Cardiac Filaminopathies: Illuminating the Divergent Role of Filamin C Mutations in Human Cardiomyopathy
Abstract
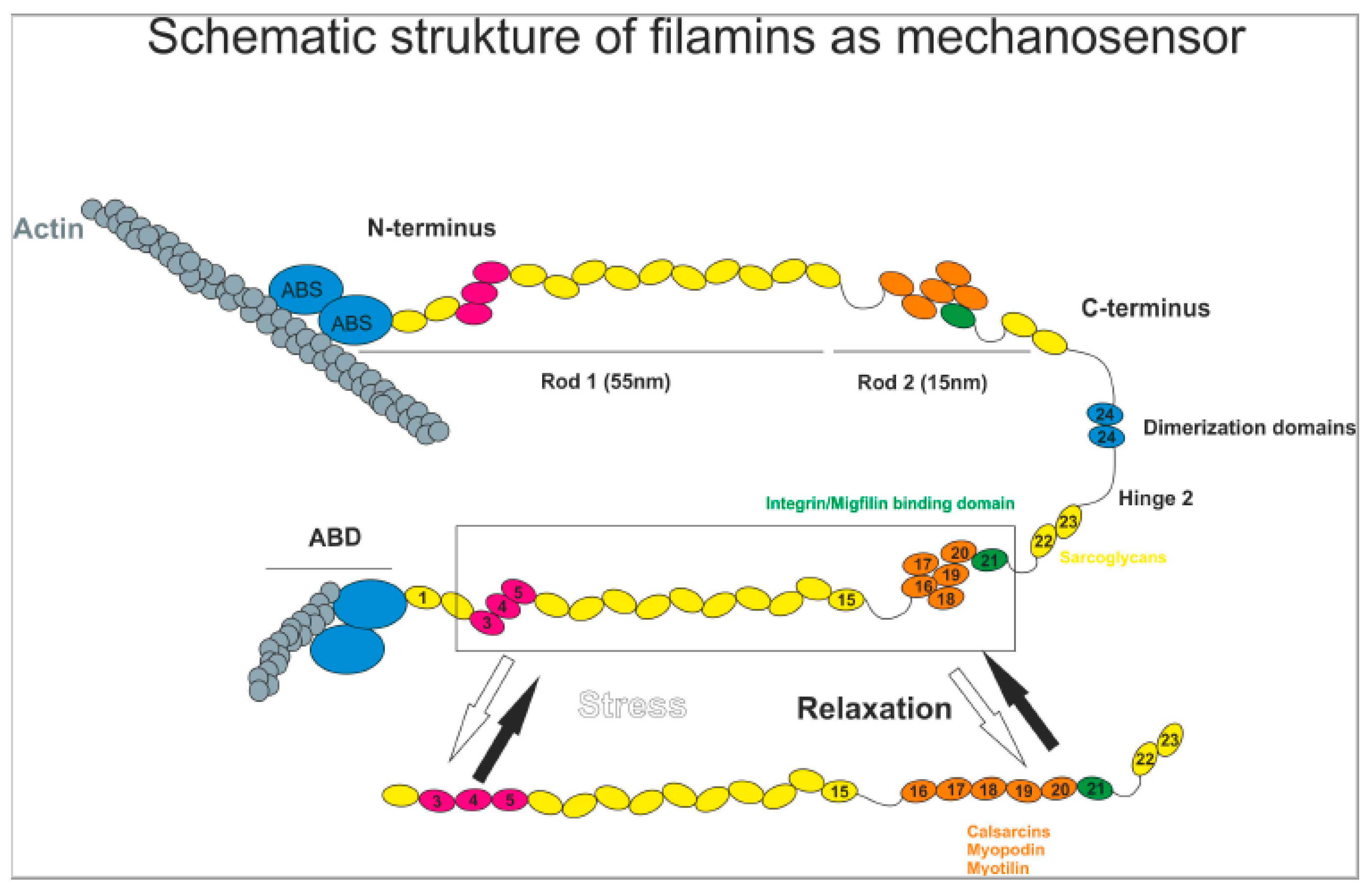
1. Introduction
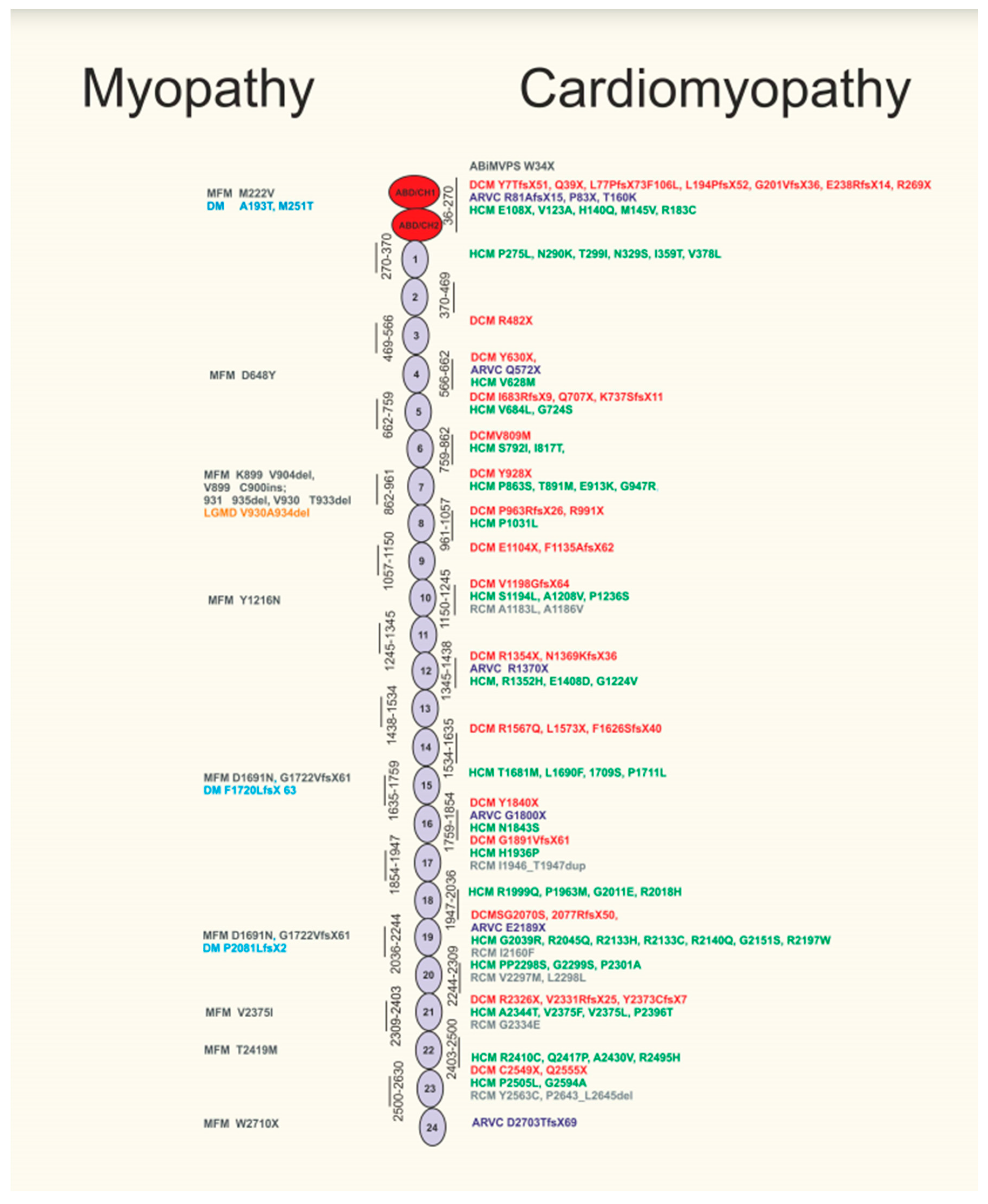
2. Filamin C Mutations Reveal a Distinct Phenotype of Human Dilated Cardiomyopathy (Dcm) with Increased Risk of Sudden Cardiac Death
3. Filamin C Mutations in Arrhythmogenic Cardiomyopathy
4. Missense Filamin C Mutations Can Result in Human Hypertrophic Cardiomyopathy
5. Filamin C Mutations in Restrictive Cardiomyopathy (RCM)
6. Filamin C Mutations in Mitral Valve Prolapse Syndrome
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burke, M.A.; Cook, S.A.; Seidman, J.G.; Seidman, C.E. Clinical and Mechanistic Insights into the Genetics of Cardiomyopathy. J. Am. Coll. Cardiol. 2016, 68, 2871–2886. [Google Scholar] [CrossRef]
- Kamisago, M.; Sharma, S.D.; DePalma, S.R.; Solomon, S.; Sharma, P.; McDonough, B.; Smoot, L.; Mullen, M.P.; Woolf, P.K.; Wigle, E.D.; et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N. Engl. J. Med. 2000, 343, 1688–1696. [Google Scholar] [CrossRef]
- Watkins, H.; Ashrafian, H.; Redwood, C. Inherited cardiomyopathies. N. Engl. J. Med. 2011, 364, 1643–1656. [Google Scholar] [CrossRef] [PubMed]
- Elliott, P.; Andersson, B.; Arbustini, E.; Bilinska, Z.; Cecchi, F.; Charron, P.; Dubourg, O.; Kuhl, U.; Maisch, B.; McKenna, W.J.; et al. Classification of the cardiomyopathies: A position statement from the European Society of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur. Heart J. 2008, 29, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Maron, B.J.; Towbin, J.A.; Thiene, G.; Antzelevitch, C.; Corrado, D.; Arnett, D.; Moss, A.J.; Seidman, C.E.; Young, J.B.; American Heart Association; et al. Contemporary definitions and classification of the cardiomyopathies: An American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 2006, 113, 1807–1816. [Google Scholar] [PubMed]
- Verdonschot, J.A.J.; Vanhoutte, E.K.; Claes, G.R.F.; Helderman-van den Enden, A.; Hoeijmakers, J.G.J.; Hellebrekers, D.; de Haan, A.; Christiaans, I.; Lekanne Deprez, R.H.; Boen, H.M.; et al. A mutation update for the FLNC gene in myopathies and cardiomyopathies. Hum. Mutat. 2020, 41, 1091–1111. [Google Scholar] [CrossRef]
