Melanoma Single-Cell Biology in Experimental and Clinical Settings
Abstract
1. Melanoma Biology, Clinics and Treatment
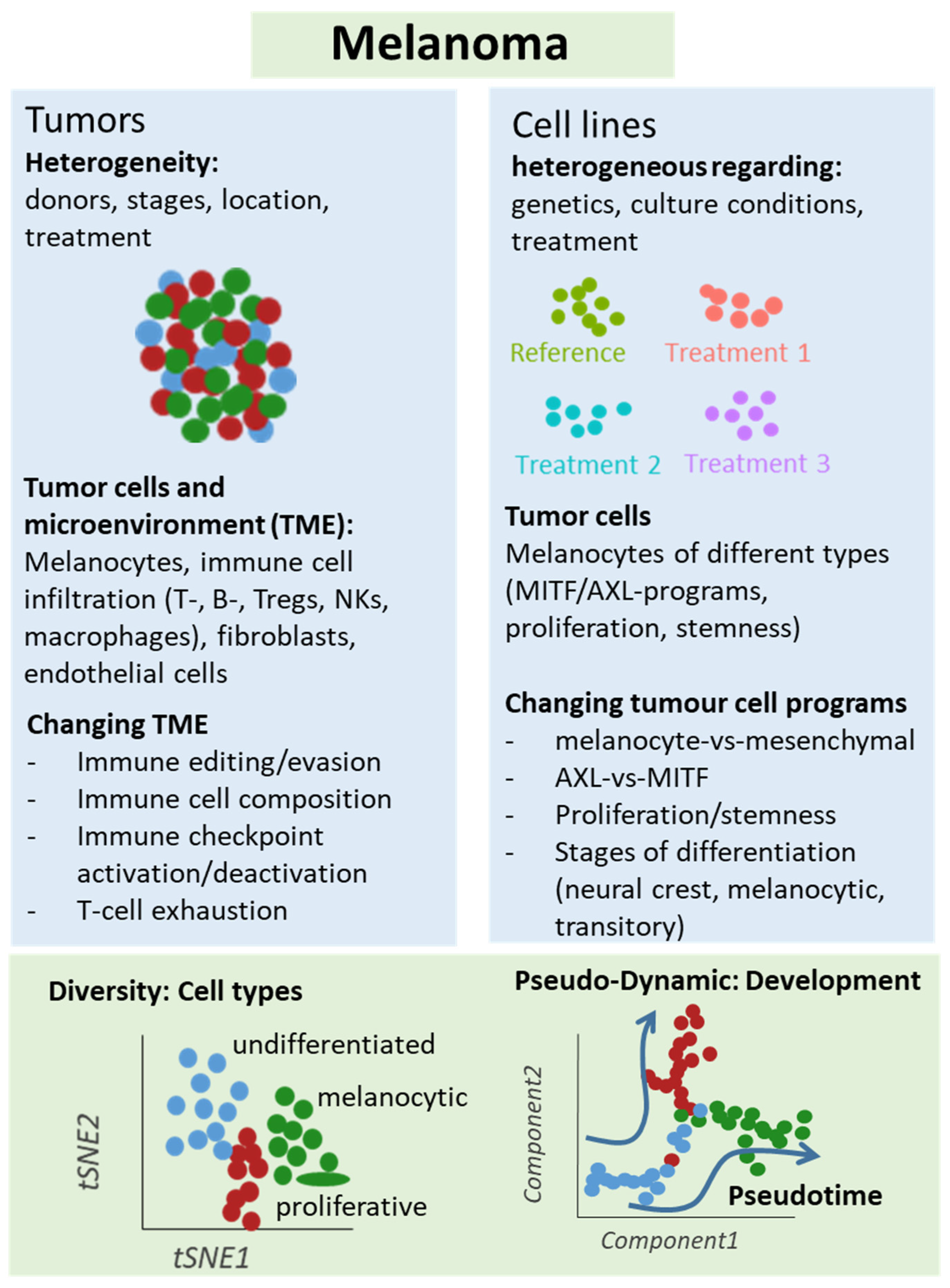
2. Clonal Heterogeneity in Melanoma
3. Single-Cell Technology
3.1. Principles of Single-Cell Individualization
3.2. Single-Cell Data Processing and Analysis
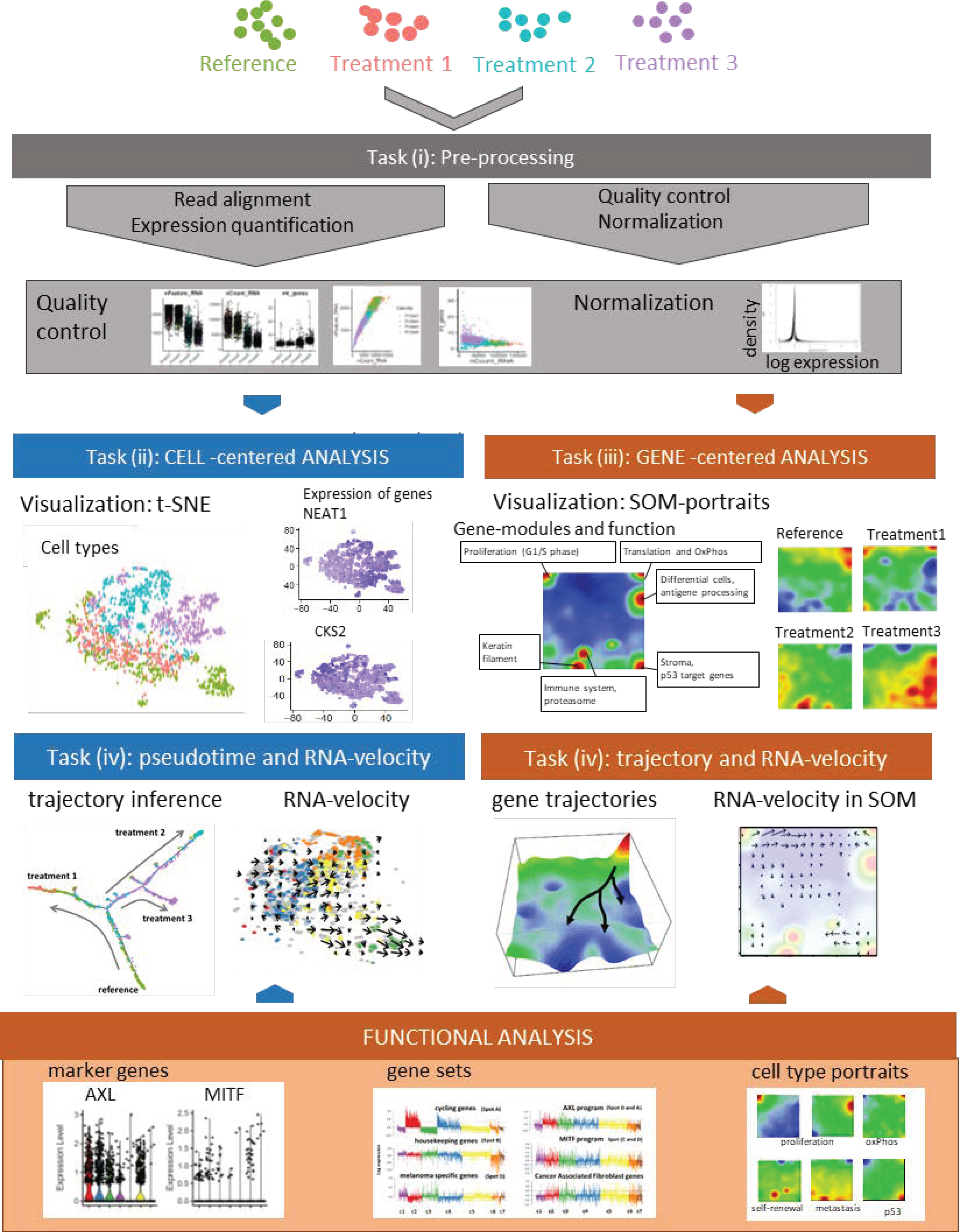
- i.
- Preprocessing aims at removing the effect of all factors without relevance for the expected biological effects. It includes read alignment, expression quantification, quality control, technical bias correction, and normalization. Mapping tools originally developed for bulk RNA-seq are mostly applicable also to scRNA-seq data. Further steps include quality control and filtering of unwanted genes and cells (e.g., cells expressing only a few genes); imputing missing values, batch correction (reducing systematic measurement biases between different runs and/or treatment groups), and normalizing gene expression (reducing unwanted variance between cells due to capture efficiency, sequencing depth, dropouts, and other technical effects). Technical noise of scRNA-seq is a common problem due to the low starting material and challenging experimental protocols. Detailed descriptions and recommendations of suited program-tools have recently been published.
- ii.
- Cell typing and diversity analysis aims at disentangling cell identities and their functional impact in the respective tissues. It includes clustering of cellular transcriptomes and their assignment to cell types (also called populations) in a supervised or unsupervised way. The former classification approach uses cell-type gene signatures taken from previous studies to assign the new data to these ‘known’ cell types. The latter class-discovery approach splits new data into de-novo groups of cells. In a second step, these new groups are related to known cell types by applying previous cell type signatures and statistical enrichment techniques, thus linking