An Overview of Healthcare Associated Infections and Their Detection Methods Caused by Pathogen Bacteria in Romania and Europe
Abstract
:1. Introduction
2. Healthcare Associated Infections
2.1. HAI in Europe
2.2. HAI in Romania
3. Testing Methods for Diagnosis Purposes of Bacteria Involved in Nosocomial Infections
4. Discussions, Future Trends, and Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sikora, A.; Zahra, F. Nosocomial Infections. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Chandra, S.; Chong, D. Health Care–Associated Infections. In Critical Care; Oropello, J.M., Pastores, S.M., Kvetan, V., Eds.; McGraw-Hill Education: New York, NY, USA, 2017. [Google Scholar]
- van Duin, D.; Paterson, D.L. Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned. Infect. Dis. Clin. 2016, 30, 377–390. [Google Scholar] [CrossRef] [Green Version]
- Voidazan, S.; Albu, S.; Toth, R.; Grigorescu, B.; Rachita, A.; Moldovan, I. Healthcare Associated Infections-A New Pathology in Medical Practice? Int. J. Environ. Res. Public Health 2020, 17, 760. [Google Scholar] [CrossRef] [Green Version]
- Public Health Timiș County. Available online: https://www.dsptimis.ro/promovare/clean_hands_19_analiza.pdf (accessed on 15 April 2021).
- World Health Organization Health Care-Associated Infections. Available online: http://www.who.int/gpsc/country_work/gpsc_ccisc_fact_sheet_en.pdf (accessed on 18 April 2021).
- Order No. 1101/2016 Regarding the Approval of the Norms of Surveillance, Prevention and Limitation of the Healthcare-Associated Infections in the Health Units. Available online: https://lege5.ro/Gratuit/geztanjsgmzq/ordinul-nr-1101-2016-privind-aprobarea-normelor-de-supraveghere-prevenire-si-limitare-a-infectiilor-asociate-asistentei-medicale-in-unitatile-sanitare (accessed on 15 April 2021).
- The National Center for the Surveillance and Control of Communicable Diseases. Guide for Diagnosis, Treatment and Prevention of Difficult Clostridium Infections. Available online: https://www.cnscbt.ro/index.php/ghiduri-si-protocoale/ghiduri/520-ghid-diagnostic-tratament-si-prevenire-clostridium-difficile/file (accessed on 12 May 2021).
- Rafila, A.; Indra, A.; Popescu, G.A.; Wewalka, G.; Allerberger, F.; Benea, S.; Badicut, I.; Aschbacher, R.; Huhulescu, S. Occurrence of Clostridium difficile Infections Due to PCR Ribotype 027 in Bucharest, Romania. J. Infect. Dev. Ctries. 2014, 8, 694–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The National Center for the Surveillance and Control of Communicable Diseases. Available online: https://www.cnscbt.ro/index.php/analiza-date-supraveghere/infectii-nosocomiale-1/2025-consumul-de-antibiotice-rezistenta-microbiana-si-infectiile-asociate-asistentei-medicale-romania-2018/file (accessed on 13 May 2021).
- Vasile, C.; Azoicăi, D.; Iancu, L.S.; Trifan, M.; Duca, E.; Fochi, M.; Manciuc, D.C.; Dumbrava, M.; Prelipceanu, M.S.; Iacob, O.; et al. Guide for the Management of Infections Associated with Healthcare, 2nd ed.; Global Arts Management Publishing House: Bucharest, Romania, 2007. [Google Scholar]
- Weinstein, R.A. Infections Acquired in Health Care Facilities. In Harrison’s Principles of Internal Medicine, 20e; Jameson, J.L., Fauci, A.S., Kasper, D.L., Hauser, S.L., Longo, D.L., Loscalzo, J., Eds.; McGraw-Hill Education: New York, NY, USA, 2018. [Google Scholar]
- Leoncio, J.M.; de Almeida, V.F.; Ferrari, R.A.P.; Capobiango, J.D.; Kerbauy, G.; Tacla, M.T.G.M. Impacto Das Infecções Relacionadas à Assistência à Saúde Nos Custos Da Hospitalização de Crianças. Rev. Esc. Enferm. USP 2019, 53, e03486. [Google Scholar] [CrossRef] [Green Version]
- Agency for Healthcare Research and Quality. Available online: https://www.ahrq.gov/sites/default/files/publications2/files/hac-cost-report2017.pdf (accessed on 29 May 2021).
- Zimlichman, E.; Henderson, D.; Tamir, O.; Franz, C.; Song, P.; Yamin, C.K.; Keohane, C.; Denham, C.R.; Bates, D.W. Health Care-Associated Infections: A Meta-Analysis of Costs and Financial Impact on the US Health Care System. JAMA Intern. Med. 2013, 173, 2039–2046. [Google Scholar] [CrossRef] [PubMed]
- Mathur, P. Prevention of Healthcare-Associated Infections in Low- and Middle-Income Countries: The “Bundle Approach”. Indian J. Med. Microbiol. 2018, 36, 155–162. [Google Scholar] [CrossRef]
- ECDC Technical Document: Core Competencies for Infection Control and Hospital Hygiene Professionals in the European Union. Available online: https://www.ecdc.europa.eu/en/publications-data/core-competencies-infection-control-and-hospital-hygiene-professionals-european (accessed on 2 June 2021).
- World Health Organisation: Guidelines on Core Components of Infection Prevention and Control Programmes at the National and Acute Health Care Facility Level. Available online: https://www.who.int/gpsc/core-components.pdf (accessed on 4 June 2021).
- World Health Organization: Infection Prevention and Control Assessment Framework at the Facility Level. Available online: https://apps.who.int/iris/handle/10665/330072 (accessed on 4 June 2021).
- Pliakos, E.E.; Andreatos, N.; Shehadeh, F.; Ziakas, P.D.; Mylonakis, E. The Cost-Effectiveness of Rapid Diagnostic Testing for the Diagnosis of Bloodstream Infections with or without Antimicrobial Stewardship. Clin. Microbiol. Rev. 2018, 31, e00095-17. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.M.; Newton, D.; Kunapuli, A.; Gandhi, T.N.; Washer, L.L.; Isip, J.; Collins, C.D.; Nagel, J.L. Impact of Rapid Organism Identification via Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Combined with Antimicrobial Stewardship Team Intervention in Adult Patients with Bacteremia and Candidemia. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2013, 57, 1237–1245. [Google Scholar] [CrossRef]
- Beganovic, M.; Costello, M.; Wieczorkiewicz, S.M. Effect of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) Alone versus MALDI-TOF MS Combined with Real-Time Antimicrobial Stewardship Interventions on Time to Optimal Antimicrobial Therapy in Patients with Posi. J. Clin. Microbiol. 2017, 55, 1437–1445. [Google Scholar] [CrossRef] [Green Version]
- Giacobbe, D.R.; Giani, T.; Bassetti, M.; Marchese, A.; Viscoli, C.; Rossolini, G.M. Rapid Microbiological Tests for Bloodstream Infections Due to Multidrug Resistant Gram-Negative Bacteria: Therapeutic Implications. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Burillo, A.; Bouza, E. Use of Rapid Diagnostic Techniques in ICU Patients with Infections. BMC Infect. Dis. 2014, 14, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Revelas, A. Healthcare—Associated Infections: A Public Health Problem. Niger. Med. J. 2012, 53, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Haque, M.; Sartelli, M.; McKimm, J.; Abu Bakar, M. Health Care-Associated Infections—An Overview. Infect. Drug Resist. 2018, 11, 2321–2333. [Google Scholar] [CrossRef] [Green Version]
- Guidelines for the Management of Adults with Hospital-Acquired, Ventilator-Associated, and Healthcare-Associated Pneumonia. Am. J. Respir. Crit. Care Med. 2005, 171, 388–416. [CrossRef] [PubMed]
- Chin-Hong, P.V.; Guglielmo, B.J. Health Care–Associated Infections. In Current Medical Diagnosis & Treatment 2021; Papadakis, M.A., McPhee, S.J., Rabow, M.W., Eds.; McGraw-Hill Education: New York, NY, USA, 2021. [Google Scholar]
- Suetens, C.; Latour, K.; Kärki, T.; Ricchizzi, E.; Kinross, P.; Moro, M.L.; Jans, B.; Hopkins, S.; Hansen, S.; Lyytikäinen, O.; et al. Prevalence of Healthcare-Associated Infections, Estimated Incidence and Composite Antimicrobial Resistance Index in Acute Care Hospitals and Long-Term Care Facilities: Results from Two European Point Prevalence Surveys, 2016 to 2017. Euro Surveill. Bull. Eur. Mal. Transm. Eur. Commun. Dis. Bull. 2018, 23, 1800516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levinson, W.; Chin-Hong, P.; Joyce, E.A.; Nussbaum, J.; Schwartz, B. Overview of the Major Pathogens & Introduction to Anaerobic Bacteria. In Review of Medical Microbiology & Immunology: A Guide to Clinical Infectious Diseases, 16e; McGraw Hill: New York, NY, USA, 2020. [Google Scholar]
- Junio, O.; Reygaert, W.C. Gram Negative Bacteria. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Healthcare Associated Infections (NSIH). European Antimicrobial Resistance Surveillance System (EARSS). Annual Report EARSS-2003. Available online: http://www.nsih.be/surv_ears/download/2004_EARSS_Annual_Report.pdf (accessed on 20 March 2021).
