.; Wolf, R.L.; Martinez-Lage, M.; Biros, G.; Alonso-Basanta, M.; O’Rourke, D.M.; Davatzikos, C. Imaging Surrogates of Infiltration Obtained Via Multiparametric Imaging Pattern Analysis Predict Subsequent Location of Recurrence of Glioblastoma. Neurosurgery 2016, 78, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.L.; Li, C.; van der Hoorn, A.; Boonzaier, N.R.; Matys, T.; Price, S.J. A Neural Network Approach to Identify the Peritumoral Invasive Areas in Glioblastoma Patients by Using MR Radiomics. Sci. Rep. 2020, 10, 9748. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Akbari, H.; Doshi, J.; Shukla, G.; Rozycki, M.; Bilello, M.; Lustig, R.; Davatzikos, C. Radiomic signature of infiltration in peritumoral edema predicts subsequent recurrence in glioblastoma: Implications for personalized radiotherapy planning. J. Med. Imaging 2018, 5, 021219. [Google Scholar] [CrossRef] [PubMed]
- Kruser, T.J.; Mehta, M.P.; Robins, H.I. Pseudoprogression after glioma therapy: A comprehensive review. Expert Rev. Neurother. 2013, 13, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Gerstner, E.R.; McNamara, M.B.; Norden, A.D.; Lafrankie, D.; Wen, P.Y. Effect of adding temozolomide to radiation therapy on the incidence of pseudo-progression. J. Neurooncol. 2009, 94, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Taal, W.; Brandsma, D.; de Bruin, H.G.; Bromberg, J.E.; Swaak-Kragten, A.T.; Smitt, P.A.; van Es, C.A.; van den Bent, M.J. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer 2008, 113, 405–410. [Google Scholar] [CrossRef]
- Qian, X.; Tan, H.; Zhang, J.; Zhao, W.; Chan, M.D.; Zhou, X. Stratification of pseudoprogression and true progression of glioblastoma multiform based on longitudinal diffusion tensor imaging without segmentation. Med. Phys. 2016, 43, 5889. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, J.; Zhang, J.; Bu, J.; Yu, Y.; Tan, Y.; Feng, Q.; Huang, M. Differentiation of Recurrence from Radiation Necrosis in Gliomas Based on the Radiomics of Combinational Features and Multimodality MRI Images. Comput. Math. Methods Med. 2019, 2019, 2893043. [Google Scholar] [CrossRef]
- Kim, J.Y.; Park, J.E.; Jo, Y.; Shim, W.H.; Nam, S.J.; Kim, J.H.; Yoo, R.E.; Choi, S.H.; Kim, H.S. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro. Oncol. 2019, 21, 404–414. [Google Scholar] [CrossRef]
- Patel, M.; Zhan, J.; Natarajan, K.; Flintham, R.; Davies, N.; Sanghera, P.; Grist, J.; Duddalwar, V.; Peet, A.; Sawlani, V. Machine learning-based radiomic evaluation of treatment response prediction in glioblastoma. Clin. Radiol. 2021, 76, 628.e17–628.e27. [Google Scholar] [CrossRef]
- Johnson, D.R.; O’Neill, B.P. Glioblastoma survival in the United States before and during the temozolomide era. J. Neurooncol. 2012, 107, 359–364. [Google Scholar] [CrossRef]
- Blumenthal, D.T.; Mendel, L.; Bokstein, F. The optimal regimen of bevacizumab for recurrent glioblastoma: Does dose matter? J. Neurooncol. 2016, 127, 493–502. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, X.; Zhang, J.; Xue, H.; Wang, L.; Jing, R.; Chen, S.; Che, F.; Heng, X.; Li, G.; et al. An MRI-based radiomics signature as a pretreatment noninvasive predictor of overall survival and chemotherapeutic benefits in lower-grade gliomas. Eur. Radiol. 2021, 31, 1785–1794. [Google Scholar] [CrossRef]
