Biomarkers of Castrate Resistance in Prostate Cancer: Androgen Receptor Amplification and T877A Mutation Detection by Multiplex Droplet Digital PCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Healthy Donor PBMC
2.2. Plasma
2.3. Genomic DNA and Plasma cfDNA Extraction
2.4. Droplet Digital PCR
2.5. Statistics
3. Results
3.1. T877A Mutation and AR Amplification Assay (AR-Amp-1/T877A)
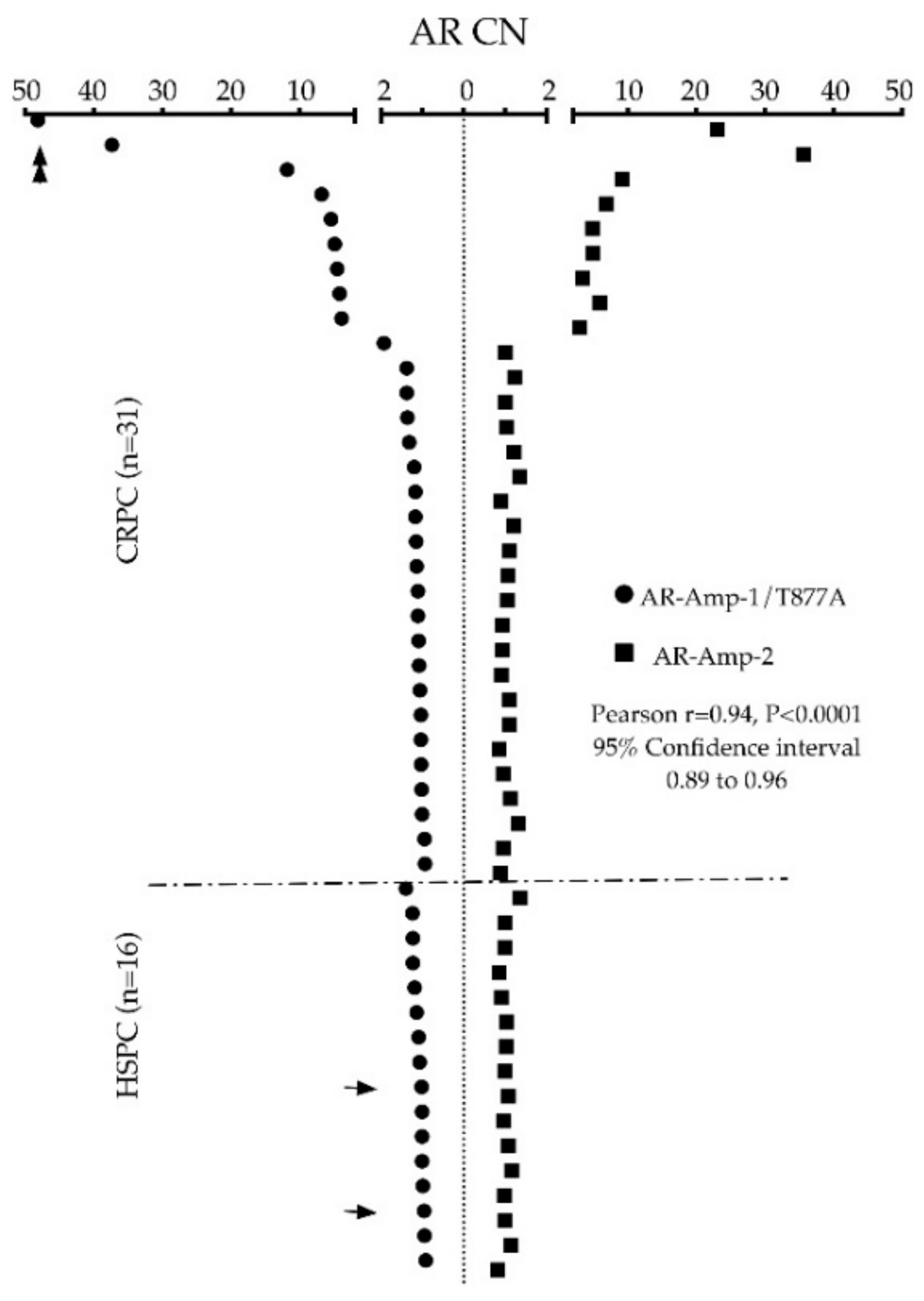
3.2. AR Amplification by AR-Amp-2 Assay
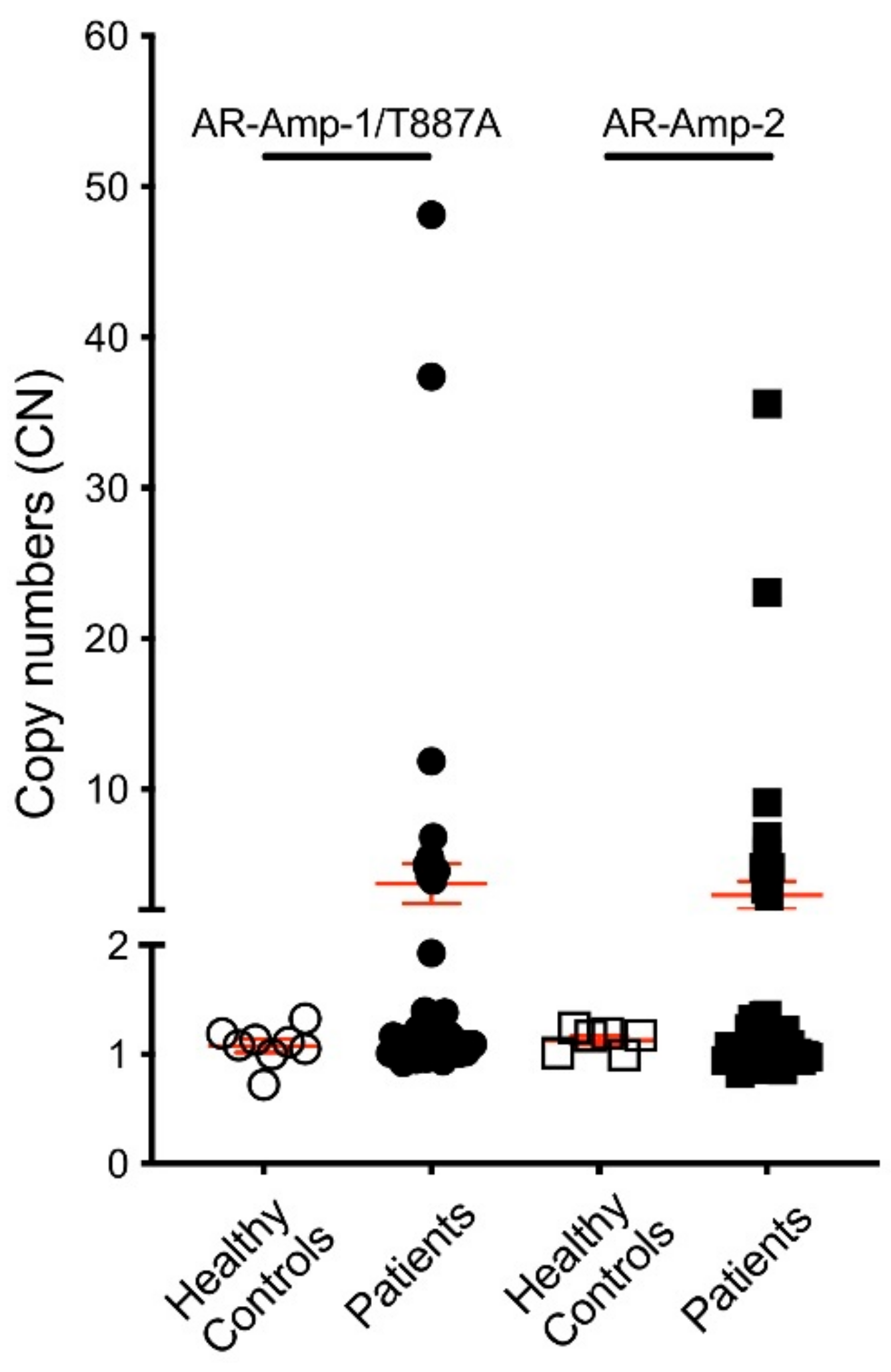
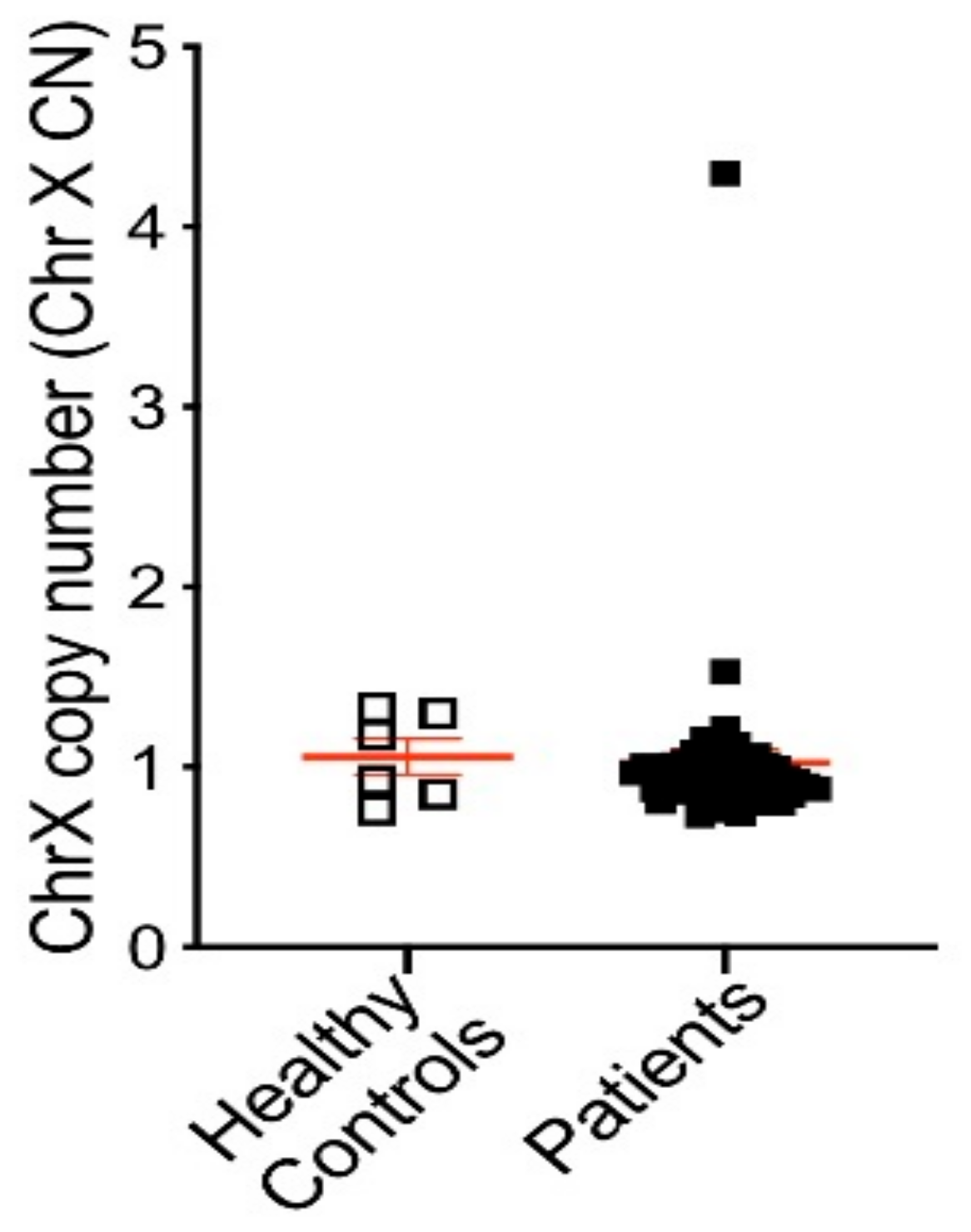
3.3. AR Alterations Detected from Patient Plasma
3.4. Association of ARamp+ and Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fontana, F.; Limonta, P. Dissecting the Hormonal Signaling Landscape in Castration-Resistant Prostate Cancer. Cells 2021, 10, 1133. [Google Scholar] [CrossRef]
- Heinlein, C.A.; Chang, C. Androgen Receptor in Prostate Cancer. Endocr. Rev. 2004, 25, 276–308. [Google Scholar] [CrossRef]
- Podolak, J.; Eilers, K.; Newby, T.; Slottke, R.; Tucker, E.; Olson, S.B.; Lue, H.-W.; Youngren, J.; Aggarwal, R.; Small, E.J.; et al. Androgen receptor amplification is concordant between circulating tumor cells and biopsies from men undergoing treatment for metastatic castration resistant prostate cancer. Oncotarget 2017, 8, 71447–71455. [Google Scholar] [CrossRef]
- Haapala, K.; Kuukasjärvi, T.; Hyytinen, E.; Rantala, I.; Helin, H.J.; Koivisto, P.A. Androgen receptor amplification is associated with increased cell proliferation in prostate cancer. Hum. Pathol. 2007, 38, 474–478. [Google Scholar] [CrossRef]