- National Nosocomial Infections Surveillance System. National Nosocomial Infections Surveillance (NNIS) System Report, Data Summary from January 1992 through June 2004, Issued October 2004. Am. J. Infect. Control 2004, 32, 470–485. [Google Scholar] [CrossRef]
- Thompson, R.L.; Cabezudo, I.; Wenzel, R.P. Epidemiology of Nosocomial Infections Caused by Methicillin-Resistant Staphylococcus aureus. Ann. Intern. Med. 1982, 97, 309–317. [Google Scholar] [CrossRef]
- Sizar, O.; Unakal, C.G. Gram Positive Bacteria. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- World Health Organisation: Guidelines for the Prevention and Control of Carbapenem-Resistant Enterobacteriaceae, Acinetobacter Baumannii and Pseudomonas Aeruginosa in Health Care Facilities. Available online: https://apps.who.int/iris/handle/10665/259462 (accessed on 14 June 2021).
- Falagas, M.E.; Tansarli, G.S.; Karageorgopoulos, D.E.; Vardakas, K.Z. Deaths Attributable to Carbapenem-Resistant Enterobacteriaceae Infections. Emerg. Infect. Dis. 2014, 20, 1170–1175. [Google Scholar] [CrossRef]
- Klevens, R.M.; Edwards, J.R.; Richards, C.L.J.; Horan, T.C.; Gaynes, R.P.; Pollock, D.A.; Cardo, D.M. Estimating Health Care-Associated Infections and Deaths in U.S. Hospitals, 2002. Public Health Rep. 2007, 122, 160–166. [Google Scholar] [CrossRef]
- Li, Y.; Gong, Z.; Lu, Y.; Hu, G.; Cai, R.; Chen, Z. Impact of Nosocomial Infections Surveillance on Nosocomial Infection Rates: A Systematic Review. Int. J. Surg. 2017, 42, 164–169. [Google Scholar] [CrossRef]
- Loveday, H.P.; Wilson, J.A.; Kerr, K.; Pitchers, R.; Walker, J.T.; Browne, J. Association between Healthcare Water Systems and Pseudomonas Aeruginosa Infections: A Rapid Systematic Review. J. Hosp. Infect. 2014, 86, 7–15. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Lee, Y.-T.; Kuo, S.-C.; Chen, T.-L.; Liu, C.-P.; Liu, C.-E. Comparison between Bacteremia Caused by Acinetobacter Pittii and Acinetobacter Nosocomialis. J. Microbiol. Immunol. Infect. 2017, 50, 62–67. [Google Scholar] [CrossRef]
- Umscheid, C.A.; Mitchell, M.D.; Doshi, J.A.; Agarwal, R.; Williams, K.; Brennan, P.J. Estimating the Proportion of Healthcare-Associated Infections That Are Reasonably Preventable and the Related Mortality and Costs. Infect. Control Hosp. Epidemiol. 2011, 32, 101–114. [Google Scholar] [CrossRef]
- Robert Wachter, K.G. Understanding Patient Safety, 3rd ed.; McGraw-Hill Professional: New York, NY, USA, 2012. [Google Scholar]
- Paradisi, F.; Corti, G.; Cinelli, R. Streptococcus Pneumoniae as an Agent of Nosocomial Infection: Treatment in the Era of Penicillin-Resistant Strains. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2001, 7, 34–42. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, J.G.; O’Keefe, P.; Tally, F.P.; Louie, T.J.; Gorbach, S.L. Bacteriology of Hospital-Acquired Pneumonia. Arch. Intern. Med. 1986, 146, 868–871. [Google Scholar] [CrossRef]
- Bassetti, M.; Vena, A.; Croxatto, A.; Righi, E.; Guery, B. How to Manage Pseudomonas Aeruginosa Infections. Drugs Context 2018, 7, 212527. [Google Scholar] [CrossRef]
- Riddle, D.J.D.; Dubberke, E.R.D. Trends in Clostridium difficile Disease: Epidemiology and Intervention. Infect. Med. 2009, 26, 211–220. [Google Scholar]
- Facciolà, A.; Pellicanò, G.F.; Visalli, G.; Paolucci, I.A.; Venanzi Rullo, E.; Ceccarelli, M.; D’Aleo, F.; Di Pietro, A.; Squeri, R.; Nunnari, G.; et al. The Role of the Hospital Environment in the Healthcare-Associated Infections: A General Review of the Literature. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 1266–1278. [Google Scholar] [CrossRef]
- Smith, H.Z.; Kendall, B. Carbapenem Resistant Enterobacteriaceae. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Sunenshine, R.H.; McDonald, L.C. Clostridium difficile-Associated Disease: New Challenges from an Established Pathogen. Cleve. Clin. J. Med. 2006, 73, 187–197. [Google Scholar] [CrossRef]
- Savidge, T.C.; Pan, W.-H.; Newman, P.; O’brien, M.; Anton, P.M.; Pothoulakis, C. Clostridium difficile Toxin B Is an Inflammatory Enterotoxin in Human Intestine. Gastroenterology 2003, 125, 413–420. [Google Scholar] [CrossRef]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile Infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef] [Green Version]
- European Centre for Disease Prevention and Control. Point Prevalence Survey of Healthcare Associated Infections and Antimicrobial Use in European Acute Care Hospitals. Available online: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/healthcare-associated-infections-antimicrobial-use-PPS.pdf (accessed on 19 June 2021).
- Pierce, J.; Markley, D.; Schellack, N.; Stevens, M.P. Guide to Infection Control in the Healthcare Setting Antimicrobial Stewardship in the Hospital Setting. Available online: https://isid.org/guide/amr/antimicrobial-stewardship/ (accessed on 18 June 2021).
- Antimicrobial Stewardship Programmes in Health-Care Facilities in Low- and Middle-Income Countries: A Who Practical toolkit. Available online: https://apps.who.int/iris/bitstream/handle/10665/329404/9789241515481-eng.pdf?sequence=1&isAllowed=y (accessed on 18 June 2021).
- Stewart, S.; Robertson, C.; Pan, J.; Kennedy, S.; Dancer, S.; Haahr, L.; Manoukian, S.; Mason, H.; Kavanagh, K.; Cook, B.; et al. Epidemiology of Healthcare-Associated Infection Reported from a Hospital-Wide Incidence Study: Considerations for Infection Prevention and Control Planning. J. Hosp. Infect. 2021, 114, 10–22. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States. Available online: https://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 16 June 2021).
- Caselli, E.; Arnoldo, L.; Rognoni, C.; D’Accolti, M.; Soffritti, I.; Lanzoni, L.; Bisi, M.; Volta, A.; Tarricone, R.; Brusaferro, S.; et al. Impact of a Probiotic-Based Hospital Sanitation on Antimicrobial Resistance and HAI-Associated Antimicrobial Consumption and Costs: A Multicenter Study. Infect. Drug Resist. 2019, 12, 501–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahmqvist, M.; Samuelsson, A.; Bastami, S.; Rutberg, H. Direct Health Care Costs and Length of Hospital Stay Related to Health Care-Acquired Infections in Adult Patients Based on Point Prevalence Measurements. Am. J. Infect. Control 2016, 44, 500–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centers for Disease Control and Prevention. Making Health Care Safer: Reducing Bloodstream Infections. Available online: http://www.cdc.gov/VitalSigns/pdf/2011-03-vitalsigns.pdf (accessed on 1 July 2021).
- World Health Organization Report on the Burden of Endemic Health Care-Associated Infection Worldwide. Clean Care Is Safer Care. A Systematic Review of the Literature. Available online: https://apps.who.int/iris/bitstream/handle/10665/80135/9789241501507_eng.pdf (accessed on 18 May 2021).
- Graves, N.; Mitchell, B.; Kiernan, J. The Cost-Effectiveness of Temporary Single Patient Rooms to Reduce Risks of Healthcare Associated Infection. J. Hosp. Infect. 2021, 116, 21–28. [Google Scholar] [CrossRef]
- Vrijens, F.; Hulstaert, F.; Devriese, S.; van de Sande, S. Hospital-Acquired Infections in Belgian Acute-Care Hospitals: An Estimation of Their Global Impact on Mortality, Length of Stay and Healthcare Costs. Epidemiol. Infect. 2012, 140, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Li, L.; Li, W.; Hou, T.; Ma, H.; Yang, Y.; Wu, A.; Liu, Y.; Wen, J.; Yang, H.; et al. Impact of Healthcare-Associated Infections on Length of Stay: A Study in 68 Hospitals in China. BioMed Res. Int. 2019, 2019, 2590563. [Google Scholar] [CrossRef] [Green Version]
- Ohannessian, R.; Gustin, M.-P.; Bénet, T.; Gerbier-Colomban, S.; Girard, R.; Argaud, L.; Rimmelé, T.; Guerin, C.; Bohé, J.; Piriou, V.; et al. Estimation of Extra Length of Stay Attributable to Hospital-Acquired Infections in Adult ICUs Using a Time-Dependent Multistate Model. Crit. Care Med. 2018, 46, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, B.G.; Ferguson, J.K.; Anderson, M.; Sear, J.; Barnett, A. Length of Stay and Mortality Associated with Healthcare-Associated Urinary Tract Infections: A Multi-State Model. J. Hosp. Infect. 2016, 93, 92–99. [Google Scholar] [CrossRef] [Green Version]
- European Academies Science Advisory Council. Healthcare-Associated Infections: The View from EASAC. Available online: http://www.easac.eu/fileadmin/PDF_s/reports_statements/Healthcare-associated.pdf (accessed on 19 June 2021).