- Kim, C.; Kim, H.S.; Shim, W.H.; Choi, C.G.; Kim, S.J.; Kim, J.H. Recurrent Glioblastoma: Combination of High Cerebral Blood Flow with MGMT Promoter Methylation Is Associated with Benefit from Low-Dose Temozolomide Rechallenge at First Recurrence. Radiology 2017, 282, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Kickingereder, P.; Wiestler, B.; Burth, S.; Wick, A.; Nowosielski, M.; Heiland, S.; Schlemmer, H.P.; Wick, W.; Bendszus, M.; Radbruch, A. Relative cerebral blood volume is a potential predictive imaging biomarker of bevacizumab efficacy in recurrent glioblastoma. Neuro. Oncol. 2015, 17, 1139–1147. [Google Scholar] [CrossRef] [Green Version]
- Kickingereder, P.; Götz, M.; Muschelli, J.; Wick, A.; Neuberger, U.; Shinohara, R.T.; Sill, M.; Nowosielski, M.; Schlemmer, H.P.; Radbruch, A.; et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin. Cancer Res. 2016, 22, 5765–5771. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Li, L.; Li, Y.; Qian, Z.; Wu, F.; He, Y.; Jiang, H.; Li, R.; Wang, D.; Zhai, Y.; et al. An MRI radiomics approach to predict survival and tumour-infiltrating macrophages in gliomas. Brain 2022, 145, 1151–1161. [Google Scholar] [CrossRef]
- Han, W.; Qin, L.; Bay, C.; Chen, X.; Yu, K.H.; Miskin, N.; Li, A.; Xu, X.; Young, G. Deep Transfer Learning and Radiomics Feature Prediction of Survival of Patients with High-Grade Gliomas. AJNR. Am. J. Neuroradiol. 2020, 41, 40–48. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, B.; Zhang, S.; Cheng, J.; Liu, X.; Wang, W.; Dong, Y.; Zhang, L.; Mo, X.; Chen, Q.; et al. Quantitative MRI-based radiomics for noninvasively predicting molecular subtypes and survival in glioma patients. NPJ Precis. Oncol. 2021, 5, 72. [Google Scholar] [CrossRef]
- Feng, X.; Tustison, N.J.; Patel, S.H.; Meyer, C.H. Brain Tumor Segmentation Using an Ensemble of 3D U-Nets and Overall Survival Prediction Using Radiomic Features. Front. Comput. Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lu, H.; Tian, Q.; Feng, N.; Yin, L.; Xu, X.; Du, P.; Liu, Y. A radiomics nomogram based on multiparametric MRI might stratify glioblastoma patients according to survival. Eur. Radiol. 2019, 29, 5528–5538. [Google Scholar] [CrossRef]
- Huang, H.; Zhang, W.; Fang, Y.; Hong, J.; Su, S.; Lai, X. Overall Survival Prediction for Gliomas Using a Novel Compound Approach. Front. Oncol. 2021, 11, 724191. [Google Scholar] [CrossRef]
| Authors and Reference No. | Year | Study Sample(n) | Imaging Method and Sequence | Feature Extraction/Software | Classification Algorithm | Main Findings |
|---|---|---|---|---|---|---|
| Chen et al. [7] | 2019 | GBMs and BM (134) | MRI CE-T1WI | Texture analysis/LifeX | Linear discriminant analysis, Logistic regression | AUC 0.80, sensitivity 69%, specificity 86%, accuracy 78% |
| Artzi et al. [8] | 2019 | GBMs (212) and BM (227) | MRI CE-T1WI | Multiple features /MatLab (R2017a) | Support vector machine | AUC 0.96, sensitivity 86%, specificity 85%, accuracy 85% |
| Bae et al. [9] | 2020 | GBMs (159) and SBM (89) | MRI (T2WI + 3D-CE-T1WI) | Multiple features /Pyradiomics 2.1.0 | Adaptive boosting, Support vector machine, Linear discriminant analysis, Deep neural network | The DNN model showed higher diagnostic performance than the traditional machine learning models, with an AUC of 0.956, sensitivity of 91%, specificity of 88%, accuracy of 89%. |
| Ortiz-Ramón et al. [10] | 2020 | GBMs (50) and BM (50) | MRI CE-T1WI | Texture analysis /MatLab (R2015b) | Support vector machine | AUC = 0.896 ± 0.067, sensitivity 82%, specificity 80% |
| Zhang et al. [11] | 2021 | GBMs (50) and SBM (50) | MRI (CE-T1WI +T2WI + DWI/ADC) + 18F-FDG PET/CT | Multiple features /Not mentioned | Random forest | The integrated radiomics model showed more efficient diagnostic performance than any other single radiomics model (AUC 0.93, sensitivity 83.5%, specificity 84.9%). |