- Edwards, J.; Krishna, N.S.; Mukherjee, R.; Watters, A.D.; Underwood, M.A.; Bartlett, J.M.S. Amplification of the androgen receptor may not explain the development of androgen-independent prostate cancer. BJU Int. 2001, 88, 633–637. [Google Scholar] [CrossRef]
- Du, M.; Huang, C.; Tan, W.; Kohli, M.; Wang, L. Multiplex Digital PCR to Detect Amplifications of Specific Androgen Receptor Loci in Cell-Free DNA for Prognosis of Metastatic Castration-Resistant Prostate Cancer. Cancers 2020, 12, 2139. [Google Scholar] [CrossRef]
- Ulz, P.; Belic, J.; Graf, R.; Auer, M.; Lafer, I.; Fischereder, K.; Webersinke, G.; Pummer, K.; Augustin, H.; Pichler, M.; et al. Whole-genome plasma sequencing reveals focal amplifications as a driving force in metastatic prostate cancer. Nat. Commun. 2016, 7, 12008. [Google Scholar] [CrossRef]
- Azad, A.A.; Volik, S.V.; Wyatt, A.W.; Haegert, A.; Le Bihan, S.; Bell, R.H.; Anderson, S.A.; McConeghy, B.; Shukin, R.; Bazov, J.; et al. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2015, 21, 2315. [Google Scholar] [CrossRef]
- Small, E.J.; Youngren, J.; Thomas, G.; Olson, S.; Toschi, A.; Foye, A.; Alumkal, J.J.; Rettig, M.; Gleave, M.E.; Evans, C.P.; et al. Androgen receptor (AR) amplification in patients with metastatic castration-resistant prostate cancer (mCRPC) refractory to therapy with abiraterone acetate or enzalutamide: Preliminary results from the SU2C/PCF/AACR West Coast Prostate Cancer Dream Team (WCDT). J. Clin. Oncol. 2014, 32, 5020. [Google Scholar] [CrossRef]
- Zaman, N.; Giannopoulos, P.N.; Chowdhury, S.; Bonneil, E.; Thibault, P.; Wang, E.; Trifiro, M.; Paliouras, M. Proteomic-coupled-network analysis of T877A-androgen receptor interactomes can predict clinical prostate cancer outcomes between White (non-Hispanic) and African-American groups. PLoS ONE 2014, 9, e113190. [Google Scholar] [CrossRef]