- Mao, Z.; Nakamura, F. Structure and Function of Filamin C in the Muscle Z-Disc. Int. J. Mol. Sci. 2020, 21, 2696. [Google Scholar] [CrossRef]
- Modarres, H.P.; Mofradt, M.R. Filamin: A structural and functional biomolecule with important roles in cell biology, signaling and mechanics. Mol. Cell. Biomech. 2014, 11, 39–65. [Google Scholar]
- Posch, M.G.; Thiemann, L.; Tomasov, P.; Veselka, J.; Cardim, N.; Garcia-Castro, M.; Coto, E.; Perrot, A.; Geier, C.; Dietz, R.; et al. Sequence analysis of myozenin 2 in 438 European patients with familial hypertrophic cardiomyopathy. Med. Sci. Monit. 2008, 14, CR372–CR374. [Google Scholar]
- Frey, N.; Olson, E.N. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J. Biol. Chem. 2002, 277, 13998–14004. [Google Scholar] [CrossRef]
- Osio, A.; Tan, L.; Chen, S.N.; Lombardi, R.; Nagueh, S.F.; Shete, S.; Roberts, R.; Willerson, J.T.; Marian, A.J. Myozenin 2 is a novel gene for human hypertrophic cardiomyopathy. Circ. Res. 2007, 100, 766–768. [Google Scholar] [CrossRef]
- Huby, A.C.; Mendsaikhan, U.; Takagi, K.; Martherus, R.; Wansapura, J.; Gong, N.; Osinska, H.; James, J.F.; Kramer, K.; Saito, K.; et al. Disturbance in Z-disk mechanosensitive proteins induced by a persistent mutant myopalladin causes familial restrictive cardiomyopathy. J. Am. Coll. Cardiol. 2014, 64, 2765–2776. [Google Scholar] [CrossRef]
- Levitas, A.; Konstantino, Y.; Muhammad, E.; Afawi, Z.; Marc Weinstein, J.; Amit, G.; Etzion, Y.; Parvari, R. D117N in Cypher/ZASP may not be a causative mutation for dilated cardiomyopathy and ventricular arrhythmias. Eur. J. Hum. Genet. 2016, 24, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Takai, E.; Akita, H.; Shiga, N.; Kanazawa, K.; Yamada, S.; Terashima, M.; Matsuda, Y.; Iwai, C.; Kawai, K.; Yokota, Y.; et al. Mutational analysis of the cardiac actin gene in familial and sporadic dilated cardiomyopathy. Am. J. Med. Genet. 1999, 86, 325–327. [Google Scholar] [CrossRef]
- Frank, D.; Frey, N. Cardiac Z-disc signaling network. J. Biol. Chem. 2011, 286, 9897–9904. [Google Scholar] [CrossRef]