unsupervised with supervised approaches. Classifying cells into types or physiological states is essential for many secondary analyses to characterize the tumor microenvironment by composition of immune cells and/or to extract varying fractions of tumor cells from different developmental stages. For this task and scRNA-seq in general, reliable reference systems with a resolution down to cell states are required. Depending on the research question, even intermediate transition states might be of interest. Reference cell atlases of cell types of different healthy and cancer tissues, of immune cells and of melanoma-related cell types extracted from previous melanoma studies, have been published in a number of reports [50,59,60] and have put an emphasis on immune cells in these settings [61,62,63].
- iii.
- Gene module and marker extraction, functional analysis, and network inference aim at understanding gene regulation of cells on the gene level including aberrant effects due to genetic defects, external stimuli leading to treatment resistance and intrinsic evolutionary adaptations on tissue level. This task analyses co-expression of groups of genes, characterizes their functional context, and infers gene networks. Gene networks affect interactions between different cell types and/or signaling pathways. Beyond simple changes in average gene expression between cell types (or across bulk-collected libraries), scRNA-seq enables a high granularity of changes in expression. Particularly, cell type-specific alterations in cell state across samples are of special interest. These analyses deliver individual marker genes and sets of signature genes characterizing the different cell types and states, and, in addition, genes reflecting interactions between the cell types in the complex microenvironments of the respective tissue type. Appropriate methods to modularize transcriptional programs are non-negative matrix normalization (NMF) [64] or self-organizing-maps (SOM) [65].