- Ho, Y.; Dehm, S.M. Androgen Receptor Rearrangement and Splicing Variants in Resistance to Endocrine Therapies in Prostate Cancer. Endocrinology 2017, 158, 1533–1542. [Google Scholar] [CrossRef]
- Liu, H.-L.; Zhong, H.-Y.; Song, T.-Q.; Li, J.-Z. A Molecular Modeling Study of the Hydroxyflutamide Resistance Mechanism Induced by Androgen Receptor Mutations. Int. J. Mol. Sci. 2017, 18, 1823. [Google Scholar] [CrossRef]
- Liu, H.; Wang, L.; Tian, J.; Li, J.; Liu, H. Molecular Dynamics Studies on the Enzalutamide Resistance Mechanisms Induced by Androgen Receptor Mutations. J. Cell. Biochem. 2017, 118, 2792–2801. [Google Scholar] [CrossRef]
- Campos-Fernández, E.; Barcelos, L.S.; de Souza, A.G.; Goulart, L.R.; Alonso-Goulart, V. Research landscape of liquid biopsies in prostate cancer. Am. J. Cancer Res. 2019, 9, 1309–1328. [Google Scholar]
- Salciccia, S.; Capriotti, A.L.; Laganà, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4637. [Google Scholar] [CrossRef]
- Bastos, D.A.; Antonarakis, E.S. CTC-derived AR-V7 detection as a prognostic and predictive biomarker in advanced prostate cancer. Expert Rev. Mol. Diagn. 2018, 18, 155–163. [Google Scholar] [CrossRef]
- Scher, H.I.; Graf, R.P.; Schreiber, N.A.; Jayaram, A.; Winquist, E.; McLaughlin, B.; Lu, D.; Fleisher, M.; Orr, S.; Lowes, L.; et al. Assessment of the Validity of Nuclear-Localized Androgen Receptor Splice Variant 7 in Circulating Tumor Cells as a Predictive Biomarker for Castration-Resistant Prostate Cancer. JAMA Oncol. 2018, 4, 1179–1186. [Google Scholar] [CrossRef]