- Frank, D.; Kuhn, C.; Katus, H.A.; Frey, N. Role of the sarcomeric Z-disc in the pathogenesis of cardiomyopathy. Future Cardiol. 2007, 3, 611–622. [Google Scholar] [CrossRef]
- Dalkilic, I.; Schienda, J.; Thompson, T.G.; Kunkel, L.M. Loss of FilaminC (FLNc) results in severe defects in myogenesis and myotube structure. Mol. Cell. Biol. 2006, 26, 6522–6534. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, Z.; Zhang, L.; Zhu, M.; Tan, C.; Zhou, X.; Evans, S.M.; Fang, X.; Feng, W.; Chen, J. Loss of Filamin C Is Catastrophic for Heart Function. Circulation 2020, 141, 869–871. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Genga, M.F.; Cuenca, S.; Dal Ferro, M.; Zorio, E.; Salgado-Aranda, R.; Climent, V.; Padron-Barthe, L.; Duro-Aguado, I.; Jimenez-Jaimez, J.; Hidalgo-Olivares, V.M.; et al. Truncating FLNC Mutations Are Associated with High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J. Am. Coll. Cardiol. 2016, 68, 2440–2451. [Google Scholar] [CrossRef]
- Fatkin, D.; MacRae, C.; Sasaki, T.; Wolff, M.R.; Porcu, M.; Frenneaux, M.; Atherton, J.; Vidaillet, H.J.; Spudich, S., Jr.; De Girolami, U.; et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999, 341, 1715–1724. [Google Scholar] [CrossRef]
- Hasselberg, N.E.; Haland, T.F.; Saberniak, J.; Brekke, P.H.; Berge, K.E.; Leren, T.P.; Edvardsen, T.; Haugaa, K.H. Lamin A/C cardiomyopathy: Young onset, high penetrance, and frequent need for heart transplantation. Eur. Heart J. 2018, 39, 853–860. [Google Scholar] [CrossRef]
- McLendon, P.M.; Robbins, J. Desmin-related cardiomyopathy: An unfolding story. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H1220–H1228. [Google Scholar] [CrossRef]
- Li, D.; Tapscoft, T.; Gonzalez, O.; Burch, P.E.; Quinones, M.A.; Zoghbi, W.A.; Hill, R.; Bachinski, L.L.; Mann, D.L.; Roberts, R. Desmin mutation responsible for idiopathic dilated cardiomyopathy. Circulation 1999, 100, 461–464. [Google Scholar] [CrossRef]