- Anderson, D.J.; Pyatt, D.G.; Weber, D.J.; Rutala, W.A. Statewide Costs of Health Care-Associated Infections: Estimates for Acute Care Hospitals in North Carolina. Am. J. Infect. Control 2013, 41, 764–768. [Google Scholar] [CrossRef] [Green Version]
- Manoukian, S.; Stewart, S.; Graves, N.; Mason, H.; Robertson, C.; Kennedy, S.; Pan, J.; Kavanagh, K.; Haahr, L.; Adil, M.; et al. Bed-Days and Costs Associated with the Inpatient Burden of Healthcare-Associated Infection in the UK. J. Hosp. Infect. 2021, 114, 43–50. [Google Scholar] [CrossRef]
- Spellberg, B.; Blaser, M.; Guidos, R.J.; Boucher, H.W.; Bradley, J.S.; Eisenstein, B.I.; Gerding, D.; Lynfield, R.; Reller, L.B.; Rex, J.; et al. Combating Antimicrobial Resistance: Policy Recommendations to Save Lives. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 52, S397–S428. [Google Scholar] [CrossRef]
- Stone, P.W. Economic Burden of Healthcare-Associated Infections: An American Perspective. Expert Rev. Pharm. Outcomes Res. 2009, 9, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Schmier, J.K.; Hulme-Lowe, C.K.; Semenova, S.; Klenk, J.A.; DeLeo, P.C.; Sedlak, R.; Carlson, P.A. Estimated Hospital Costs Associated with Preventable Health Care-Associated Infections If Health Care Antiseptic Products Were Unavailable. Clinicoecon. Outcomes Res. 2016, 8, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Shepard, J.; Frederick, J.; Wong, F.; Madison, S.; Tompkins, L.; Hadhazy, E. Could the Prevention of Health Care-Associated Infections Increase Hospital Cost? The Financial Impact of Health Care-Associated Infections from a Hospital Management Perspective. Am. J. Infect. Control 2020, 48, 255–260. [Google Scholar] [CrossRef]
- Lamarsalle, L.; Hunt, B.; Schauf, M.; Szwarcensztein, K.; Valentine, W.J. Evaluating the Clinical and Economic Burden of Healthcare-Associated Infections during Hospitalization for Surgery in France. Epidemiol. Infect. 2013, 141, 2473–2482. [Google Scholar] [CrossRef] [Green Version]
- Arefian, H.; Hagel, S.; Heublein, S.; Rissner, F.; Scherag, A.; Brunkhorst, F.M.; Baldessarini, R.J.; Hartmann, M. Extra Length of Stay and Costs Because of Health Care-Associated Infections at a German University Hospital. Am. J. Infect. Control 2016, 44, 160–166. [Google Scholar] [CrossRef]
- World Population Review. Available online: https://worldpopulationreview.com/country-rankings/low-income-countries (accessed on 27 April 2022).
- European Centre for Disease Prevention and Control Point Prevalence Survey of Healthcare Associated Infections and Antimicrobial Use in European Acute Care Hospitals—ECDC PPS Validation Protocol Version 3.1.2. Available online: https://www.ecdc.europa.eu/en/publications-data/point-prevalence-survey-healthcare-associated-infections-and-antimicrobial-use-4 (accessed on 14 March 2021).
- Gastmeier, P.; Geffers, C.; Brandt, C.; Zuschneid, I.; Sohr, D.; Schwab, F.; Behnke, M.; Daschner, F.; Rüden, H. Effectiveness of a Nationwide Nosocomial Infection Surveillance System for Reducing Nosocomial Infections. J. Hosp. Infect. 2006, 64, 16–22. [Google Scholar] [CrossRef]
- Bénet, T.; Ecochard, R.; Voirin, N.; Machut, A.; Lepape, A.; Savey, A.; Vanhems, P. Effect of Standardized Surveillance of Intensive Care Unit-Acquired Infections on Ventilator-Associated Pneumonia Incidence. Infect. Control Hosp. Epidemiol. 2014, 35, 1290–1293. [Google Scholar] [CrossRef]
- Bouza, E.; San Juan, R.; Muñoz, P.; Voss, A.; Kluytmans, J. A European Perspective on Nosocomial Urinary Tract Infections II. Report on Incidence, Clinical Characteristics and Outcome (ESGNI-004 Study). European Study Group on Nosocomial Infection. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2001, 7, 532–542. [Google Scholar] [CrossRef] [Green Version]
- Rosenthal, V.D.; Bijie, H.; Maki, D.G.; Mehta, Y.; Apisarnthanarak, A.; Medeiros, E.A.; Leblebicioglu, H.; Fisher, D.; Álvarez-Moreno, C.; Khader, I.A.; et al. International Nosocomial Infection Control Consortium (INICC) Report, Data Summary of 36 Countries, for 2004–2009. Am. J. Infect. Control 2012, 40, 396–407. [Google Scholar] [CrossRef] [PubMed]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable Deaths and Disability-Adjusted Life-Years Caused by Infections with Antibiotic-Resistant Bacteria in the EU and the European Economic Area in 2015: A Population-Level Modelling Analysis. Lancet. Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, A.W. Control of Hospital Acquired Infections and Antimicrobial Resistance in Europe: The Way to Go. Wien. Med. Wochenschr. 2019, 169, 25–30. [Google Scholar] [CrossRef] [Green Version]
- Boccella, M.; Santella, B.; Pagliano, P.; De Filippis, A.; Casolaro, V.; Galdiero, M.; Borrelli, A.; Capunzo, M.; Boccia, G.; Franci, G. Prevalence and Antimicrobial Resistance of Enterococcus Species: A Retrospective Cohort Study in Italy. Antibiotics 2021, 10, 1552. [Google Scholar] [CrossRef] [PubMed]
- Colgan, R.; Powers, J.H. Appropriate Antimicrobial Prescribing: Approaches That Limit Antibiotic Resistance. Am. Fam. Physician 2001, 64, 999–1004. [Google Scholar]
- CDC: Core Elements of Hospital Antibiotic Stewardship Programs. Available online: https://www.cdc.gov/antibiotic-use/core-elements/hospital.html (accessed on 21 June 2021).
- Lautenbach, E.; LaRosa, L.A.; Marr, A.M.; Nachamkin, I.; Bilker, W.B.; Fishman, N.O. Changes in the Prevalence of Vancomycin-Resistant Enterococci in Response to Antimicrobial Formulary Interventions: Impact of Progressive Restrictions on Use of Vancomycin and Third-Generation Cephalosporins. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2003, 36, 440–446. [Google Scholar] [CrossRef] [Green Version]
- Lawton, R.M.; Fridkin, S.K.; Gaynes, R.P.; McGowan, J.E.J. Practices to Improve Antimicrobial Use at 47 US Hospitals: The Status of the 1997 SHEA/IDSA Position Paper Recommendations. Society for Healthcare Epidemiology of America/Infectious Diseases Society of America. Infect. Control Hosp. Epidemiol. 2000, 21, 256–259. [Google Scholar] [CrossRef]
- The National Center for the Surveillance and Control of Communicable Diseases: Methodology for Surveillance of Nosocomial Infections in the Sentinel System and Microbial Resistance. Available online: https://www.cnscbt.ro/index.php/metodologii/infectii-nosocomiale/167-metodologie-2013-sentinela-nosocomiala-amr/file (accessed on 21 June 2021).
- Collective Movie. Available online: https://www.collectivemovie.com/ (accessed on 12 February 2022).
- Pirii, L.E.; Friedrich, A.W.; Rossen, J.W.A.; Vogels, W.; Beerthuizen, G.I.J.M.; Nieuwenhuis, M.K.; Kooistra-Smid, A.M.D.; Bathoorn, E. Extensive Colonization with Carbapenemase-Producing Microorganisms in Romanian Burn Patients: Infectious Consequences from the Colectiv Fire Disaster. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 175–183. [Google Scholar] [CrossRef] [Green Version]
- Remus, C.; O’Brien, T. Corruption and Conflagration: (In)Justice and Protest in Bucharest after the Colectiv Fire. Urban Geogr. 2020, 41, 368–388. [Google Scholar] [CrossRef]
- Ágnes, P.; Zahorán, C.C. Victims of Health Care: Lesson from the Documentary Colectiv. Cent. Eur. Horiz. 2020, 1, 33–37. [Google Scholar]
- Grundmann, H.; Glasner, C.; Albiger, B.; Aanensen, D.M.; Tomlinson, C.T.; Andrasević, A.T.; Cantón, R.; Carmeli, Y.; Friedrich, A.W.; Giske, C.G.; et al. Occurrence of Carbapenemase-Producing Klebsiella Pneumoniae and Escherichia Coli in the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE): A Prospective, Multinational Study. Lancet. Infect. Dis. 2017, 17, 153–163. [Google Scholar] [CrossRef] [Green Version]
- 2012/506/EU: Commission Implementing Decision of 8 August 2012 Amending Decision 2002/253/EC Laying down Case Definitions for Reporting Communicable Diseases to the Community Network under Decision No 2119/98/EC of the European Parliament and of the Counc. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32012D0506 (accessed on 28 June 2021).
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2013. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net). Available online: https://www.ecdc.europa.eu/en/publications-data/antimicrobial-resistance-surveillance-europe-2013 (accessed on 14 April 2021).
- The National Center for the Surveillance and Control of Communicable Diseases: Guide on Carbapenemase-Producing Enterobacteriaceae: Diagnosis, Prevention of Interhuman Transmission and Treatment. Available online: https://www.cnscbt.ro/index.php/ghiduri-si-protocoale/ghiduri/522-ghid-privind-enterobacteriaceae-producatoare-de-carbapenemaze-diagnosticul-prevenirea-transmiterii-interumane-si-tratamentul-infectiilor-produse/file (accessed on 22 June 2021).