- iv.
- Analysis of developmental trajectories in terms of pseudotime and RNA velocity aim at deducing time-dependent aspects of tissue development and cancer progression from cross-sectional scRNA-seq data. scRNA-seq experiments provide snapshot data, which resolves the molecular heterogeneity of cell cultures and tissues with single cell resolution under static conditions (see task (ii)). Given, that each cell is measured only once, one needs computational methods to deduce developmental trajectories on cellular level from time-independent data. The pseudotime model assumes that single-cell transcriptomes can be understood as a series of microscopic states of cellular development that exist in parallel at the same (real) time in the cell culture or tissue under study. Moreover, the model assumes that the temporal development smoothly and continuously changes transcriptional states in small and densely distributed steps so that the similarity of transcriptional characteristics can serve as a proxy of time, called pseudotime. It scales development in units of values between zero and unities for the start and end points, respectively. The pseudotime algorithm typically aligns the cells along a trajectory in reduced multi-dimensional space where a large variety of projection algorithms can be applied, differing regarding criteria such as cellular ordering, topology, scalability and usability [66]. Each method has its own characteristics in terms of the underlying algorithm, produced outputs and regarding the topology of the pseudotime trajectory (e.g., predefined linear, multibranched, cyclic, or ‘inferred from the data’). ‘RNA-velocity’ provides another independent approach to infer developmental trajectories from static scRNA-seq data [67,68]. It directly ‘forecasts’ the transcriptional state of a cell based on the relation between spliced and unspliced mRNA in terms of a directional change of cell state in cell-diversity space (task (v)). RNA-velocity provides a vector-field reflecting transcriptional changes of each cell, which can be transferred into developmental trajectories joining sources and sinks of mRNA abundance in cell-state and gene-state space.
- v.
- Dimension reduction, visualization of cell- and gene-state space, and data portrayals aim at enabling the intuitive perception of complex data in order to extract ‘hidden’ information and to support hypothesis development and testing. scRNA-seq data are high-dimensional data (ten-thousands of transcripts multiplied with ten-to-hundred thousand of cells multiplied with a multitude of biological conditions), which is difficult to visualize in its original form. Dimension reduction and appropriate visualization are therefore important challenges in all four tasks listed above. Conceptually, two perpendicular types of information have to be considered, namely cell- and gene-centered views on the scRNA transcriptomes as addressed in tasks II and III, respectively [61,69]. For the view on cell diversity, different methods projecting multidimensional cell-transcriptomes data into two dimensions are in use, such as Principal Component Analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP) [70,71]. These methods produce point clouds in cell similarity space visualizing mutual similarities between the single-cell transcriptomes in terms of colored clusters of cells (task II) and/or of colored expression levels of selected gene markers and signatures in the individual cell transcriptomes (task III). For visualization of transcriptomic landscapes in gene state space, we developed the expression portrayal method based on self-organizing map (SOM) machine learning. Such landscapes support identification of modules of co-expressed genes, of their mutual network topology and of their functional context [72,73].
3.3. Single-Cell Exome-Seq
4. Single-Cell Analyses in Melanoma
4.1. Primary Melanomas, Lymph Node Metastases and Cell Lines
4.2. Treatment Resistance Under Immune Checkpoint Inhibition
4.3. Treatment Resistance under Targeted Treatment
5. Spatial Sequencing in Melanoma
6. Single-Cell Sequencing of Copy Number Variations
7. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bai, X.; Flaherty, K.T. Targeted and immunotherapies in BRAF mutant melanoma: Where we stand and what to expect. Br. J. Dermatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Schadendorf, D.; Van Akkooi, A.C.J.; Berking, C.; Griewank, K.G.; Gutzmer, R.; Hauschild, A.; Stang, A.; Roesch, A.; Ugurel, S. Melanoma. Lancet 2018, 392, 971–984. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef] [PubMed]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.-P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef]