| Causans et al. [12] | 2021 | GBMs (71) and BM (72) | MRI 3D-CE-T1WI | Multiple features /PyRadiomics 2.1.2 | Logistic regression | AUC 0.85, sensitivity 75%, specificity 86%, accuracy 80% |
| Su et al. [13] | 2021 | GBMs (157) and SBM (98) | MRI CE-T1WI | Multiple features/AK software version 3.2.0 | Logistic regression | AUC 0.81, sensitivity 85.3%, specificity 72.3%, accuracy 76.3% |
| Sartoretti et al. [14] | 2021 | GBMs (21) and BM (27) | MRI APTWI | Multiple features/3D Slicer (v. 4.10.2) with PyRadiomics package | Multiple perceptron | AUC 0.836, sensitivity 81.3%, specificity 81.1% |
| Marginean et al. [15] | 2022 | HGGs (17) and SBM (19) | CT CECT | Texture analysis /MaZda version 5 | Multiple regression | Seven texture parameters were able to differentiate between HGGs and BMs with variable sensitivity (56.67–96.67%) and specificity (69.23–100%). |
| Cao et al. [16] | 2022 | GBMs (50) and SBM (50) | MRI (CE-T1WI + T2WI) + 18F-FDG PET/CT | Multiple features /Python’s PyRadiomics package | Support vector machine, Logistic regression, K nearest neighbors, Random forest, Adaptive boosting | The model set based on MRI combined with 18F-FDG-PET had the highest average AUC (0.93) compared with isolated MRI or 18F-FDG-PET. |
| Authors and Reference No. | Year | Study Sample(n) | Imaging Method/Sequence | Feature Extraction/Software | Classification Algorithm | Main Findings |
|---|---|---|---|---|---|---|
| Chen et al. [21] | 2018 | HGGs (220) and LGGs (54) | T1WI + CE-T1WI + T2WI + T2-FLAIR | Multiple features /Pyradiomics | Support vector machine | Accuracy 91.27%, weighted macroprecision 91.27%, weighted macrorecall 91.27% |
| Tian et al. [22] | 2018 | HGGs (111) and LGGs (42) | T1WI + CE-T1WI + T2WI + DWI/ADC + 3D-ASL | Texture analysis /MatLab (R2012b) | Support vector machine | AUC 0.987, accuracy 96.8% for classifying LGGs from HGGs; AUC 0.992, accuracy 98.1% for classifying grades III from IV. |
| Jeong et al. [23] | 2019 | HGGs (13) and LGGs (12) | DSC-MRI | Multiple features /Imaging Biomarker Explorer | Random forest | AUC was 0.94 and the mean prediction accuracy was 0.950 ± 0.091 for HGG and 0.850 ± 0.255 for LGG. |
| Park et al. [24] | 2019 | LGGs 204 | CE-T1WI + T2WI + T2-FLAIR | Multiple features /Pyradiomics 1.2.0 | Elastic net, Random forest, Gradient boosting machine, Linear discriminant analysis | The performance of the best classifier was good in the internal validation set (AUC, 0.85) and fair in the external validation set (AUC, 0.72) to predict the LGG grade. |
| Nakamoto et al. [25] | 2019 | HGGs 224 (WHO III 77, IV 147) | CE-T1WI + T2WI | Multiple features /Open-source MATLAB code | Logistic regression, Support vector machine, Standard neural network, Random forest, Naïve Bayes | The mean AUC value for all prediction models constructed by the machine learning algorithms in the LOOCV of the primary dataset was 0.902 ± 0.024. In the independent validation, the mean AUC value for all prediction models was 0.747 ± 0.034. |
| Haubold et al. [26] | 2020 | Gliomas 30 (WHO 1 1, 2 13, 3 7, 4 9) | 18F-FET PET-MRI | Multiple features /Pyradiomics | Support vector machine, Random forest | The AUC of differentiating low-grade glioma vs. high-grade glioma was 85.2%. |
| Zhang et al. [27] | 2020 | HGGs (65) and LGGs (43) | DTI | Multiple features /Matlab 2016b | Support vector machine | AUC 0.93, accuracy 0.94, sensitivity 0.98, and specificity 0.86 in classifying LGG from HGG, while AUC 0.99, accuracy 0.98, sensitivity 0.98, and specificity 1.00 in classifying grade III from IV. |