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038. [Google Scholar] [CrossRef]
- Nimir, M.; Ma, Y.; Jeffreys, S.A.; Opperman, T.; Young, F.; Khan, T.; Ding, P.; Chua, W.; Balakrishnar, B.; Cooper, A.; et al. Detection of AR-V7 in Liquid Biopsies of Castrate Resistant Prostate Cancer Patients: A Comparison of AR-V7 Analysis in Circulating Tumor Cells, Circulating Tumor RNA and Exosomes. Cells 2019, 8, 688. [Google Scholar] [CrossRef]
- Armstrong, A.J.; Luo, J.; Nanus, D.M.; Giannakakou, P.; Szmulewitz, R.Z.; Danila, D.; Healy, P.; Anand, M.; Berry, W.; Zhang, T.; et al. Prospective Multicenter Study of Circulating Tumor Cell AR-V7 and Taxane versus Hormonal Treatment Outcomes in Metastatic Castration-Resistant Prostate Cancer. JCO Precis. Oncol. 2020, 4, 1285–1301. [Google Scholar] [CrossRef]
- Sciarra, A.; Gentilucci, A.; Silvestri, I.; Salciccia, S.; Cattarino, S.; Scarpa, S.; Gatto, A.; Frantellizzi, V.; Von Heland, M.; Ricciuti, G.P.; et al. Androgen receptor variant 7 (AR-V7) in sequencing therapeutic agents for castratrion resistant prostate cancer: A critical review. Medicine 2019, 98, e15608. [Google Scholar] [CrossRef]
- Kohli, M.; Li, J.; Du, M.; Hillman, D.W.; Dehm, S.M.; Tan, W.; Carlson, R.; Campion, M.B.; Wang, L.; Wang, L.; et al. Prognostic association of plasma cell-free DNA-based androgen receptor amplification and circulating tumor cells in pre-chemotherapy metastatic castration-resistant prostate cancer patients. Prostate Cancer Prostatic Dis. 2018, 21, 411–418. [Google Scholar] [CrossRef]
- Sumiyoshi, T.; Mizuno, K.; Yamasaki, T.; Miyazaki, Y.; Makino, Y.; Okasho, K.; Li, X.; Utsunomiya, N.; Goto, T.; Kobayashi, T.; et al. Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer. Sci. Rep. 2019, 9, 4030. [Google Scholar] [CrossRef]