- Krauthammer, M.; Kong, Y.; Bacchiocchi, A.; Evans, P.; Pornputtapong, N.; Wu, C.; McCusker, J.P.; Ma, S.; Cheng, E.; Straub, R.; et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat. Genet. 2015, 47, 996–1002. [Google Scholar] [CrossRef]
- Davies, M.A.; Flaherty, K.T. Melanoma in 2017: Moving treatments earlier to move further forwards. Nat. Rev. Clin. Oncol. 2018, 15, 75–76. [Google Scholar] [CrossRef]
- Grimaldi, A.M.; Simeone, E.; Ascierto, P.A. The role of MEK inhibitors in the treatment of metastatic melanoma. Curr. Opin. Oncol. 2014, 26, 196–203. [Google Scholar] [CrossRef]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Chiarion Sileni, V.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef]
- Menzies, A.M.; Long, G.V. Systemic treatment for BRAF-mutant melanoma: Where do we go next? Lancet Oncol. 2014, 15, e371–e381. [Google Scholar] [CrossRef]
- Dummer, R.; Brase, J.C.; Garrett, J.; Campbell, C.D.; Gasal, E.; Squires, M.; Gusenleitner, D.; Santinami, M.; Atkinson, V.; Mandalà, M.; et al. Adjuvant dabrafenib plus trametinib versus placebo in patients with resected, BRAFV600-mutant, stage III melanoma (COMBI-AD): Exploratory biomarker analyses from a randomised, phase 3 trial. Lancet Oncol. 2020, 21, 358–372. [Google Scholar] [CrossRef]
- Amaria, R.N.; Menzies, A.M.; Burton, E.M.; Scolyer, R.A.; Tetzlaff, M.T.; Antdbacka, R.; Ariyan, C.; Bassett, R.; Carter, B.; Daud, A.; et al. Neoadjuvant systemic therapy in melanoma: Recommendations of the International Neoadjuvant Melanoma Consortium. Lancet Oncol. 2019, 20, e378–e389. [Google Scholar] [CrossRef]
- Moriceau, G.; Hugo, W.; Hong, A.; Shi, H.; Kong, X.; Yu, C.C.; Koya, R.C.; Samatar, A.A.; Khanlou, N.; Braun, J.; et al. Tunable-combinatorial Mechanisms of Acquired Resistance Limit the Efficacy of BRAF/MEK Co-targeting but Result in Melanoma Drug Addiction. Cancer Cell 2015, 27, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Fung, C.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Hyman, J.; Shahheydari, H.; Tembe, V.; Thompson, J.F.; Saw, R.P.; et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014, 5, 5694. [Google Scholar] [CrossRef] [PubMed]
- Wagle, N.; Van Allen, E.M.; Treacy, D.J.; Frederick, D.T.; Cooper, Z.A.; Taylor-Weiner, A.; Rosenberg, M.; Goetz, E.M.; Sullivan, R.J.; Farlow, D.N.; et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014, 4, 61–68. [Google Scholar] [CrossRef]
- Rizos, H.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Fung, C.; Hyman, J.; Haydu, L.E.; Mijatov, B.; Becker, T.M.; Boyd, S.C.; et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: Spectrum and clinical impact. Clin. Cancer Res. 2014, 20, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Hugo, W.; Kong, X.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef]
- Hugo, W.; Shi, H.; Sun, L.; Piva, M.; Song, C.; Kong, X.; Moriceau, G.; Hong, A.; Dahlman, K.B.; Johnson, D.B.; et al. Non-genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015, 162, 1271–1285. [Google Scholar] [CrossRef]
- Sun, C.; Wang, L.; Huang, S.; Heynen, G.J.J.E.; Prahallad, A.; Robert, C.; Haanen, J.; Blank, C.; Wesseling, J.; Willems, S.M.; et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014, 508, 118–122. [Google Scholar] [CrossRef]
- Müller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.; Kong, X.; Possik, P.A.; Cornelissen-Steijger, P.D.M.; Geukes Foppen, M.H.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 5712. [Google Scholar] [CrossRef]
- Johannessen, C.M.; Boehm, J.S.; Kim, S.Y.; Thomas, S.R.; Wardwell, L.; Johnson, L.A.; Emery, C.M.; Stransky, N.; Cogdill, A.P.; Barretina, J.; et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010, 468, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014, 4, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.-K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Kunz, M.; Hölzel, M. The impact of melanoma genetics on treatment response and resistance in clinical and experimental studies. Cancer Metastasis Rev. 2017, 36, 53–75. [Google Scholar] [CrossRef]
- Hirsch, L.; Zitvogel, L.; Eggermont, A.; Marabelle, A. PD-Loma: A cancer entity with a shared sensitivity to the PD-1/PD-L1 pathway blockade. Br. J. Cancer 2019, 120, 3–5. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Rutkowski, P.; Lao, C.D.; Cowey, C.L.; Schadendorf, D.; Wagstaff, J.; Dummer, R.; et al. Five-Year Survival with Combined Nivolumab and Ipilimumab in Advanced Melanoma. N. Engl. J. Med. 2019, 381, 1535–1546. [Google Scholar] [CrossRef]
- Bagchi, S.; Yuan, R.; Engleman, E.G. Immune Checkpoint Inhibitors for the Treatment of Cancer: Clinical Impact and Mechanisms of Response and Resistance. Annu. Rev. Pathol. 2020. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Snyder, A.; Wolchok, J.D.; Chan, T.A. Genetic basis for clinical response to CTLA-4 blockade. N. Engl. J. Med. 2015, 372, 783. [Google Scholar] [CrossRef]