| Gutta et al. [28] | 2021 | Gliomas 237 (WHO I 17, II 59, III 46, IV 115) | T1WI + CE-T1WI + T2WI + T2-FLAIR | Multiple features /Pyradiomics | Convolutional neural networks, Support vector machine, Random forests, Gradient boosting | Using learned features extracted from the convolutional neural network achieved an average accuracy of 87%, outperforming the methods considering radiomic features alone. |
| Su et al. [29] | 2021 | Gliomas 139 (WHO I 2, II 67, III 36, IV 34) | FLAIR + DWI/ADC + DKI | Multiple features /MATLAB platform (v13.0) | Adjusted-imbalanced Logistic regression | The combination of all multi-parameter MRI radiomics features performed the best predictive AUC (0.853) for differentiating low-/high-grade gliomas. |
| Cheng et al. [30] | 2021 | HGGs (210) and LGGs (75) | T1WI + CE-T1WI + T2WI + T2-FLAIR | Multiple features /PyRadiomics toolbox | Logistic regression, Support vector machine, Random forest, XGBoost | The radiomic signatures utilizing the features of intratumoral volume and peritumoral volume both showed a high accuracy in predicting glioma grade, with AUCs reaching 0.968. |
| Ning et al. [31] | 2021 | HGGs (211) and LGGs (356) | CE-T1WI + T2-FLAIR | Multiple features /Python 3.6 | Support vector machine | The AUC, sensitivity, and specificity of the model based on a combination of radiomics and deep features were 0.94, 86%, and 92%, respectively, for the validation cohort. |
| Ding et al. [32] | 2022 | HGGs (68) and LGGs (83) | CE-T1WI | Multiple features /PyRadiomics 3.0.1 | Support vector machine, Random forest, Logistic regression | The optimal model was a random forest model that combined radiomic features and VGG16 deep learning features derived from multiplanar CE-T1W MPR images, which achieved an AUC of 0.847 in the training cohort and 0.898 in the test cohort. |
| Lin et al. [33] | 2022 | HGGs (50) and LGGs (50) | T1WI + CE-T1WI + T2WI + DWI/ADC + 1H-MRS + DTI | Multiple features /Analysis-Kit | Logistic regression | CE-T1WI exhibited the highest grading efficacy among single sequences (AUC 0.92; sensitivity 0.89; specificity 0.85), but the efficacy of the combined model was higher (AUC 0.97; sensitivity 0.94; specificity 0.91). |
| Authors and Reference No. | Year | Study Sample (n) | Clinical Information Included | Imaging Method/Sequence | Feature Extraction/Software | Classification Algorithm | Main Findings |
|---|---|---|---|---|---|---|---|
| Lohmann et al. [39] | 2018 | Gliomas 84 (IDH mut 26, IDH wt 58) | No | FET-PET MRI | Texture analysis /LIFEx 2.2 | Logistic regression | The overall accuracy of the model (combination of standard PET parameters with textural features) was 82% after 5-fold cross-validation and 86% after 10-fold cross-validation. |
| Li et al. [40] | 2018 | IDH1 mut (20), IDH1 wt (205) | Yes | T1WI + CE-T1WI + T2WI + T2-FLAIR | Multiple features/In-house Matlab program | Random forest | The model combining all-region imaging features with age achieved the best performance of accuracy of 97%, AUC 0.96. |
| Li et al. [41] | 2019 | IDH mut (51), IDH wt (76) | Yes | 18F-FDG PET/CT | Multiple features /PyRadiomics | Logistic regression | The generated radiomic signature with the incorporation of age and type of tumor metabolism achieved AUCs of 0.911 and 0.900 in the training and validation cohorts, respectively. |
| Liu et al. [42] | 2019 | LGGs 158 (IDH mut 118, IDH wt 40) | No | T2WI | Multiple features /MATLAB 2014a | Logistic regression | Using a classification model of 86 radiomic features, the enrolled patients were correctly classified into the IDH wt and the IDH mut groups (AUC = 1.00). |