- Doi, Y.; Paterson, D.L. Carbapenemase-Producing Enterobacteriaceae. Semin. Respir. Crit. Care Med. 2015, 36, 74–84. [Google Scholar] [CrossRef] [Green Version]
- Baroiu, L.; Alina, I.; Drăgănescu, M.; Debita, M.; Arbune, M.; Rugina, S. Actualităţi În Etiopatogenia, Epidemiologia Şi Diagnosticul Infecţiei Cu Clostridium difficile. Rom. J. Infect. Dis. 2014, 21, 184–189. [Google Scholar] [CrossRef]
- The National Center for the Surveillance and Control of Communicable Diseases. Data Analysis 2019. Available online: https://www.cnscbt.ro/index.php/analiza-date-supraveghere/infectii-nosocomiale-1/2704-consumul-de-antibiotice-rezistenta-microbiana-si-infectii-asociate-asistentei-medicale-in-romania-2019/file (accessed on 12 August 2021).
- Arbune, M.; Gurau, G.; Niculet, E.; Iancu, A.V.; Lupasteanu, G.; Fotea, S.; Vasile, M.C.; Tatu, A.L. Prevalence of Antibiotic Resistance of ESKAPE Pathogens Over Five Years in an Infectious Diseases Hospital from South-East of Romania. Infect. Drug Resist. 2021, 14, 2369–2378. [Google Scholar] [CrossRef] [PubMed]
- Petca, R.-C.; Mareș, C.; Petca, A.; Negoiță, S.; Popescu, R.-I.; Boț, M.; Barabás, E.; Chibelean, C.B. Spectrum and Antibiotic Resistance of Uropathogens in Romanian Females. Antibiotics 2020, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC). Data from the ECDC Surveillance Atlas—Antimicrobial Resistance. Reports 2005–2019. Available online: https://ecdc.europa.eu/en/antimicrobial-resistance/surveillance-and-disease-data/data-ecdc (accessed on 8 January 2022).
- Ionescu, R.; Mediavilla, J.R.; Chen, L.; Grigorescu, D.O.; Idomir, M.; Kreiswirth, B.N.; Roberts, R.B. Molecular Characterization and Antibiotic Susceptibility of Staphylococcus aureus from a Multidisciplinary Hospital in Romania. Microb. Drug Resist. 2010, 16, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Monecke, S.; Müller, E.; Dorneanu, O.S.; Vremeră, T.; Ehricht, R. Molecular Typing of MRSA and of Clinical Staphylococcus aureus Isolates from Iaşi, Romania. PLoS ONE 2014, 9, e97833. [Google Scholar] [CrossRef] [Green Version]
- WHO. Antimicrobial Resistance Surveillance in Europe 2022–2020 Data (2022). Available online: https://www.euro.who.int/en/health-topics/disease-prevention/antimicrobial-resistance/publications/2022/antimicrobial-resistance-surveillance-in-europe-2022-2020-data-2022 (accessed on 1 March 2022).
- Gavriliu, L.C.; Benea, O.E.; Benea, S. Antimicrobial Resistance Temporal Trend of Klebsiella Pneumoniae Isolated from Blood. J. Med. Life 2016, 9, 419–423. [Google Scholar] [PubMed]
- Allocati, N.; Masulli, M.; Alexeyev, M.F.; Di Ilio, C. Escherichia Coli in Europe: An Overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef]
- Cambrea, S. Antibiotic Susceptibility of Escherichia Coli Strains Isolated in a Pediatric Population from South Eastern Romania. J. Pediatr. Infect. Dis. 2014, 9, 157–162. [Google Scholar] [CrossRef] [Green Version]
- Ministry of Health: The Government of Romania Ministry of Health Activity Report for the Year 2019. Available online: http://www.ms.ro/wp-content/uploads/2020/12/Raport-de-activitate-pentru-anul-2019-1.pdf (accessed on 25 June 2021).
- World Health Organization. Improving Infection Prevention and Control at the Health Facility: Interim Practical Manual Supporting Implementation of the WHO Guidelines on Core Components of Infection Prevention and Control Programmes 2018. Available online: https://apps.who.int/iris/handle/10665/279788 (accessed on 25 June 2021).
- World Health Organization. Minimum Requirements for Infection Prevention and Control Programmes. World Health Organization 2019. Available online: https://apps.who.int/iris/handle/10665/330080 (accessed on 25 June 2021).
- Dick, A.W.; Perencevich, E.N.; Pogorzelska-Maziarz, M.; Zwanziger, J.; Larson, E.L.; Stone, P.W. A Decade of Investment in Infection Prevention: A Cost-Effectiveness Analysis. Am. J. Infect. Control 2015, 43, 4–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClung, L.; Obasi, C.; Knobloch, M.J.; Safdar, N. Health Care Worker Perspectives of Their Motivation to Reduce Health Care-Associated Infections. Am. J. Infect. Control 2017, 45, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Zeitler, K.; Narayanan, N. The Present and Future State of Antimicrobial Stewardship and Rapid Diagnostic Testing: Can One Ideally Succeed Without the Other? Curr. Treat. Options Infect. Dis. 2019, 11, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Ferone, M.; Gowen, A.; Fanning, S.; Scannell, A. Microbial Detection and Identification Methods: Bench Top Assays to Omics Approaches. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3106–3129. [Google Scholar] [CrossRef]
- Parsons, B.D.; Zelyas, N.; Berenger, B.M.; Chui, L. Detection, Characterization, and Typing of Shiga Toxin-Producing Escherichia Coli. Front. Microbiol. 2016, 7, 478. [Google Scholar] [CrossRef] [Green Version]
- Harbarth, S.; Hawkey, P.M.; Tenover, F.; Stefani, S.; Pantosti, A.; Struelens, M.J. Update on Screening and Clinical Diagnosis of Meticillin-Resistant Staphylococcus aureus (MRSA). Int. J. Antimicrob. Agents 2011, 37, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Hassoun, A.; Linden, P.K.; Friedman, B. Incidence, Prevalence, and Management of MRSA Bacteremia across Patient Populations-a Review of Recent Developments in MRSA Management and Treatment. Crit. Care 2017, 21, 211. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid Methods for the Detection of Foodborne Bacterial Pathogens: Principles, Applications, Advantages and Limitations. Front. Microbiol. 2014, 5, 770. [Google Scholar] [CrossRef] [Green Version]
- Lui, C.; Cady, N.C.; Batt, C.A. Nucleic Acid-Based Detection of Bacterial Pathogens Using Integrated Microfluidic Platform Systems. Sensors 2009, 9, 3713–3744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sin, M.L.Y.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and Challenges in Biosensor-Based Diagnosis of Infectious Diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.W.; Ha, D.; Lee, J.; Lee, S.K.; Kim, T. Review of Micro/Nanotechnologies for Microbial Biosensors. Front. Bioeng. Biotechnol. 2015, 3, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zhu, Y.; Wu, X.; Hoffmann, M.R. Rapid Detection Methods for Bacterial Pathogens in Ambient Waters at the Point of Sample Collection: A Brief Review. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, S84–S90. [Google Scholar] [CrossRef]
- Lee, C.-R.; Lee, J.H.; Park, K.S.; Kim, Y.B.; Jeong, B.C.; Lee, S.H. Global Dissemination of Carbapenemase-Producing Klebsiella Pneumoniae: Epidemiology, Genetic Context, Treatment Options, and Detection Methods. Front. Microbiol. 2016, 7, 895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, M.-C.; Kuo, A.-J.; Liu, K.-L.; Wen, Y.-H.; Chia, J.-H.; Chang, P.-Y.; Lee, M.-H.; Wu, T.-L.; Chang, S.-C.; Lu, J.-J. Routine Identification of Microorganisms by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry: Success Rate, Economic Analysis, and Clinical Outcome. J. Microbiol. Immunol. Infect. 2017, 50, 662–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, A.T.; Tallent, S.M. From Commensal to Consumer: Staphylococcus aureus Toxins, Diseases, and Detection Methods. J. AOAC Int. 2018, 101, 1127–1134. [Google Scholar] [CrossRef]
- Capatina, D.; Feier, B.; Hosu, O.; Tertis, M.; Cristea, C. Analytical Methods for the Characterization and Diagnosis of Infection with Pseudomonas Aeruginosa: A Critical Review. Anal. Chim. Acta 2022, 1204, 339696. [Google Scholar] [CrossRef] [PubMed]
- Faron, M.L.; Ledeboer, N.A.; Buchan, B.W. Resistance Mechanisms, Epidemiology, and Approaches to Screening for Vancomycin-Resistant Enterococcus in the Health Care Setting. J. Clin. Microbiol. 2016, 54, 2436–2447. [Google Scholar] [CrossRef] [Green Version]
- Alizadeh, N.; Rezaee, M.A.; Kafil, H.S.; Barhaghi, M.H.S.; Memar, M.Y.; Milani, M.; Hasani, A.; Ghotaslou, R. Detection of Carbapenem-Resistant Enterobacteriaceae by Chromogenic Screening Media. J. Microbiol. Methods 2018, 153, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Hou, Y.; Peters, B.M.; Chen, D.; Li, B.; Li, L.; Shirtliff, M.E. Chromogenic Media for MRSA Diagnostics. Mol. Biol. Rep. 2016, 43, 1205–1212. [Google Scholar] [CrossRef]
- Suwantarat, N.; Roberts, A.; Prestridge, J.; Seeley, R.; Speser, S.; Harmon, C.; Zhang, C.; Henciak, S.; Stamper, P.D.; Ross, T.; et al. Comparison of Five Chromogenic Media for Recovery of Vancomycin-Resistant Enterococci from Fecal Samples. J. Clin. Microbiol. 2014, 52, 4039–4042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samani, S.S.; Khojastehnezhad, A.; Ramezani, M.; Alibolandi, M.; Yazdi, F.T.; Mortazavi, S.A.; Khoshbin, Z.; Abnous, K.; Taghdisi, S.M. Ultrasensitive Detection of Micrococcal Nuclease Activity and Staphylococcus aureus Contamination Using Optical Biosensor Technology—A Review. Talanta 2021, 226, 122168. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, L.; Huang, Q.; Tong, T.; Zhou, Y.; Li, Z.; Bai, Q.; Liang, H.; Chen, L. Recent Advances on Aptamer-Based Biosensors for Detection of Pathogenic Bacteria. World J. Microbiol. Biotechnol. 2021, 37, 45. [Google Scholar] [CrossRef]
- Pourakbari, R.; Shadjou, N.; Yousefi, H.; Isildak, I.; Yousefi, M.; Rashidi, M.-R.; Khalilzadeh, B. Recent Progress in Nanomaterial-Based Electrochemical Biosensors for Pathogenic Bacteria. Microchim. Acta 2019, 186, 820. [Google Scholar] [CrossRef] [PubMed]
- Nagy, E. What Do We Know about the Diagnostics, Treatment and Epidemiology of Clostridioides (Clostridium) difficile Infection in Europe? J. Infect. Chemother. Off. J. Japan Soc. Chemother. 2018, 24, 164–170. [Google Scholar] [CrossRef] [Green Version]
- Schweitzer, L.; Gervais, P.; Paquet-Bolduc, B.; Loo, V.G.; Longtin, Y. Detection of Free Toxin B in the Stool of Asymptomatic Clostridioides difficile Carriers by the Cell Cytotoxicity Neutralization Assay. Open Forum Infect. Dis. 2021, 8, ofab209. [Google Scholar] [CrossRef]
- Gateau, C.; Couturier, J.; Coia, J.; Barbut, F. How to: Diagnose Infection Caused by Clostridium difficile. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2018, 24, 463–468. [Google Scholar] [CrossRef] [Green Version]
- Brown, D.F.J.; Edwards, D.I.; Hawkey, P.M.; Morrison, D.; Ridgway, G.L.; Towner, K.J.; Wren, M.W.D. Guidelines for the Laboratory Diagnosis and Susceptibility Testing of Methicillin-Resistant Staphylococcus aureus (MRSA). J. Antimicrob. Chemother. 2005, 56, 1000–1018. [Google Scholar] [CrossRef] [Green Version]
- Brooks, L.R.K.; Mias, G.I. Streptococcus Pneumoniae’s Virulence and Host Immunity: Aging, Diagnostics, and Prevention. Front. Immunol. 2018, 9, 1366. [Google Scholar] [CrossRef]
- Buonomini, A.R.; Riva, E.; Di Bonaventura, G.; Gherardi, G. Rapid Detection of Methicillin-Resistant Staphylococcus aureus Directly from Blood for the Diagnosis of Bloodstream Infections: A Mini-Review. Diagnostics 2020, 10, 830. [Google Scholar] [CrossRef] [PubMed]
- Manual for the Laboratory Identification and Antimicrobial Susceptibility Testing of Bacterial Pathogens of Public Health Importance in the Developing World. Available online: https://www.who.int/csr/resources/publications/drugresist/en/IAMRmanual.pdf?ua=1 (accessed on 12 May 2021).