- Hugo, W.; Zaretsky, J.M.; Sun, L.; Song, C.; Moreno, B.H.; Hu-Lieskovan, S.; Berent-Maoz, B.; Pang, J.; Chmielowski, B.; Cherry, G.; et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016, 165, 35–44. [Google Scholar] [CrossRef]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.-L.; Roh, W.; Reuben, A.; Cooper, Z.A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Bassett, R.L.; Gopalakrishnan, V.; Wani, K.; et al. Analysis of Immune Signatures in Longitudinal Tumor Samples Yields Insight into Biomarkers of Response and Mechanisms of Resistance to Immune Checkpoint Blockade. Cancer Discov. 2016, 6, 827–837. [Google Scholar] [CrossRef] [PubMed]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martín-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Schilling, B.; Liu, D.; Sucker, A.; Livingstone, E.; Jerby-Arnon, L.; Zimmer, L.; Gutzmer, R.; Satzger, I.; Loquai, C.; et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 2019, 25, 1916–1927. [Google Scholar] [CrossRef]
- Auslander, N.; Zhang, G.; Lee, J.S.; Frederick, D.T.; Miao, B.; Moll, T.; Tian, T.; Wei, Z.; Madan, S.; Sullivan, R.J.; et al. Robust prediction of response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2018, 24, 1545–1549. [Google Scholar] [CrossRef]
- Carter, J.A.; Gilbo, P.; Atwal, G.S. IMPRES does not reproducibly predict response to immune checkpoint blockade therapy in metastatic melanoma. Nat. Med. 2019, 25, 1833–1835. [Google Scholar] [CrossRef]
- Effern, M.; Glodde, N.; Braun, M.; Liebing, J.; Boll, H.N.; Yong, M.; Bawden, E.; Hinze, D.; Van den Boorn-Konijnenberg, D.; Daoud, M.; et al. Adoptive T Cell Therapy Targeting Different Gene Products Reveals Diverse and Context-Dependent Immune Evasion in Melanoma. Immunity 2020, 53, 564–580.e9. [Google Scholar] [CrossRef]
- Landsberg, J.; Kohlmeyer, J.; Renn, M.; Bald, T.; Rogava, M.; Cron, M.; Fatho, M.; Lennerz, V.; Wölfel, T.; Hölzel, M.; et al. Melanomas resist T-cell therapy through inflammation-induced reversible dedifferentiation. Nature 2012, 490, 412–416. [Google Scholar] [CrossRef]
- Yeon, M.; Kim, Y.; Jung, H.S.; Jeoung, D. Histone Deacetylase Inhibitors to Overcome Resistance to Targeted and Immuno Therapy in Metastatic Melanoma. Front. Cell Dev. Biol. 2020, 8, 486. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Nowell, P.C. The clonal evolution of tumor cell populations. Science 1976, 194, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Barrett, M.T.; Lenkiewicz, E.; Evers, L.; Holley, T.; Ruiz, C.; Bubendorf, L.; Sekulic, A.; Ramanathan, R.K.; Von Hoff, D.D. Clonal evolution and therapeutic resistance in solid tumors. Front. Pharmacol. 2013, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, S.; Caldas, C. The implications of clonal genome evolution for cancer medicine. N. Engl. J. Med. 2013, 368, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Yancovitz, M.; Litterman, A.; Yoon, J.; Ng, E.; Shapiro, R.L.; Berman, R.S.; Pavlick, A.C.; Darvishian, F.; Christos, P.; Mazumdar, M.; et al. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS ONE 2012, 7, e29336. [Google Scholar] [CrossRef] [PubMed]
- Harbst, K.; Lauss, M.; Cirenajwis, H.; Isaksson, K.; Rosengren, F.; Törngren, T.; Kvist, A.; Johansson, M.C.; Vallon-Christersson, J.; Baldetorp, B.; et al. Multiregion Whole-Exome Sequencing Uncovers the Genetic Evolution and Mutational Heterogeneity of Early-Stage Metastatic Melanoma. Cancer Res. 2016, 76, 4765–4774. [Google Scholar] [CrossRef]
- Sanna, A.; Harbst, K.; Johansson, I.; Christensen, G.; Lauss, M.; Mitra, S.; Rosengren, F.; Häkkinen, J.; Vallon-Christersson, J.; Olsson, H.; et al. Tumor genetic heterogeneity analysis of chronic sun-damaged melanoma. Pigment Cell Melanoma Res. 2020, 33, 480–489. [Google Scholar] [CrossRef]
- Obenauf, A.C.; Zou, Y.; Ji, A.L.; Vanharanta, S.; Shu, W.; Shi, H.; Kong, X.; Bosenberg, M.C.; Wiesner, T.; Rosen, N.; et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 2015, 520, 368–372. [Google Scholar] [CrossRef]
- Shaffer, S.M.; Emert, B.L.; Reyes Hueros, R.A.; Cote, C.; Harmange, G.; Schaff, D.L.; Sizemore, A.E.; Gupte, R.; Torre, E.; Singh, A.; et al. Memory Sequencing Reveals Heritable Single-Cell Gene Expression Programs Associated with Distinct Cellular Behaviors. Cell 2020, 182, 947–959.e17. [Google Scholar] [CrossRef]