| Tan et al. [43] | 2019 | Astrocytomas 105 (IDH mut 51, IDH wt 54) | Yes | CE-T1WI + T2-FLAIR + DWI/ADC | Multiple features /Not mentioned | Support vector machine | The radiomics nomogram based on the radiomics signature and age performed better than the clinico-radiological model (training cohort, AUC = 0.913 and 0.817; validation cohort, AUC = 0.900 and 0.804). |
| Wu et al. [44] | 2019 | Gliomas 126 (IDH mut 39, IDH wt 87) | No | T1WI + CE-T1WI + T2WI + T2-FLAIR | Multiple features /R software (version 3.3.1) | Support vector machine, Random forest, Adaptive boosting, Naive Bayes, Flexible discriminant analysis, k-Nearest neighbors, Neural network | Random forest showed the highest predictive performance (accuracy 0.885 ± 0.041, AUC 0.931 ± 0.036). |
| Park et al. [45] | 2020 | LGGs 168 (IDH mut 113, IDH wt 55) | No | DTI + CE-T1WI + T2WI + T2-FLAIR | Multiple features /PyRadiomics | Random forest | Adding DTI radiomics to conventional radiomics significantly improved the accuracy of IDH status subtyping (AUC 0.900, p = 0.006). |
| Peng et al. [46] | 2020 | IDH mut (50), IDH wt (55) | No | CE-T1WI + T2WI + ASL | Multiple features /Pyradiomics | Support vector machine | The accuracy and AUC of the classifier, which combines the features of all three sequences, achieved 82.3% and 0.770 (p < 0.05), respectively. |
| Niu et al. [47] | 2020 | HGGs 182 (IDH mut 79, IDH wt 103) | No | CE-T1WI | Multiple features /Analysis Kit | Logistic regression | The radiomic model showed good discrimination in both the primary dataset (AUC 0.87, sensitivity 85.5%, specificity 75.4%) and the validation dataset (AUC 0.86, sensitivity 91.3%, specificity 69.0%). |
| Tan et al. [48] | 2020 | Astrocytomas 62 (IDH mut 30, IDH wt 32) | Yes | DKI + DTI | Multiple features /Not mentioned | Logistic regression | The radiomics model built using the three most informative radiomics features for each genotype yielded an AUC of 0.831 for predicting IDH genotype. |
| Manikis et al. [49] | 2021 | IDH mut (41), IDH wt (119) | No | DSC-MRI | Multiple features /Pyradiomics | Support vector machine, Random forest, K-nearest neighbor, Logistic regression, L1 norm penalties, Decision trees | The maximum performance of the IDH mutation status prediction on the validation set had an accuracy of 70.6% (AUC 0.667, sensitivity 60%, specificity 73.6%) when dynamic-based standardization of the images was performed prior to the radiomics. |
| Choi et al. [50] | 2021 | Gliomas 1166 (grades II–IV) | No | CE-T1WI + T2WI + T2-FLAIR | Multiple features /PyRadiomics 2.2.0 | Convolutional neural network | The hybrid model achieved accuracies of 93.8%, 87.9%, and 78.8%, with AUCs of 0.96, 0.94, and 0.86 in the internal test, SNUH, and TCIA sets, respectively. |
| Zaragori et al. [51] | 2022 | Gliomas 72 (IDH mut 43, IDH wt 29) | No | 18F-FDOPA PET/CT | Multiple features /Pyradiomics | Logistic regression, Neural networks, Random forest, Support vector machine | The combination of logistic regression with L2 regularization and 5 selected features was the best-performing model for predicting IDH mutations and yielded an AUC of 0.831. |
| Authors and Reference No. | Year | Study Sample (n) | Clinical Information Included | Imaging Method/Sequence | Feature Extraction/Software | Classification Algorithm | Main Findings |
|---|---|---|---|---|---|---|---|
| Xi et al. [53] | 2018 | GBMs 98 (MGMT methylated 48, unmethylated 50) | No | T1WI + CE-T1WI + T2WI | Multiple features /MatLab 2014a | Support vector machine | The best classification system for predicting MGMT promoter methylation status originated from the combination of 36 T1WI, T2WI, and CE-T1WI image features, with an accuracy of 86.59%. |