- Varghese, R.; Jayaraman, R.; Veeraraghavan, B. Current Challenges in the Accurate Identification of Streptococcus Pneumoniae and Its Serogroups/Serotypes in the Vaccine Era. J. Microbiol. Methods 2017, 141, 48–54. [Google Scholar] [CrossRef]
- Tarr, G.A.M.; Lin, C.Y.; Vandermeer, B.; Lorenzetti, D.L.; Tarr, P.I.; Chui, L.; Hartling, L.; Freedman, S.B.; on behalf of the Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE). Diagnostic Test Accuracy of Commercial Tests for Detection of Shiga Toxin–Producing Escherichia Coli: A Systematic Review and Meta-Analysis. Clin. Chem. 2020, 66, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, D.; Giron, M. Enterobacter Infections. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Fleece, M.E.; Pholwat, S.; Mathers, A.J.; Houpt, E.R. Molecular Diagnosis of Antimicrobial Resistance in Escherichia Coli. Expert Rev. Mol. Diagn. 2018, 18, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Strategies for Identification of Carbapenemase-Producing Enterobacteriaceae. J. Antimicrob. Chemother. 2013, 68, 487–489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Willems, R.J.L.; Friedrich, A.W.; Rossen, J.W.A.; Bathoorn, E. Enterococcus Faecium: From Microbiological Insights to Practical Recommendations for Infection Control and Diagnostics. Antimicrob. Resist. Infect. Control 2020, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, C.; Maquelin, K.; Pina, P.; Ngo Thi, N.A.; Choo-Smith, L.P.; Sockalingum, G.D.; Sandt, C.; Ami, D.; Orsini, F.; Doglia, S.M.; et al. Classification and Identification of Enterococci: A Comparative Phenotypic, Genotypic, and Vibrational Spectroscopic Study. J. Clin. Microbiol. 2001, 39, 1763–1770. [Google Scholar] [CrossRef] [Green Version]
- Willey, B.M.; Jones, R.N.; McGeer, A.; Witte, W.; French, G.; Roberts, R.B.; Jenkins, S.G.; Nadler, H.; Low, D.E. Practical Approach to the Identification of Clinically Relevant Enterococcus Species. Diagn. Microbiol. Infect. Dis. 1999, 34, 165–171. [Google Scholar] [CrossRef]
- UK Standards for Microbiology Investigations Identification of Pseudomonas Species and Other NonGlucose Fermenters NHS. Available online: https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/422699/ID_17i3.pdf (accessed on 14 June 2021).
- Dortet, L.; Poirel, L.; Errera, C.; Nordmann, P.; Patel, R. CarbAcineto NP Test for Rapid Detection of Carbapenemase-Producing Acinetobacter spp. J. Clin. Microbiol. 2014, 52, 2359–2364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayakumar, S.; Biswas, I.; Veeraraghavan, B. Accurate Identification of Clinically Important Acinetobacter spp.: An Update. Futur. Sci. OA 2019, 5, FSO395. [Google Scholar] [CrossRef] [Green Version]
- Sivaranjani, V.; Umadevi, S.; Srirangaraj, S.; Kali, A.; Seetha, K. Multi-Drug Resistant Acinetobacter Species from Various Clinical Samples in a Tertiary Care Hospital from South India. Australas. Med. J. 2013, 6, 697–700. [Google Scholar] [CrossRef]
- Kulkarni, S.S.; Madalgi, R.; Ajantha, G.S.; Kulkarni, R.D. Identification of Genus Acinetobacter: Standardization of in-House PCR and Its Comparison with Conventional Phenotypic Methods. J. Lab. Physicians 2017, 9, 279–282. [Google Scholar] [CrossRef]
- Vila, J.; Gómez, M.D.; Salavert, M.; Bosch, J. Rapid Diagnostic Methods in Clinical Microbiology: Clinical Needs. Infect. Dis. Clin. Microbiol. 2017, 35, 41–46. [Google Scholar]
- Cercenado, E. Utility of Rapid Microbiological Techniques for the Diagnosis of Severe Infections. Rev. Esp. Quimioter. Publ. Of. Soc. Esp. Quimioter. 2017, 30, 52–55. [Google Scholar]
- Ho, C.-S.; Jean, N.; Hogan, C.A.; Blackmon, L.; Jeffrey, S.S.; Holodniy, M.; Banaei, N.; Saleh, A.A.E.; Ermon, S.; Dionne, J. Rapid Identification of Pathogenic Bacteria Using Raman Spectroscopy and Deep Learning. Nat. Commun. 2019, 10, 4927. [Google Scholar] [CrossRef] [PubMed]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.-F. Deep Learning and Artificial Intelligence Methods for Raman and Surface-Enhanced Raman Scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
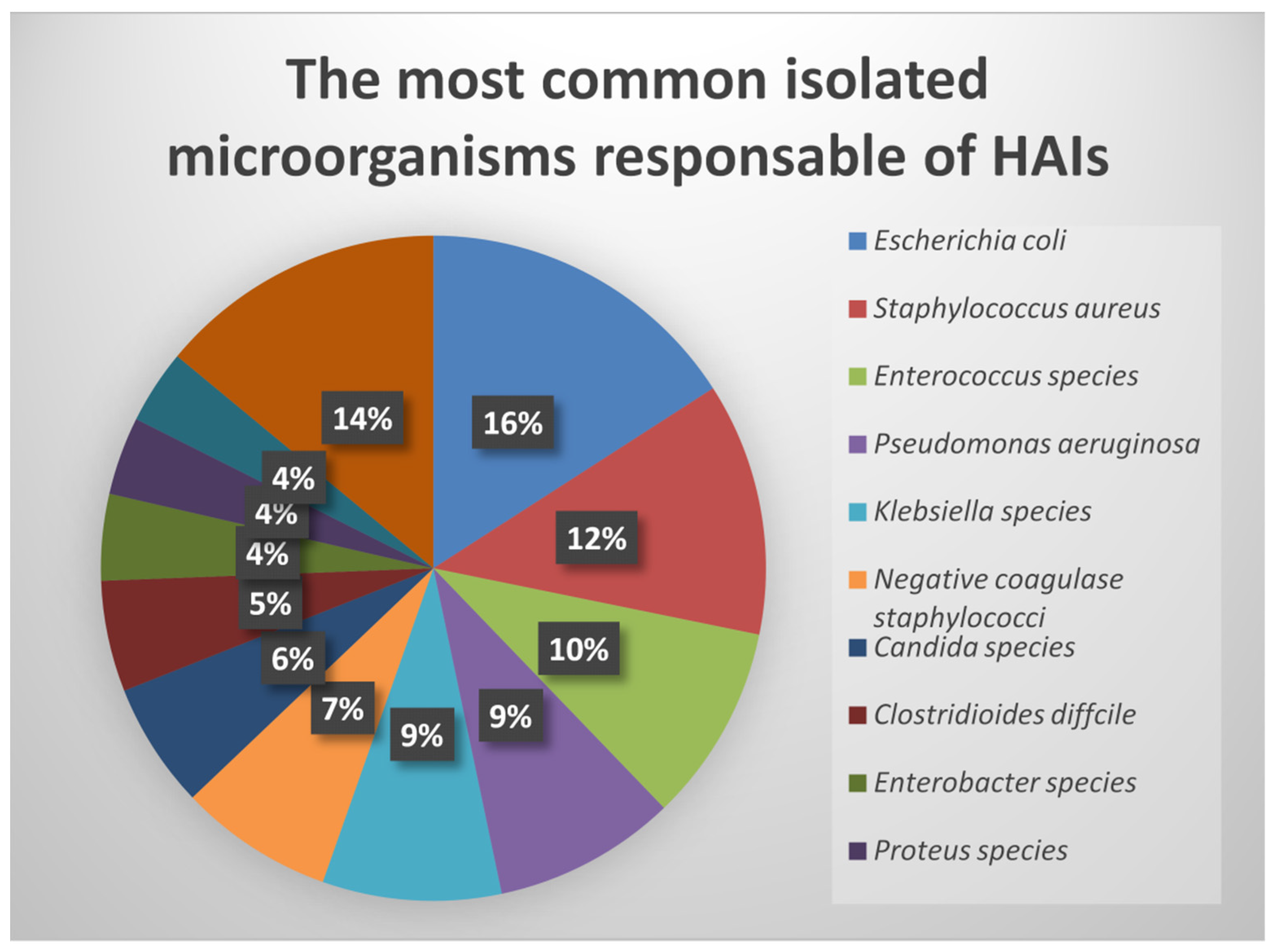
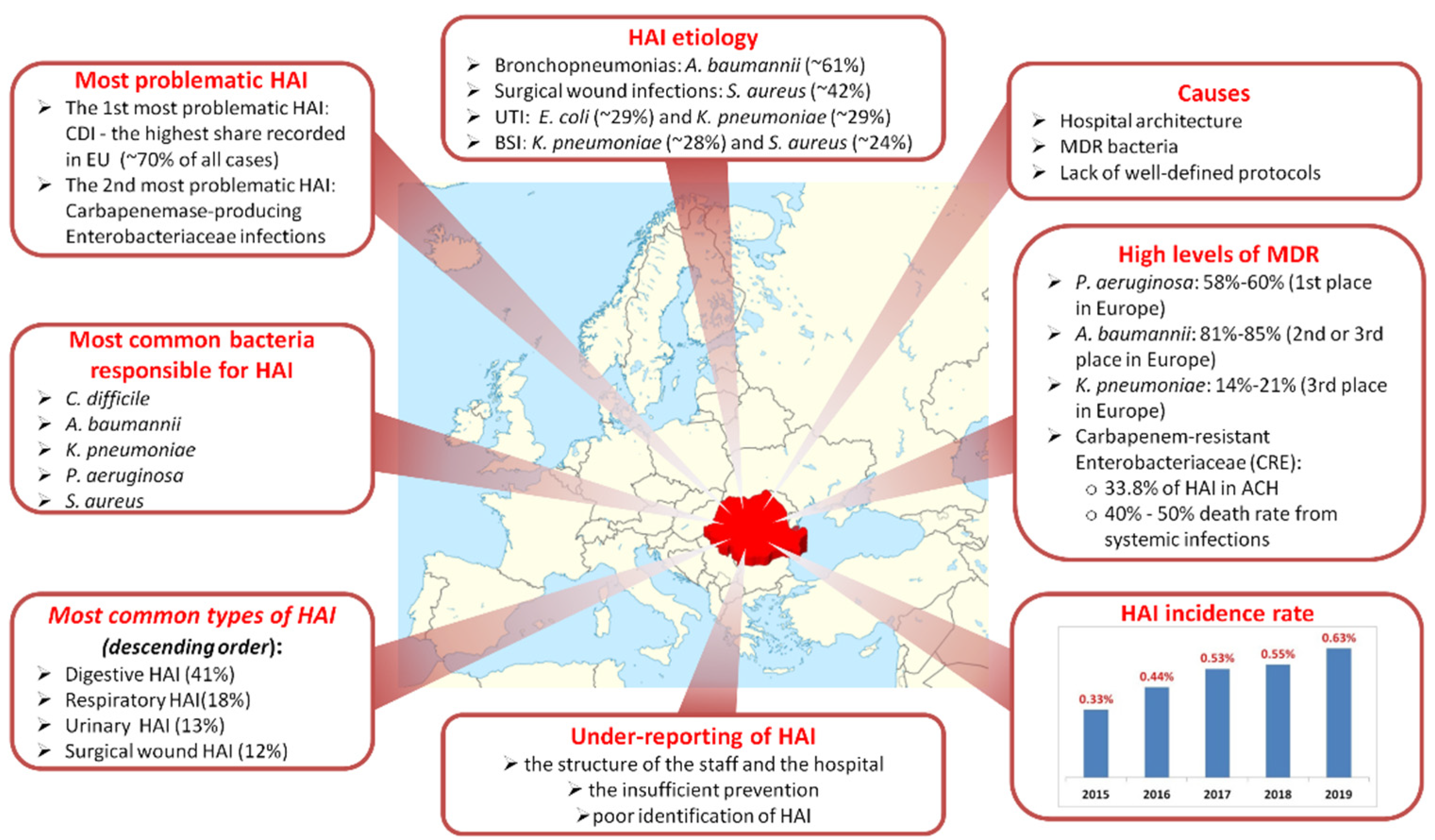
| Characteristics | % of HAI | Causative Organisms | Mortality | Preventable |
|---|---|---|---|---|
| Central line-associated bloodstream infections (CLABSI) | ||||
| 90% associated with a catheter in the bloodstream [2] | 10–15 [38] | S. aureus (23%) Candida spp. (13%) Coagulase-negative Staphylococcus (12%) Enterococcus spp. (12%) Streptococcus spp. (12%) E. coli (8%) Bacteroides spp. (6%) [1,2,39] | 12–25% [2,39] P. aeruginosa: 50% [40] A. baumannii: 29.8–36.9% [41] | 65–70% [42] |
| Catheter-associated urinary tract infections (CAUTI) | ||||
| Associated with preceding instrumentation or indwelling bladder catheters [1,39] | 30–40% [2,43] | E. coli Klebsiella pneumonia/oxytoca Enterococcus spp. Pseudomonas aeruginosa Candida spp. [1] | 13,000 deaths per year in USA [2] | 65–70% [42] |
| Surgical site infections (SSI). Skin and soft tissue infections (SSTI) | ||||
| Skin, gastrointestinal tract, and female genital tract serve as a reservoir of the healthy flora that may contaminate the surgical site (1) MDR pathogens are increasing [2] | 20–24% [2,39,43] | Occasionally are due to airborne spread of skin squames [39] E. coli S. aureus: most common cause of postsurgical wound infections [30] Klebsiella spp. Enterobacter spp. Enterococcus spp. Streptococcus spp. Coagulase-negative Staphylococcus [1,2,39] | Over one-third of postoperative deaths [2] | 40–60% [2,43] |
| Ventilator-associated pneumonia (VAP) and Hospital acquired pneumonia (HAP) | ||||
| 5 to 26 % of mechanically ventilated patients develop VAP, after 48 h of intubation [1,2,39] HAP develops after 48 h of admission [1,2] In elderly patients, up to 38% of HAP is due to S. pneumoniae [44] MDR pathogens are increasing in VAP [1] | 24–27% [2,39] | S. aureus: common in HAP [44] and in VAP in intensive care units (ICU) [30] P. aeruginosa Candida spp. Klebsiella oxytoca and K. pneumoniae Streptococcus spp. Enterobacter spp. [1,2] | Up to 50% [2] Pneumococcal pneumonia: 19% [45] P. aeruginosa: >30% [46] | 55% [42] |
| Gastrointestinal infections with Clostridioides difficile | ||||
| Most common nosocomial cause of diarrhea [47,48] It colonizes the large intestine of approximately 3% of the general population and up to 30–40% of hospitalized patients [48,49,50] Only toxigenic strains, producing toxin A and/or B, are pathogenic [51] CDI risks: use of multiple antibiotics, broader-spectrum agents, and longer duration of therapy [2] Spores infect patients via the fecal-oral route [2] | 12% [2] | Clostridioides difficile | Up to 7.2% [43] Up to 29,600 deaths per year in the USA [2,52] and up to 3700 deaths per year in Europe [53] | No data |