- Brodehl, A.; Pour Hakimi, S.A.; Stanasiuk, C.; Ratnavadivel, S.; Hendig, D.; Gaertner, A.; Gerull, B.; Gummert, J.; Paluszkiewicz, L.; Milting, H. Restrictive Cardiomyopathy is Caused by a Novel Homozygous Desmin (DES) Mutation p.Y122H Leading to a Severe Filament Assembly Defect. Genes (Basel) 2019, 10, 918. [Google Scholar] [CrossRef]
- Oomen, A.; Jones, K.; Yeates, L.; Semsarian, C.; Ingles, J.; Sy, R.W. Rare desmin variant causing penetrant life-threatening arrhythmic cardiomyopathy. Heart Rhythm. Case Rep. 2018, 4, 318–323. [Google Scholar] [CrossRef] [PubMed]
- Harada, H.; Hayashi, T.; Nishi, H.; Kusaba, K.; Koga, Y.; Koga, Y.; Nonaka, I.; Kimura, A. Phenotypic expression of a novel desmin gene mutation: Hypertrophic cardiomyopathy followed by systemic myopathy. J. Hum. Genet. 2018, 63, 249–254. [Google Scholar] [CrossRef]
- Begay, R.L.; Tharp, C.A.; Martin, A.; Graw, S.L.; Sinagra, G.; Miani, D.; Sweet, M.E.; Slavov, D.B.; Stafford, N.; Zeller, M.J.; et al. FLNC Gene Splice Mutations Cause Dilated Cardiomyopathy. JACC Basic Transl. Sci. 2016, 1, 344–359. [Google Scholar] [CrossRef] [PubMed]
- Reinstein, E.; Gutierrez-Fernandez, A.; Tzur, S.; Bormans, C.; Marcu, S.; Tayeb-Fligelman, E.; Vinkler, C.; Raas-Rothschild, A.; Irge, D.; Landau, M.; et al. Congenital dilated cardiomyopathy caused by biallelic mutations in Filamin C. Eur. J. Hum. Genet. 2016, 24, 1792–1796. [Google Scholar] [CrossRef]
- Marcus, F.I.; McKenna, W.J.; Sherrill, D.; Basso, C.; Bauce, B.; Bluemke, D.A.; Calkins, H.; Corrado, D.; Cox, M.G.; Daubert, J.P.; et al. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: Proposed modification of the Task Force Criteria. Eur. Heart J. 2010, 31, 806–814. [Google Scholar] [CrossRef]
- Towbin, J.A.; McKenna, W.J.; Abrams, D.J.; Ackerman, M.J.; Calkins, H.; Darrieux, F.C.C.; Daubert, J.P.; de Chillou, C.; DePasquale, E.C.; Desai, M.Y.; et al. 2019 HRS expert consensus statement on evaluation, risk stratification, and management of arrhythmogenic cardiomyopathy: Executive summary. Heart Rhythm 2019, 16, e373–e407. [Google Scholar] [CrossRef]
- Corrado, D.; Basso, C.; Judge, D.P. Arrhythmogenic Cardiomyopathy. Circ. Res. 2017, 121, 784–802. [Google Scholar] [CrossRef] [PubMed]