- Naderi, A. C1orf64 is a novel androgen receptor target gene and coregulator that interacts with 14-3-3 protein in breast cancer. Oncotarget 2017, 8, 57907–57933. [Google Scholar] [CrossRef][Green Version]
- Ma, Y.; Luk, A.; Young, P.F.; Lynch, D.; Chua, W.; Balakrishnar, B.; De Souza, P.; Becker, M.T. Droplet Digital PCR Based Androgen Receptor Variant 7 (AR-V7) Detection from Prostate Cancer Patient Blood Biopsies. Int. J. Mol. Sci. 2016, 17, 1264. [Google Scholar] [CrossRef]
- Ding, P.N.; Becker, T.; Bray, V.; Chua, W.; Ma, Y.; Xu, B.; Lynch, D.; de Souza, P.; Roberts, T. Plasma next generation sequencing and droplet digital PCR-based detection of epidermal growth factor receptor (EGFR) mutations in patients with advanced lung cancer treated with subsequent-line osimertinib. Thorac. Cancer 2019, 10, 1879–1884. [Google Scholar] [CrossRef]
- Thorek, D.L.J.; Ku, A.T.; Mitsiades, N.; Veach, D.; Watson, P.A.; Metha, D.; Strand, S.-E.; Sharma, S.K.; Lewis, J.S.; Abou, D.S.; et al. Harnessing Androgen Receptor Pathway Activation for Targeted Alpha Particle Radioimmunotherapy of Breast Cancer. Clin. Cancer Res. 2018, 25, 881–891. [Google Scholar] [CrossRef]
- Song, B.; Park, S.-H.; Zhao, J.C.; Fong, K.-W.; Li, S.; Lee, Y.; Yang, Y.A.; Sridhar, S.; Lu, X.; Abdulkadir, S.A.; et al. Targeting FOXA1-mediated repression of TGF-β signaling suppresses castration-resistant prostate cancer progression. J. Clin. Investig. 2019, 129, 569–582. [Google Scholar] [CrossRef]