| Group | Selected Antimicrobials | Characteristics |
|---|---|---|
| Access group | Amikacin Amoxicillin Amoxicillin + clavulanic acid Ampicillin Benzathine benzylpenicillin Benzylpenicillin Cefalexin Cefazolin Chloramphenicol Clindamycin Cloxacillin Doxycycline Gentamicin Metronidazole Nitrofurantoin Phenoxymethylpenicillin Procaine benzylpenicillin Spectinomycin Sulfamethoxazole + trimethoprim | This group includes antimicrobials and antimicrobials classes that have activity against a wide range of commonly encountered susceptible pathogens while showing lower resistance potential than antibiotics in the Watch and Reserve groups. Access antimicrobials should be widely available, affordable, and quality assured to improve access and promote appropriate use. Selected Access group antimicrobials (shown here) are included on the WHO Essential Medicine List (EML) as essential first-choice or second-choice empirical treatment options for specific infectious syndromes. |
| Watch group | Azithromycin Cefixime Cefotaxime Ceftazidime Ceftriaxone Cefuroxime Ciprofloxacin Clarithromycin Meropenem Piperacillin + tazobactam Vancomycin | This group includes antimicrobials and antimicrobials classes that have higher resistance potential. This group includes most of the highest priority agents among the Critically Important Antimicrobials for Human Medicine and/or antimicrobials that are at relatively high risk of selection of bacterial resistance. Watch group antimicrobials should be prioritized as key targets of national and local stewardship programs and monitoring. Selected Watch group antimicrobials (shown here) are included on the WHO EML as essential first-choice or second-choice empirical treatment options for a limited number of specific infectious syndromes. |
| Reserve group | Azithromycin Cefixime Cefotaxime Ceftazidime Ceftriaxone Cefuroxime Ciprofloxacin Clarithromycin Meropenem Piperacillin + tazobactam Vancomycin | This group includes antimicrobials and antimicrobials classes that should be reserved for treatment of confirmed or suspected infections due to MDRorganisms and treated as “last-resort” options. Their use should be tailored to highly specific patients and settings when all alternatives have failed or are not suitable. They could be protected and prioritized as key targets of national and international stewardship programs, involving monitoring and utilization reporting, to preserve their effectiveness. Selected Reserve group antibiotics (shown here) are included on the WHO EML when they have a favourable risk-benefit profile and proven activity against “Critical Priority” or “High Priority” pathogens identified by the WHO Priority Pathogens List, notablyCRE. |
| Type of HAI | Country/Region | Excess LOS (Days) | Estimated Costs | References |
|---|---|---|---|---|
| CLABSI | USA | 7–15 15 for MRSA | $31,000–65,000 per episode $2.7 billion per year | [12,14,15,39,43] |
| Europe | 4–14 | €4200–13,030 per episode | [61] | |
| Australia | 5.33 | $245,371 per 100,000 occupied bed-days | [62] | |
| Scotland | 11.4 | £9109 per case | [56] | |
| Belgium | 10.2 | [63] | ||
| India | 5 | $14,818 per case | [61] | |
| China | 12.8 | [64] | ||
| CAUTI | USA | $13,000 per episode $340 million each year | [2,14] | |
| France | 1.5 | [65] | ||
| Belgium | 4.6 | [63] | ||
| Australia | 2–5 | $85,081 per 100,000 occupied bed-days | [62,66] | |
| India | 8 | [61] | ||
| China | 10.3 | [64] | ||
| SSI | USA | 11 23 for MRSA | $3000–29,000 per episode. $43,000 per episode for MRSA. Up to 10 billion per year. | [2,12,14,15,67,68] |
| Australia | 4–8 | $508,243 per 100,000 occupied bed-days | [62] | |
| Scotland | 9.8 | £7830 per case | [56,69] | |
| Belgium | 5.9 | [63] | ||
| China | 11.8 | [64] | ||
| VAP and HAP | USA | 9.1 in the ICU for VAP Up to 9 for HAP | $47,000 per episode for VAP $40,000 per episode for HAP | [2,14,39] |
| Australia | 2.82 | $276,469 per 100,000 occupied bed-days | [62] | |
| France | 6.5 | [65] | ||
| Scotland | 16.3 | £13,024 per case | [56,69] | |
| India | 11 | [61] | ||
| CDI | USA | 3 | Up to $17,000 per episode | [14,15] |
| Australia | 0.5 | $8782 per 100,000 occupied bed-days | [62] | |
| Belgium | 12.1 | [63] |
| Characteristics | Advantage | Disadvantage | References |
|---|---|---|---|
| Nucleic acid-Based Methods | |||
| Detects specific DNA sequences in the target bacteria Hybridisation of the nucleic acid to a synthetic oligonucleotide Examples: PCR, mPCR, RT-qPCR, LAMP, NASBA, Oligonucleotide DNA Microarray. | Sensitive Specific Faster than culture growth Low detection limit Automated | Require pure samples Prone to contamination Sample processing takes several hours High cost Need for trained personnel Complexity No distinction between viable and not viable bacteria | [120,121] |
| Biosensor-Based Methods | |||
| Can be used for the detection of the whole-cell bacterium, virulence factors, different metabolites, or quorum sensing molecules Examples: electrochemical, optical, piezoelectric biosensors | Low limit of detection Small sample volume Real-time and rapid detection High sensitivity Cost effective method Miniaturization and portability | Sensitive to sample matrix effects | [120,122,123,124] |
| Immunological-Based Methods | |||
| Based on antibody-antigen interactions The sensitivity and specificity are determined by the binding strength of an antibody to its antigen Examples: ELISA, ICA | Sensitive Specific Automated Detection of bacterial toxins | Low sensitivity Prone to false negative results Cross-reactivity with similar antigens Need for specific antibodies High cost | [116,120] |
| Mass spectrometry-basedmethods | |||
| Examples: MALDI TOF MS, HPLC(UPLC)-MS/MS | Very specific and sensitive Rapid and accurate detection Capable of characterizing carbapenemase-producing bacteria Allows the detection of bacterial isolates in biofilms | High acquisition costs for the equipment High annual maintenance costs The need for an extraction or concentration step | [125,126,127] |
| Chromogenic Media | Bacteria | Colour for the Positive Results | Observations | References |
|---|---|---|---|---|
| CHROM agar KPC | E. coli | red | Enhanced with mediators that prevent the growth of the carbapenem-sensitive organisms Limitation in the detection of KPC-2-producing K. pneumoniae isolates | [130] |
| Klebsiella spp., Enterobacter spp., Citrobacter spp. | metallic blue | |||
| P. aeruginosa | translucent cream | |||
| CHROMagar™ MRSA | MRSA | mauve-colored | Contains a powder base (agar, peptones, yeast extract, salts and chromogenic mix) and a proprietary supplement (powder form qsf 20 L) | [131] |