- Mestroni, L.; Sbaizero, O. Arrhythmogenic Cardiomyopathy: Mechanotransduction Going Wrong. Circulation 2018, 137, 1611–1613. [Google Scholar] [CrossRef] [PubMed]
- Begay, R.L.; Graw, S.L.; Sinagra, G.; Asimaki, A.; Rowland, T.J.; Slavov, D.B.; Gowan, K.; Jones, K.L.; Brun, F.; Merlo, M.; et al. Filamin C Truncation Mutations Are Associated with Arrhythmogenic Dilated Cardiomyopathy and Changes in the Cell-Cell Adhesion Structures. JACC Clin. Electrophysiol. 2018, 4, 504–514. [Google Scholar] [CrossRef]
- Brun, F.; Gigli, M.; Graw, S.L.; Judge, D.P.; Merlo, M.; Murray, B.; Calkins, H.; Sinagra, G.; Taylor, M.R.; Mestroni, L.; et al. FLNC truncations cause arrhythmogenic right ventricular cardiomyopathy. J. Med. Genet. 2020, 57, 254–257. [Google Scholar] [CrossRef]
- Ader, F.; De Groote, P.; Réant, P.; Rooryck-Thambo, C.; Dupin-Deguine, D.; Rambaud, C.; Khraiche, D.; Perret, C.; Pruny, J.F.; Mathieu-Dramard, M.; et al. FLNC pathogenic variants in patients with cardiomyopathies: Prevalence and genotype-phenotype correlations. Clin. Genet. 2019, 96, 317–329. [Google Scholar] [CrossRef] [PubMed]
- Valdes-Mas, R.; Gutierrez-Fernandez, A.; Gomez, J.; Coto, E.; Astudillo, A.; Puente, D.A.; Reguero, J.R.; Alvarez, V.; Moris, C.; Leon, D.; et al. Mutations in filamin C cause a new form of familial hypertrophic cardiomyopathy. Nat. Commun. 2014, 5, 5326. [Google Scholar] [CrossRef] [PubMed]
- Gómez, J.; Lorca, R.; Reguero, J.R.; Morís, C.; Martín, M.; Tranche, S.; Alonso, B.; Iglesias, S.; Alvarez, V.; Díaz-Molina, B.; et al. Screening of the Filamin C Gene in a Large Cohort of Hypertrophic Cardiomyopathy Patients. Circ. Cardiovasc. Genet. 2017, 10, e001584. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; Wang, J.; Zhang, C.; Wu, G.; Zhu, C.; Tang, B.; Zou, Y.; Huang, X.; Hui, R.; Song, L.; et al. Mutation profile of FLNC gene and its prognostic relevance in patients with hypertrophic cardiomyopathy. Mol. Genet. Genom. Med. 2018, 6, 1104–1113. [Google Scholar] [CrossRef]
- Walsh, R.; Buchan, R.; Wilk, A.; John, S.; Felkin, L.E.; Thomson, K.L.; Chiaw, T.H.; Loong, C.C.W.; Pua, C.J.; Raphael, C.; et al. Defining the genetic architecture of hypertrophic cardiomyopathy: Re-evaluating the role of non-sarcomeric genes. Eur. Heart J. 2017, 38, 3461–3468. [Google Scholar] [CrossRef] [PubMed]