- Lin, Z.; Meng, X.; Wen, J.; Corral, J.M.; Andreev, D.; Kachler, K.; Schett, G.; Chen, X.; Bozec, A. Intratumor Heterogeneity Correlates With Reduced Immune Activity and Worse Survival in Melanoma Patients. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Wolf, Y.; Bartok, O.; Patkar, S.; Eli, G.B.; Cohen, S.; Litchfield, K.; Levy, R.; Jiménez-Sánchez, A.; Trabish, S.; Lee, J.S.; et al. UVB-Induced Tumor Heterogeneity Diminishes Immune Response in Melanoma. Cell 2019, 179, 219–235.e21. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Lin, Y.; Navin, N. Advancing Cancer Research and Medicine with Single-Cell Genomics. Cancer Cell 2020, 37, 456–470. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, S.; Teichmann, S.A. Single cell transcriptomics comes of age. Nat. Commun. 2020, 11, 4307. [Google Scholar] [CrossRef] [PubMed]
- Renaud, G.; Stenzel, U.; Maricic, T.; Wiebe, V.; Kelso, J. deML: Robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics 2015, 31, 770–772. [Google Scholar] [CrossRef] [PubMed]
- Renaud, G.; Kircher, M.; Stenzel, U.; Kelso, J. freeIbis: An efficient basecaller with calibrated quality scores for Illumina sequencers. Bioinformatics 2013, 29, 1208–1209. [Google Scholar] [CrossRef]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef]
- Ji, F.; Sadreyev, R.I. Single-Cell RNA-seq: Introduction to Bioinformatics Analysis. Curr. Protoc. Mol. Biol. 2019, 127, e92. [Google Scholar] [CrossRef]
- Lähnemann, D.; Köster, J.; Szczurek, E.; McCarthy, D.J.; Hicks, S.C.; Robinson, M.D.; Vallejos, C.A.; Campbell, K.R.; Beerenwinkel, N.; Mahfouz, A.; et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020, 21, 31. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef]
- Uhlén, M.; Hallström, B.M.; Lindskog, C.; Mardinoglu, A.; Pontén, F.; Nielsen, J. Transcriptomics resources of human tissues and organs. Mol. Syst. Biol. 2016, 12, 862. [Google Scholar] [CrossRef]
- Hackl, H.; Charoentong, P.; Finotello, F.; Trajanoski, Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nat. Rev. Genet. 2016, 17, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Nieto, P.; Elosua-Bayes, M.; Trincado, J.L.; Marchese, D.; Massoni-Badosa, R.; Salvany, M.; Henriques, A.; Mereu, E.; Moutinho, C.; Ruiz, S.; et al. A Single-Cell Tumor Immune Atlas for Precision Oncology. bioRxiv 2020. [Google Scholar] [CrossRef]
- Xie, X.; Liu, M.; Zhang, Y.; Wang, B.; Zhu, C.; Wang, C.; Li, Q.; Huo, Y.; Guo, J.; Xu, C.; et al. Single-cell transcriptomic landscape of human blood cells. Natl. Sci. Rev. 2020. [Google Scholar] [CrossRef]
- Ho, Y.-J.; Anaparthy, N.; Molik, D.; Mathew, G.; Aicher, T.; Patel, A.; Hicks, J.; Hammell, M.G. Single-cell RNA-seq analysis identifies markers of resistance to targeted BRAF inhibitors in melanoma cell populations. Genome Res. 2018, 28, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Gerber, T.; Willscher, E.; Loeffler-Wirth, H.; Hopp, L.; Schadendorf, D.; Schartl, M.; Anderegg, U.; Camp, G.; Treutlein, B.; Binder, H.; et al. Mapping heterogeneity in patient-derived melanoma cultures by single-cell RNA-seq. Oncotarget 2017, 8, 846–862. [Google Scholar] [CrossRef] [PubMed]
- Saelens, W.; Cannoodt, R.; Todorov, H.; Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 2019, 37, 547–554. [Google Scholar] [CrossRef]
- Bergen, V.; Lange, M.; Peidli, S.; Wolf, F.A.; Theis, F.J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 2020, 38, 1408–1414. [Google Scholar] [CrossRef]
- La Manno, G.; Soldatov, R.; Zeisel, A.; Braun, E.; Hochgerner, H.; Petukhov, V.; Lidschreiber, K.; Kastriti, M.E.; Lönnerberg, P.; Furlan, A.; et al. RNA velocity of single cells. Nature 2018, 560, 494–498. [Google Scholar] [CrossRef]
- Schmidt, M.; Loeffler-Wirth, H.; Binder, H. Developmental scRNAseq Trajectories in Gene- and Cell-State Space-The Flatworm Example. Genes (Basel) 2020, 11, 1214. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. Available online: http://arxiv.org/pdf/1802.03426v3 (accessed on 18 September 2020).
- Kobak, D.; Berens, P. The art of using t-SNE for single-cell transcriptomics. Nat. Commun. 2019, 10, 5416. [Google Scholar] [CrossRef] [PubMed]
- Wirth, H.; Von Bergen, M.; Binder, H. Mining SOM expression portraits: Feature selection and integrating concepts of molecular function. BioData Min. 2012, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Hopp, L.; Wirth, H.; Fasold, M.; Binder, H. Portraying the expression landscapes of cancer subtypes. Syst. Biomed. 2013, 1, 99–121. [Google Scholar] [CrossRef]
- Xu, X.; Hou, Y.; Yin, X.; Bao, L.; Tang, A.; Song, L.; Li, F.; Tsang, S.; Wu, K.; Wu, H.; et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 2012, 148, 886–895. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc. Natl. Acad. Sci. USA. 2014, 111, 17947–17952. [Google Scholar] [CrossRef]
- Dey, S.S.; Kester, L.; Spanjaard, B.; Bienko, M.; Van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 2015, 33, 285–289. [Google Scholar] [CrossRef]
- Leung, M.L.; Wang, Y.; Waters, J.; Navin, N.E. SNES: Single nucleus exome sequencing. Genome Biol. 2015, 16, 55. [Google Scholar] [CrossRef]