| BBL™ CHROMagar™ MRSA | MRSA | mauve colored | Contains inhibitory agents and cefoxitin | [131] |
| CHROMagarTM STEC | STEC | - | Able to detect most of the STEC serotypes | [117] |
| CHROMagarTM O157 | O157 STEC | mauve | Is not able to detect most non-O157 STEC | [117] |
| Other strains of E. coli | blue | |||
| CHROMagarTM STEC O104 | O104:H4 STEC | - | Specifically designed during the 2011 E. coli O104:H4 outbreak in Europe | [117] |
| ChromID ESBL | Enterobacteriaceae | - | Contains cefpodoxime for the isolation of ESBLs Limitation in detection of OXA-48-like producing isolates that are susceptible to cefpodoxime in the absence of co-production of an ESBL | [130] |
| ChromID Pa | P. aeruginosa | purple | Contains ß-alanyl 6 pentylresorufamine | [128] |
| ChromID™ MRSA | MRSA | green | Contains α-glucosidase and cefoxitin | [131] |
| ChromID VRE | E. faecium | violet | For VRE isolation | [132] |
| E. faecalis | blue to green | |||
| ChromID OXA-48 | CPE | Direct isolation from clinical samples | [125] | |
| Brilliance CRE Agar | E. coli | pink | Contains a modified carbapenem for the isolation of CRE | [130] |
| Klebsiella spp., Enterobacter spp., Citrobacter spp | blue | |||
| Brilliance ESBL-agar | Enterobacteriaceae | Detection of ESBLs producing Enterobacteriaceae | [130] | |
| Oxoid Brilliance™ MRSA | MRSA | blue | Result in 18 h Allows the differentiation between MRSA and MSSA | [131] |
| Colorex KPC, ID Carba | CRE | Can prevent the growth of Gram-positive and non-carbapenemase producers bacteria | [130] | |
| Rapidec Carba NP test and Rapid CARB Screen | Enterobacteriaceae | For rapid and efficient detection of KPC, NDM, and OXA-48-like producers | [125] | |
| MRSASelect | MRSA | strong pink | Contains antimicrobial and antifungal inhibitors | [131] |
| InTray Colorex VRE | E. faecium E. faecalis | pink to mauve, no differentiation between E. faecium and E. faecalis | Contain vancomycin For VRE isolation | [132] |
| VRESelect | E. faecium | pink | ||
| E. faecalis | blue | |||
| HardyCHROM VRE | E. faecium | dark blue | ||
| E. faecalis | dark red | |||
| Spectra VRE | E. faecium | navy blue to pink | ||
| E. faecalis | light blue |
| Bacteria | Testing Methods | Observations | Biological Sample |
|---|---|---|---|
| C. difficile | TC: isolate the strain on a selective media and detect the toxin production CCCNA: detection of the free toxin directly from the stool sample The test uses Vero cell line to detect the presence of a cytopathic effect neutralized by C. difficile antitoxin B. [136] | The diagnosis is complicated because of the phenomenon of asymptomatic carriage of toxigenic C. difficile [137] TC and CCCNA are the “gold standards” for C. difficile CCCNA is specific, but it requires long time to obtain the results [136] | Liquid or unformed stool |
| RT-PCR targeting specific genes: 16S rRNA, the toxin B gene (tcdB), binary toxin genes (cdtA and cdtB), and tcdC gene [136,138] | NAAT are rapid, sensitive Several commercial NAAT are available for direct detection of toxin genes in routine laboratories [136] | ||
| ELISA and ICA (many commercially available kits) for the detection of GDH and toxins Multi step method: Initial screening for GDH, detects the presence of C. difficile [136] Detection of the two free toxins (toxin A–TcdA, toxin B–TcdB) (stool CTA) [136] | ELISA test is rapid, but is not sensitive enough. GDH is a very sensitive assay, but not so specific: it indicates if the bacteria are present, but not if the bacteria are producing toxins [136] | ||
| MRSA | Broth enrichment culture prior to inoculation of selective agar media (including chromogenic media)—the standard method in most European laboratories [118] | Speciation of isolates is essential to distinguish S. aureus from coagulase-negative staphylococci [139] | Nasal swab Wound swab Skin lesion swab Purulent respiratory secretion (sputum cultures) Blood [140] |
| PCR methods that target a DNA segment where the MRSA-specific SCCmec gene meets the S. aureus orfX gene [119] GeneXpert MRSA/SA BC Assay targeting the gene encoding staphylococcal protein A (spa), mecA, and the junction region between orfX chromosome and the SCCmec element BD Max StaphSR assay (automated qualitative in vitro diagnostic test) [141] LAMP method: eazyplex® MRSA (a commercial test) | The assays can be performed rapidly, with results available in 1–3 h [118] Allow the differentiation between MSSA and MRSA Eazyplex® MRSA can detect S. aureus, S. epidermidis, mecA, and mecC from nasal and pharyngeal swabs | ||
| ICA tests: LAT, based on a monoclonal antibody against a protein produced by the mecA gene (PBP2a) and double gel immunodiffusion/microslide Clearview Exact—a PBP2a-antibody test BinaxNOW Staphylococcus aureus Test—does not identify MRSA specifically, but can rule out other staphylococci and Gram-positive cocci [119,127,139] | LAT can distinguish MRSA from MSSA, even in low-level samples Clearview Exact has short analysis time (6 min), similar performances with LAT, but requires fewer steps [119] | ||
| Aptamer-based sensors, immunosensors for direct detection Fluorescent, phosphorescent and colorimetric biosensors for micrococcal nuclease (MNase) detection [133,134] | Simple and cost effective methods The colorimetric method can be monitored by the naked eye or smartphone camera | ||
| Toxins detection by HPLC-MS/MS [127] | No isolation of toxin required [127] | ||
| S. pneumoniae | The routine culture-based identification of S. pneumoniae involves bile solubility and optochin susceptibility testing Cultures: inoculation of blood agar (and/or chocolate agar) plates, chromogenic media [140,142] | Pneumococci can be differentiated from other catalase-negative viridans streptococci by their susceptibility to optochin and solubility in bile salts [142] | Blood Urine Cerebrospinal fluid Respiratory secretions (nose, lungs) Sputum |
| Molecular methods (PCR, RT-PCR, mPCR, multi locus sequence typing) using an array of pneumococcal specific targets: pneumolysin (ply), autolysin (lytA), pneumococcal surface antigen A (psaA), manganese-dependent superoxide dismutase (sodA) and penicillin binding protein (pbp) [140,143] | The culture-independent method for the detection of pneumococci recommended by the WHO is RT-PCR targeting lytA developed by CDC There is a genetic similarity between S. pneumoniae and other closely related species such as S. mitis, S. oralis, and S. pseudopneumoniae [143] | ||
| E. coli | Culture-based methods (using chromogenic media, Rainbow R Agar O157, MacConkey agar and sorbitol-MacConkey medium for non-sorbitol fermenting E. coli) [117,144] Indole testing [145] | There is no culture medium available for the detection of all STEC serotypes [117] | Fecal specimens Enriched stool cultures Blood cultures Urine [117,146] |