| Li et al. [54] | 2018 | GBMs 193 (MGMT methylated 86, unmethylated 107) | Yes | T1WI + CE-T1WI + T2WI + T2-FLAIR | Multiple features /R package Boruta | Random forest | The radiomics model with six all-relevant features allowed pretreatment prediction of MGMT methylation (AUC = 0.88, accuracy = 80%). |
| Jiang et al. [55] | 2019 | LGGs 122 (MGMT methylated 86, unmethylated 107) | No | 3D CE-T1WI + T2WI | Multiple features /PyRadiomics 2.1.0 | Support vector machine, Random forest, AdaBoost | The fusion radiomics model, which was constructed from the concatenation of both series, displayed the best performance, with an accuracy of 84.9% and an AUC of 0.970 in the training dataset, and an accuracy of 88.6% and an AUC of 0.898 in the validation dataset. |
| Wei et al. [56] | 2019 | Astrocytomas 105 (MGMT methylated 73, unmethylated 32) | Yes | CE-T1WI + T2-FLAIR + ADC | Multiple features /PyRadiomics | Logistic regression | The fusion radiomics signature exhibited supreme power for predicting MGMT promoter methylation, with AUCs of 0.925 in the training cohort and 0.902 in the validation cohort. |
| Kong et al. [57] | 2019 | Gliomas 107 (MGMT methylated 59, unmethylated 48) | Yes | 18F-FDG-PET/CT | Multiple features /PyRadiomics | Support vector machine, Logistic regression | The radiomics signature displayed the best performance with AUCs reaching 0.94 and 0.86 in the primary and validation cohorts, respectively, which outweigh the performances of the clinical signature and fusion signature. |
| Crisi et al. [58] | 2020 | GBMs 59 (MGMT methylated 20, unmethylated 39) | No | DSC-MRI | Multiple features /LIFEx | Naive Bayes, Decision trees, Multilayer perceptron | The model formulated by multilayer perceptron machine learning methods performed well with 75% sensitivity, 85% specificity, and an AUC of 0.84. |
| Qian et al. [59] | 2020 | GBMs 69 (MGMT methylated 26, unmethylated 43) | No | 18F-DOPA-PET/CT | Multiple features /PyRadiomics | Extra trees, Support vector machine, Random forest, XGBoost, Neural network | The Random Forest model based on features extracted HGG contour alone achieved 80% ± 10% accuracy for 95% confidence level in predicting MGMT status. |
| Huang et al. [60] | 2021 | Gliomas 53 (MGMT methylated 21, unmethylated 32) | Yes | T1WI + CE-T1WI + T2WI + T2-FLAIR | Texture analysis /Analysis Kit | Logistic regression | The AUCs for the combined model based on Radscores were 0.818, with 90.5% sensitivity and 72.7% specificity, in the GBM dataset, and 0.833, with 70.2% sensitivity and 90.6% specificity, in the overall gliomas dataset. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, P.; Chen, H.; Lv, K.; Geng, D. A Survey of Radiomics in Precision Diagnosis and Treatment of Adult Gliomas. J. Clin. Med. 2022, 11, 3802. https://doi.org/10.3390/jcm11133802
Du P, Chen H, Lv K, Geng D. A Survey of Radiomics in Precision Diagnosis and Treatment of Adult Gliomas. Journal of Clinical Medicine. 2022; 11(13):3802. https://doi.org/10.3390/jcm11133802
Chicago/Turabian StyleDu, Peng, Hongyi Chen, Kun Lv, and Daoying Geng. 2022. "A Survey of Radiomics in Precision Diagnosis and Treatment of Adult Gliomas" Journal of Clinical Medicine 11, no. 13: 3802. https://doi.org/10.3390/jcm11133802
APA StyleDu, P., Chen, H., Lv, K., & Geng, D. (2022). A Survey of Radiomics in Precision Diagnosis and Treatment of Adult Gliomas. Journal of Clinical Medicine, 11(13), 3802. https://doi.org/10.3390/jcm11133802