- Leung, M.L.; Davis, A.; Gao, R.; Casasent, A.; Wang, Y.; Sei, E.; Vilar, E.; Maru, D.; Kopetz, S.; Navin, N.E. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res. 2017, 27, 1287–1299. [Google Scholar] [CrossRef]
- Albertí-Servera, L.; Demeyer, S.; Govaerts, I.; Swings, T.; De Bie, J.; Gielen, O.; Brociner, M.; Michaux, L.M.; Maertens, J.; Uyttebroeck, A.; et al. Single-cell DNA amplicon sequencing reveals clonal heterogeneity and evolution in T-cell acute lymphoblastic leukemia. Blood 2020. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Liang, S.; Wang, F.; Han, J.; Chen, K. Latent periodic process inference from single-cell RNA-seq data. Nat. Commun. 2020, 11, 1441. [Google Scholar] [CrossRef] [PubMed]
- Wouters, J.; Kalender-Atak, Z.; Minnoye, L.; Spanier, K.I.; De Waegeneer, M.; Bravo González-Blas, C.; Mauduit, D.; Davie, K.; Hulselmans, G.; Najem, A.; et al. Robust gene expression programs underlie recurrent cell states and phenotype switching in melanoma. Nat. Cell Biol. 2020, 22, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Sade-Feldman, M.; Yizhak, K.; Bjorgaard, S.L.; Ray, J.P.; De Boer, C.G.; Jenkins, R.W.; Lieb, D.J.; Chen, J.H.; Frederick, D.T.; Barzily-Rokni, M.; et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018, 175, 998–1013.e20. [Google Scholar] [CrossRef] [PubMed]
- Jerby-Arnon, L.; Shah, P.; Cuoco, M.S.; Rodman, C.; Su, M.-J.; Melms, J.C.; Leeson, R.; Kanodia, A.; Mei, S.; Lin, J.-R.; et al. A Cancer Cell Program Promotes T Cell Exclusion and Resistance to Checkpoint Blockade. Cell 2018, 175, 984–997.e24. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Van der Leun, A.M.; Yofe, I.; Lubling, Y.; Gelbard-Solodkin, D.; Van Akkooi, A.C.J.; Van den Braber, M.; Rozeman, E.A.; Haanen, J.B.A.G.; Blank, C.U.; et al. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell 2019, 176, 775–789.e18. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Rogiers, A.; Marin-Bejar, O.; Aibar, S.; Femel, J.; Dewaele, M.; Karras, P.; Brown, D.; Chang, Y.H.; Debiec-Rychter, M.; et al. Toward Minimal Residual Disease-Directed Therapy in Melanoma. Cell 2018, 174, 843–855.e19. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Lim, H.; Chun, H.; Lee, J.H.; Bang, D. Single-cell analysis of a mutant library generated using CRISPR-guided deaminase in human melanoma cells. Commun. Biol. 2020, 3, 154. [Google Scholar] [CrossRef]
- Su, Y.; Wei, W.; Robert, L.; Xue, M.; Tsoi, J.; Garcia-Diaz, A.; Homet Moreno, B.; Kim, J.; Ng, R.H.; Lee, J.W.; et al. Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc. Natl. Acad. Sci. USA. 2017, 114, 13679–13684. [Google Scholar] [CrossRef]
- Su, Y.; Ko, M.E.; Cheng, H.; Zhu, R.; Xue, M.; Wang, J.; Lee, J.W.; Frankiw, L.; Xu, A.; Wong, S.; et al. Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line. Nat. Commun. 2020, 11, 2345. [Google Scholar] [CrossRef]
- Cojoc, M.; Mäbert, K.; Muders, M.H.; Dubrovska, A. A role for cancer stem cells in therapy resistance: Cellular and molecular mechanisms. Semin. Cancer Biol. 2015, 31, 16–27. [Google Scholar] [CrossRef]
- Lawson, D.A.; Bhakta, N.R.; Kessenbrock, K.; Prummel, K.D.; Yu, Y.; Takai, K.; Zhou, A.; Eyob, H.; Balakrishnan, S.; Wang, C.-Y.; et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature 2015, 526, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Thibaut, R.; Bost, P.; Milo, I.; Cazaux, M.; Lemaître, F.; Garcia, Z.; Amit, I.; Breart, B.; Cornuot, C.; Schwikowski, B.; et al. Bystander IFN-γ activity promotes widespread and sustained cytokine signaling altering the tumor microenvironment. Nat. Cancer 2020, 1, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Mojtahedi, M.; Skupin, A.; Zhou, J.; Castaño, I.G.; Leong-Quong, R.Y.Y.; Chang, H.; Trachana, K.; Giuliani, A.; Huang, S. Cell Fate Decision as High-Dimensional Critical State Transition. PLoS Biol. 2016, 14, e2000640. [Google Scholar] [CrossRef] [PubMed]
- Thrane, K.; Eriksson, H.; Maaskola, J.; Hansson, J.; Lundeberg, J. Spatially Resolved Transcriptomics Enables Dissection of Genetic Heterogeneity in Stage III Cutaneous Malignant Melanoma. Cancer Res. 2018, 78, 5970–5979. [Google Scholar] [CrossRef] [PubMed]
- Crosetto, N.; Bienko, M.; Van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet. 2015, 16, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstråhle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 497–514.e22. [Google Scholar] [CrossRef]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Velazquez-Villarreal, E.I.; Maheshwari, S.; Sorenson, J.; Fiddes, I.T.; Kumar, V.; Yin, Y.; Webb, M.G.; Catalanotti, C.; Grigorova, M.; Edwards, P.A.; et al. Single-cell sequencing of genomic DNA resolves sub-clonal heterogeneity in a melanoma cell line. Commun. Biol. 2020, 3, 318. [Google Scholar] [CrossRef]