| Immunoassays that detect Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2) antigens Example: Premier R EHEC microwell immunoassay, ProSpectTM Shiga Toxin E. coli assay, BioStar R SHIGATOX (optical immunoassay), the Duopath Verotoxin-testTM (ICA test), Premier R EHEC and the Shiga Toxin ChekTM assay (microwell immunoassays) [117,144] | These methods have a prognostic value because there is a clear correlation of Stx2 with the clinical severity of the infection [117] Duopath Verotoxin-testTM differentiates between Stx1-and Stx2-producing STEC | ||
| Molecular methods, especially mPCR and RT-PCR, and also LAMP, targeting genes encoding Stx1 and Stx2 (stx1 and stx2) and other virulence genes such as the intimin gene, eae, and hemolysin gene, ehx4 [117,144] | Can rapidly detect STEC regardless of the serogroup [144] | ||
| Aptamer-based biosensors (fluorescent, electrochemical, SERS) [134,135] | Sensitive and specific methods | ||
| K. pneumoniae | Gram stain Culture-based methods using chromogenic media Phenotypic tests: The carbapenem inactivation method (CIM), Hodge test combined with an EDTA disk test or a disk test using boronic acid compounds [125] Susceptibility testing for carbapenemase producers [147] Indole testing [145] | Can be used for the detection of KPC-producing K. pneumoniae isolates Detection of carbapenemase producers based only on minimum inhibitory concentration values may lack sensitivity [147] | Respiratory secretions Blood cultures |
| Molecular methods: mPCR, RT-PCR targeting the carbapenemase gene, DNA microarray, LAMP [49,125] | Molecular methods can detect almost all bla genes, including KPCs, NDMs, OXA48-likes, present in bacterial pathogens | ||
| Electrochemical aptamer-based biosensors [135] | |||
| Enterobacter spp. | Gram stain The gold standard: cultures, with at least two sets of blood cultures, one aerobic and one anaerobic bottle MacConkey agar to determine if the specimen is lactose fermentin Indole testing [145] | Enterobacter spp are motile, in contrast to Klebsiella, which is not motile [145] | Respiratory secretions Wound swab Skin lesion swab Urine Blood |
| Enterococcus spp. and VRE | The most common standard methods: MIC determination, disk diffusion, and the breakpoint agar method [148] Culture-based methods: Growth on bile esculin agar and in 6.5% salt broth A positive esculin in combination with a positive PYR reaction to presumptive identification [149] Bile esculin azide agar, triphenyl-tetrazolium chloride azide agar, or an equivalent medium, supplemented with 6 μg/mL of vancomycin, will grow most VRE within 24- to 72-h incubation at 37 °C [150] | E. faecalis and E. faecium are usually easily identified by most commercial systems With respect to vancomycin intermediate or resistant strains, two key characteristics are motility and pigment E. casseliflavus is both motile and possesses a yellow pigment E. gallinarum is also motile but non-pigmented E. faecalis and E. faecium demonstrate neither characteristic [149] | Urine Blood Stool specimens Perirectal cultures Purulent secretions |
| PCR-based methods targeting vanA and vanB Commercially available molecular tests: the Roche LightCycler VRE (vanA/vanB) detection 337 assay, the Cepheid Xpert vanA/vanB assay, and the Cepheind Xpert vanA assay [129,148] | Nine different van ligase genes have been described in enterococci, but only the genes encoding vanA and vanB are usually targeted [148] | ||
| P. aeruginosa | Culture based methods: Gram stain, aerobic incubation on nutrient agar (Pseudomonas isolation agar, agars with cetrimide, chromID Pseudomonas, acetamide-based agar) [128] | Incubation for 24–48 h at 37 °C on Pseudomonas selective agars in air at 35–37 °C [151] | Exhaled breath condensates Sputum Nasal or throat swab Wound swab Skin lesion swab Skin biopsy Blood Urine [105] |
| Molecular methods: PCR (rapid and reliable identification), LAMP and polymerase spiral reaction, targeting several genes (16S rRNA, ecfX, oprL, gyrB, toxA, etc.) [128,151] | Allows the detection of P. aeruginosa growing in biofilms | ||
| Immunoassays: ELISA (commercial kits, such as IgG ELISA kit with three P. aeruginosa antigens) and ICA (most often being used the sandwich format that uses gold nanoparticle labeled anti-P. aeruginosa monoclonal antibody) and for its QS molecules [128] | Commercial ELISA kits for P. aeruginosa are widely used for routine testing in some European countries | ||
| Biosensors (electrochemical, optical, piezoelectric) for the detection of whole-cell bacterium or for the detection of metabolites and QS molecules [128] | High sensitivity, with low limits of detection | ||
| MS-based methods: MALDI-TOF-MS and HPLC-MS for the detection of QS molecules and virulence factors (pyocyanin) [128] | Allows the detection of bacterial isolates in biofilms Sample preparation is very important for the detection of bacteria using this method | ||
| Acinetobacter spp. | Preliminary and biochemical tests: Gram stain, catalase test, oxidase test, and hanging drop preparation for motility, CarbAcineto NP test (rapid detection of carbapenemase-producing Acinetobacter spp.) [152] | Identification of Acinetobacter at species level remains complicated and the traditional phenotypic methods such as culture and biochemical identification are slow, unreliable, and less efficient [153] | Pus Endotracheal aspirate Urine Blood Sputum [154] |
| Molecular methods: PCR [152,155] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szabó, S.; Feier, B.; Capatina, D.; Tertis, M.; Cristea, C.; Popa, A. An Overview of Healthcare Associated Infections and Their Detection Methods Caused by Pathogen Bacteria in Romania and Europe. J. Clin. Med. 2022, 11, 3204. https://doi.org/10.3390/jcm11113204
Szabó S, Feier B, Capatina D, Tertis M, Cristea C, Popa A. An Overview of Healthcare Associated Infections and Their Detection Methods Caused by Pathogen Bacteria in Romania and Europe. Journal of Clinical Medicine. 2022; 11(11):3204. https://doi.org/10.3390/jcm11113204
Chicago/Turabian StyleSzabó, Sándor, Bogdan Feier, Denisa Capatina, Mihaela Tertis, Cecilia Cristea, and Adina Popa. 2022. "An Overview of Healthcare Associated Infections and Their Detection Methods Caused by Pathogen Bacteria in Romania and Europe" Journal of Clinical Medicine 11, no. 11: 3204. https://doi.org/10.3390/jcm11113204
APA StyleSzabó, S., Feier, B., Capatina, D., Tertis, M., Cristea, C., & Popa, A. (2022). An Overview of Healthcare Associated Infections and Their Detection Methods Caused by Pathogen Bacteria in Romania and Europe. Journal of Clinical Medicine, 11(11), 3204. https://doi.org/10.3390/jcm11113204