- Merson, S.; Yang, Z.H.; Brewer, D.; Olmos, D.; Eichholz, A.; McCarthy, F.; Fisher, G.; Kovacs, G.; Berney, D.M.; Foster, C.S.; et al. Focal amplification of the androgen receptor gene in hormone-naive human prostate cancer. Br. J. Cancer 2014, 110, 1655–1662. [Google Scholar] [CrossRef]
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018, 8, 444. [Google Scholar] [CrossRef]
- González-Billalabeitia, E.; Conteduca, V.; Wetterskog, D.; Jayaram, A.; Attard, G. Circulating tumor DNA in advanced prostate cancer: Transitioning from discovery to a clinically implemented test. Prostate Cancer Prostatic Dis. 2019, 22, 195–205. [Google Scholar] [CrossRef]
- Edwards, J.; Krishna, N.S.; Grigor, K.M.; Bartlett, J.M.S. Androgen receptor gene amplification and protein expression in hormone refractory prostate cancer. Br. J. Cancer 2003, 89, 552–556. [Google Scholar] [CrossRef]
- Röpke, A.; Erbersdobler, A.; Hammerer, P.; Palisaar, J.; John, K.; Stumm, M.; Wieacker, P. Gain of androgen receptor gene copies in primary prostate cancer due to X chromosome polysomy. Prostate 2004, 59, 59–68. [Google Scholar] [CrossRef]
- Zhang, X.; Hong, S.-Z.; Lin, E.-J.; Wang, D.A.Y.; Li, Z.-J.; Chen, L.I. Amplification and protein expression of androgen receptor gene in prostate cancer cells: Fluorescence in situ hybridization analysis. Oncol. Lett. 2015, 9, 2617–2622. [Google Scholar] [CrossRef]
- Romanel, A.; Tandefelt, D.G.; Conteduca, V.; Jayaram, A.; Casiraghi, N.; Wetterskog, D.; Salvi, S.; Amadori, D.; Zafeiriou, Z.; Rescigno, P.; et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015, 7, 312re310. [Google Scholar] [CrossRef]
- Katsanis, S.H.; Katsanis, N. Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 2013, 14, 415–426. [Google Scholar] [CrossRef]
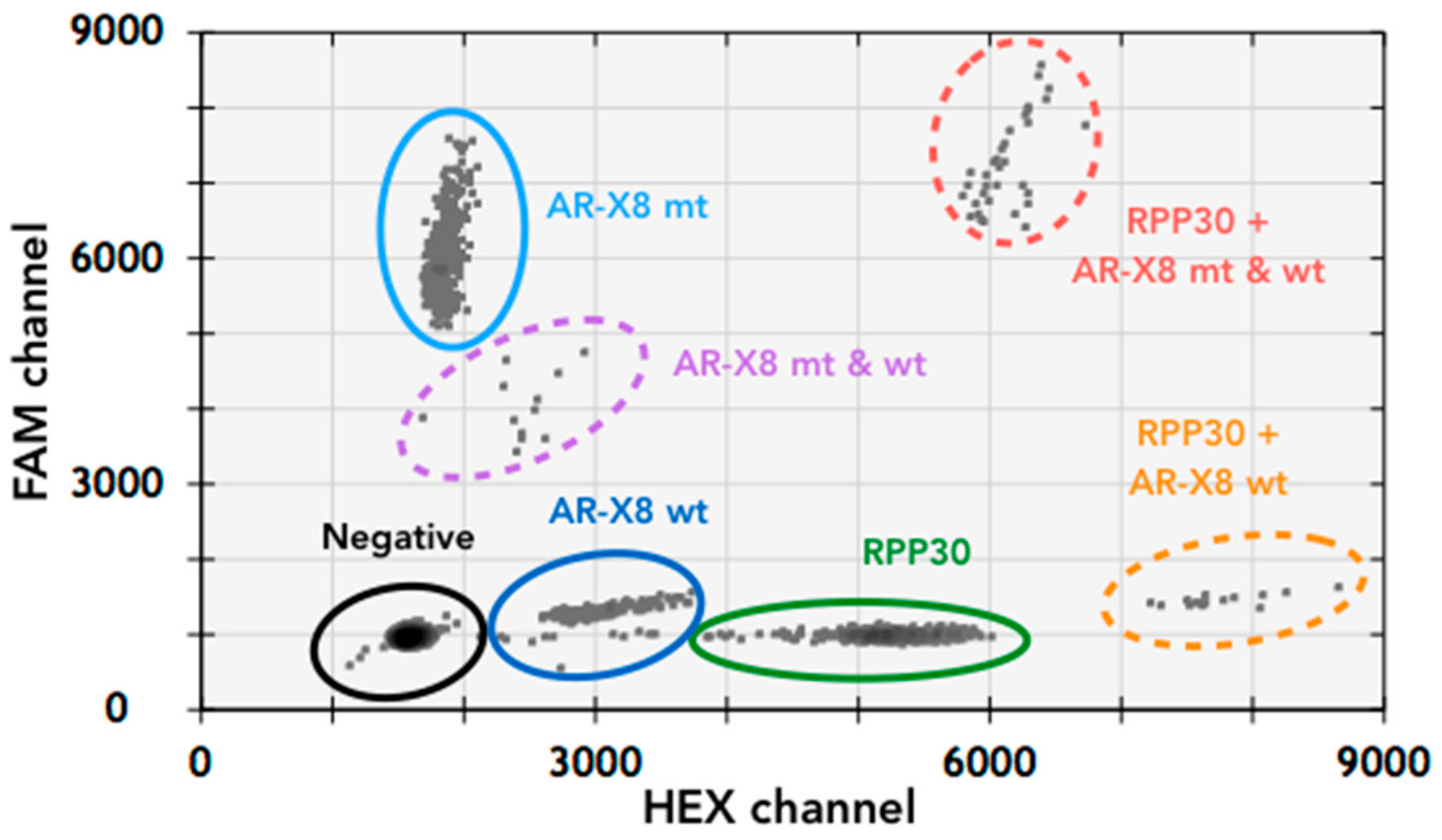