- Slyper, M.; Porter, C.B.M.; Ashenberg, O.; Waldman, J.; Drokhlyansky, E.; Wakiro, I.; Smillie, C.; Smith-Rosario, G.; Wu, J.; Dionne, D.; et al. Author Correction: A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med. 2020, 26, 1307. [Google Scholar] [CrossRef]
| No | Melanoma Samples | Experimental or Clinical Set-Up | Characteristics of Clonal Structure | Main Findings | References |
|---|---|---|---|---|---|
| 1. | Primary melanomas and metastases (n = 19) | Untreated | Clonal signatures of cell cycle, spatial context, drug-resistance programs | Presence of AXL-high/MITF-low population in a AXL-low/ MITF-high cluster; single-cell signatures with prognostic relevance | [80] |
| 2. | Melanoma cell lines representing different stages of differentiation (n = 8) | Untreated | Cell clones with SOX9 and SOX10 high expression and transitional cells, knockdown of SOX10 affects clonal structure | Transition between gene networks instead of selection of individual clones (transcriptional plasticity) | [82] |
| 3. | Melanoma short-term cultures (BRAF and/or NRAS mutant) (n = 3) | Untreated | Clonal structure of cell cycle, stromal, OxPhos, pigmentation genes | Four different clonal structures with additional subclonal structures and stem cell-like subclones | [65] |
| 4. | Samples from 32 metastatic melanoma patients (n = 48) | Anti-PD1 inhibitor treatment of patients, either alone or in combination with anti-CTLA4 treatment | CD8+ T cells clones consisted of memory/survival (TCF7+) and exhaustion (CD38+) clones, respectively | TCF7+/CD8+ T cells are crucial for treatment response | [83] |
| 5. | Human melanoma samples (n = 33) | Clinical samples under anti-CTLA4 treatment | Clonal immune exclusion program: CDK4/CDK6 expression, JAK-STAT3 signaling, TNF pathway, senescence-associated programs, Myc targets | CDK4/CDK6 inhibitor treatment of resistant clones improved survival of mice in a murine melanoma model | [84] |
| 6. | Human melanoma samples (n = 25) | Anti-PD-1 inhibitor treatment of patients, either alone or in combination with anti-CTLA4 treatment | CD4+/CD8+ T cells with clusters of resting, transitional and exhausted T cells | Dysfunctional (exhausted) CD8+ T cells are still proliferative and showed tumor reactivity ex vivo | [85] |
| 7. | Tumor tissue of melanoma cell line mouse xenografts (minimal residual disease) (n = 3) | Murine xenograft model, BRAFi treatment | Minimal residual disease with 4 different transcriptional subpopulations (pigmented, SMC, NCSC, invasive cells) | Enrichment of neuronal stem cells population after BRAFi treatment; successful treatment with retinoid receptor inhibitor | [86] |
| 8. | A375 and 451Lu melanoma cell lines (n = 2) | BRAFi treatment | Patterns of resistance are present in parental cells and vice versa | Identification of a pre-resistant state at the tip of the parental population | [64] |
| 9. | Melanoma cell line A375 (n = 1) | BRAFi treatment after CRISPR/Cas interference with MAPK pathway | Clonal selection of treatment resistant clones | Resistance-mediating positions in MAPK genes were mostly located around MEK1E203K or KRASQ61 | [87] |
| 10. | BRAF-mutant melanoma cell lines (n = 3) | BRAFi treatment; testing of 13 different proteomic markers with single-cell barcode chip technology | Increased clonal heterogeneity under treatment | Activation of MEK/ERK and NF-κB p65 signaling in resitant clones; NF-κB inhibitor increased sensitivity of cells | [88] |
| 11. | BRAF-mutant melanoma cell line (n = 1) | BRAFi treatment; testing of 19 different proteomic markers with single-cell barcode chip technology | Drug-induced clonal cell states changes with NGFR/AXL or MITF, MART1 patterns | Two different trajectories of treatment resistance of MITF-high and MITF- low cells | [89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Binder, H.; Schmidt, M.; Loeffler-Wirth, H.; Mortensen, L.S.; Kunz, M. Melanoma Single-Cell Biology in Experimental and Clinical Settings. J. Clin. Med. 2021, 10, 506. https://doi.org/10.3390/jcm10030506
Binder H, Schmidt M, Loeffler-Wirth H, Mortensen LS, Kunz M. Melanoma Single-Cell Biology in Experimental and Clinical Settings. Journal of Clinical Medicine. 2021; 10(3):506. https://doi.org/10.3390/jcm10030506
Chicago/Turabian StyleBinder, Hans, Maria Schmidt, Henry Loeffler-Wirth, Lena Suenke Mortensen, and Manfred Kunz. 2021. "Melanoma Single-Cell Biology in Experimental and Clinical Settings" Journal of Clinical Medicine 10, no. 3: 506. https://doi.org/10.3390/jcm10030506
APA StyleBinder, H., Schmidt, M., Loeffler-Wirth, H., Mortensen, L. S., & Kunz, M. (2021). Melanoma Single-Cell Biology in Experimental and Clinical Settings. Journal of Clinical Medicine, 10(3), 506. https://doi.org/10.3390/jcm10030506