- Elliott, P.; Andersson, B.; Arbustini, E.; Bilinska, Z.; Cecchi, F.; Charron, P.; Dubourg, O.; Kuhl, U.; Maisch, B.; McKenna, W.J.; et al. Classification of the cardiomyopathies. Kardiol. Pol. 2008, 66, 533–540. [Google Scholar] [CrossRef]
- Tucker, N.R.; McLellan, M.A.; Hu, D.; Ye, J.; Parsons, V.A.; Mills, R.W.; Clauss, S.; Dolmatova, E.; Shea, M.A.; Milan, D.J.; et al. Novel Mutation in FLNC (Filamin C) Causes Familial Restrictive Cardiomyopathy. Circ. Cardiovasc. Genet. 2017, 10, e001780. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Wei, Q.; Wu, B.; Liu, X.; Mading, A.; Yang, L.; Li, Y.; Liu, F.; Pan, X.; Wang, H. Clinical exome sequencing revealed that FLNC variants contribute to the early diagnosis of cardiomyopathies in infant patients. Transl. Pediatr. 2020, 9, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Bains, S.; Tester, D.J.; Asirvatham, S.J.; Noseworthy, P.A.; Ackerman, M.J.; Giudicessi, J.R. A Novel Truncating Variant in FLNC-Encoded Filamin C May Serve as a Proarrhythmic Genetic Substrate for Arrhythmogenic Bileaflet Mitral Valve Prolapse Syndrome. Mayo Clin. Proc. 2019, 94, 906–913. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eden, M.; Frey, N. Cardiac Filaminopathies: Illuminating the Divergent Role of Filamin C Mutations in Human Cardiomyopathy. J. Clin. Med. 2021, 10, 577. https://doi.org/10.3390/jcm10040577
Eden M, Frey N. Cardiac Filaminopathies: Illuminating the Divergent Role of Filamin C Mutations in Human Cardiomyopathy. Journal of Clinical Medicine. 2021; 10(4):577. https://doi.org/10.3390/jcm10040577
Chicago/Turabian StyleEden, Matthias, and Norbert Frey. 2021. "Cardiac Filaminopathies: Illuminating the Divergent Role of Filamin C Mutations in Human Cardiomyopathy" Journal of Clinical Medicine 10, no. 4: 577. https://doi.org/10.3390/jcm10040577
APA StyleEden, M., & Frey, N. (2021). Cardiac Filaminopathies: Illuminating the Divergent Role of Filamin C Mutations in Human Cardiomyopathy. Journal of Clinical Medicine, 10(4), 577. https://doi.org/10.3390/jcm10040577