| Primers | Probes | |
|---|---|---|
| AR Exon 8 (AR-X8) | FP:5′-CCCTACAGATTGCGAGAGAGC-3′(500 nM) RP: 5′-GAAAGTCCACGCTCACCATGT-3′(500 nM) | MT:5′-[6FAM]ATCAGTTCGCTTTTGACCTG[BHQ1]-3′(250 nM) WT: 5′-[HEX]ATCAGTTCACTTTTGACCTG[BHQ1]-3′(250 nM) |
| RPP30 | FP:5′-GATTTGGACCTGCGAGCG-3′(500 nM) RP: 5′-GCGGCTGTCTCCACAAGT-3′(500 nM) | 5′-[HEX]TCTGACCTGAAGGCTCTG[BHQ1]-3′ (250 nM) |
| AR Exon 1 (AR-X1) | FP:5′-CCTATGCAAATGCCTGCCTG-3′(500 nM) RP: 5′-GCACTCTGCATTCGTTTCCC-3′(500 nM) | 5′-[6FAM]AAGTCCGGTACAAAGCCAG[BHQ1]-3′(125 nM) |
| AR Exon 2 (AR-X2) | FP:5′-TTTCCACCCCAGAAGACCTG-3′(500 nM) RP: 5′-AAGACCTTGCAGCTTCCACA-3′(500 nM) | 5′-[6FAM]CACCCAGAAGCTTCATCTC[BHQ1]-3′(250 nM) |
| MYM | FP:5′-ACAGGGAACAGAACAAGCTGGTCTT-3′(1000 nM) RP:5′-GCAAGACCCTGTGTAAGAACTTTGA-3′(1000 nM) | 5′-[HEX]CATTACGATCCACATGTGATAG[BHQ]-3′(250 nM) |
| TBP | FP:5′-ACAGAAGTTGGGTTTTCCAGC-3′(500 nM) RP: 5′-TCACATCACAGCTCCCCAC-3′(500 nM) | 5′-[HEX]TCTTGGACTTCAAGATTCAG[BHQ1]-3′(500 nM) |
| Cell Sample (gDNA) | T877A | WT | RPP30 | AR:RPP30 | AR CN |
|---|---|---|---|---|---|
| PBMC-M | 0 | 45.4 | 90 | 0.51:1 | 1 |
| PBMC-F | 0 | 81 | 77 | 1.05:1 | 2 |
| MFM-223 | 0 | 198 | 51.5 | 3.84:1 | 8 |
| VCaP | 0 | 721 | 52.5 | 13.73:1 | 28 |
| LNCaP | 114 | 0 | 228 | 0.50:1 | 1 |
| LNCaP + PBMC-M (2:1) | 30.1 | 16.2 | 89.3 | 0.52:1 | 1.0 |
| LNCaP + MFM-223 (2:1) | 29 | 60.7 | 68.7 | 1.31:1 | 2.6 |
| LNCaP + VCaP (2:1) | 20.4 | 191 | 61.7 | 3.43:1 | 6.9 |
| Cell Sample (gDNA) | AR-X1 | AR-X2 | TBP | MYM* | AR CN |
| PBMC-M | 45.9 | 42 | 87 | 84.6 | 1 |
| PBMC-F | 73 | 72.6 | 73.1 | 75 | 2 |
| MFM-223 | 173 | 176 | 46.3 | 51.8 | 7 |
| VCaP | 696 | 709 | 54 | 69 | 23 |
| LNCaP | 97 | 100 | 189 | 194 | 1 |
| Treatment Type | |||||||
|---|---|---|---|---|---|---|---|
| Chem+ | Chem− | Enz/Abi+ | Enz/Abi− | Chem and Enz/Abi+ | Others | Total | |
| High AR CN | 5 | 4 | 4 | 5 | 2 | 7 | 9 |
| Normal AR CN | 7 | 14 | 10 | 11 | 2 | 19 | 21 |
| Total | 12 | 18 | 14 | 16 | 4 | 26 | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, F.P.; Becker, T.M.; Nimir, M.; Opperman, T.; Chua, W.; Balakrishnar, B.; de Souza, P.; Ma, Y. Biomarkers of Castrate Resistance in Prostate Cancer: Androgen Receptor Amplification and T877A Mutation Detection by Multiplex Droplet Digital PCR. J. Clin. Med. 2022, 11, 257. https://doi.org/10.3390/jcm11010257
Young FP, Becker TM, Nimir M, Opperman T, Chua W, Balakrishnar B, de Souza P, Ma Y. Biomarkers of Castrate Resistance in Prostate Cancer: Androgen Receptor Amplification and T877A Mutation Detection by Multiplex Droplet Digital PCR. Journal of Clinical Medicine. 2022; 11(1):257. https://doi.org/10.3390/jcm11010257
Chicago/Turabian StyleYoung, Francis P., Therese M. Becker, Mohammed Nimir, Thomas Opperman, Wei Chua, Bavanthi Balakrishnar, Paul de Souza, and Yafeng Ma. 2022. "Biomarkers of Castrate Resistance in Prostate Cancer: Androgen Receptor Amplification and T877A Mutation Detection by Multiplex Droplet Digital PCR" Journal of Clinical Medicine 11, no. 1: 257. https://doi.org/10.3390/jcm11010257
APA StyleYoung, F. P., Becker, T. M., Nimir, M., Opperman, T., Chua, W., Balakrishnar, B., de Souza, P., & Ma, Y. (2022). Biomarkers of Castrate Resistance in Prostate Cancer: Androgen Receptor Amplification and T877A Mutation Detection by Multiplex Droplet Digital PCR. Journal of Clinical Medicine, 11(1), 257. https://doi.org/10.3390/jcm11010257







