Abstract
Atopic dermatitis is frequently associated with the onset of other allergic conditions, such as asthma, rhino-conjunctivitis and food allergy. The etiology of atopic dermatitis is marginally understood in spite of the number of predisposing factors, above all, mutations in the Filaggrin gene (FLG). In this study, the association between loss-of-function variants in the FLG gene and other allergic manifestations, in particular food allergy, was evaluated in an Italian pediatric population affected by atopic dermatitis. The 10 more frequently mutated loci in the FLG gene were genotyped in 238 children affected by atopic dermatitis and tested for association with clinical features of allergic disorders by a multivariate logistic regression model. R501X and 2282del4 were the only two mutations identified; 12.2% of children carry one of these variants, corresponding to an allelic frequency of 6.5%. According to multivariate statistical analysis, loss-of-function variants in the FLG gene represent a risk factor for the onset of severe manifestations of food allergy (OR = 8.9; CI: 3.1–28.3). Peanut and hazelnut were identified as high-risk foods in patients with FLG mutations. This study demonstrates that atopic children carrying FLG mutations represent a high-risk population due to their predisposition to develop severe food allergy reactions, such as anaphylaxis.
1. Introduction
Atopic dermatitis (AD), also known as atopic eczema, is a relapsing inflammatory skin disease with a considerable social and economic burden. It is the most common inflammatory skin disease during childhood with various manifestations over the years and a progression frequently associated with the onset of allergic conditions, especially food allergy (FA), asthma and rhino-conjunctivitis (RC) [1,2].
The complex pathogenesis leading to AD is not yet completely understood, but many hypotheses support the idea of a multifactorial disease involving genetic and immune system alterations along with environmental factors. Since the discovery by Palmer et al. of a strong association between loss-of-function mutations of the Filaggrin (FLG) gene and AD and asthma [3], the concept of a skin barrier genetic defect causing AD has replaced the “inside–outside” hypothesis. In the European population, the frequency of the most common FLG gene alterations (R501X and 2282del4) varies among different regions, depending on their ethnic group [4,5].
AD and FA are often associated in children: FA may occur in about one third of patients with AD and AD has been associated with earlier and persistent allergic sensitization to foods [6,7]. In children with AD, the early sensitization to food allergens (in particular cow’s milk and hen’s egg) and/or to inhalant allergens represents a risk factor for a more persistent and severe disease and asthma development [8]. Many studies have indicated that transcutaneous allergic sensitization may be facilitated by FLG null-mutations, and that both peanut allergy and AD are strongly associated with FLG loss-of-function variants [9,10,11,12], as well as other cutaneous conditions [13,14]. In an English population-based birth cohort, the exposure to environmental peanuts early in life (within the first year of life) was associated with peanut sensitization and allergy in children carrying a FLG gene mutation [10]. In support of the role of the skin barrier in driving AD, allergic disease and food sensitization, a retrospective follow-up study suggested that the improvement of the skin management in children with AD could reduce or delay the appearance of allergic respiratory diseases [8]. Meanwhile, a recent pilot study with a lipid-rich skin emollient from birth was found to improve skin barrier function and reduce food allergen sensitization [15].
ImmunoglobulinE (IgE)-associated AD is more frequent in childhood than non-IgE-associated AD, and these two phenotypes generally differ in their onset age and remission pattern. Therefore, it is important, especially in early life, to distinguish these two phenotypes in order to evaluate possible allergy-related conditions and to predict the occurrence of other allergic phenotypes later in childhood [16,17].
Considering the wide spectrum of phenotypes possibly associated to FLG mutations, this study aimed to identify the association of FLG loss-of-function variants with allergic phenotypes, in particular with severe food allergy.
2. Experimental Section
2.1. Study Population
Patients suffering from AD evaluated at the Pediatric Allergology and Pediatric Dermatology Outpatient Clinics of the S. Orsola-Malpighi Hospital of Bologna (Bologna, Italy) were recruited for this study. All patients were followed up with clinical examinations at least once a year, or more frequently depending on the severity of their disease and clinical symptoms. The inclusion criteria were: informed consent signed by parents or legal tutors of the child, Italian ancestry, clinical diagnosis of AD within pediatric age (<14 years), formulated on the basis of Hanifin and Rajka criteria [18,19]. Older patients do not imply persistent dermatologic disease, since they were all taken in charge by the Pediatric Allergology Unit.
2.2. Ethics
The study was conducted in accordance with the approved guidelines of the Declaration of Helsinki. The study protocol was approved by the Ethical Committee of S.Orsola-Malpighi Hospital of Bologna (code 040/2011/U/Tess). Written informed consent was obtained from all study participants or their legal guardians.
2.3. Clinical and Immunologic Evaluation
Clinical data of patients were collected retrospectively, and AD severity was assessed by using the SCORAD index [20]. Atopic sensitization of patients was evaluated by skin prick test (SPT) and by the determination of the total serum IgE and specific IgE. Food allergy was diagnosed through open-label oral food challenge (OFC), following reported guidelines [21], apart from children who had exhibited clinically proven severe food allergy reactions or anaphylaxis episodes.
SPTs (Lofarma, Milan, Italy) were performed for the most common foods (milk and egg) and airborne allergens but also for additional allergens according to the history. The test was considered positive or negative by comparing the allergen wheal with that of histamine. The analysis of sera samples was performed by immune-enzymatic method (ImmunoCAP, ThermoFisher Scientific, Uppsala, Sweden). Total serum IgE levels were defined higher or normal according to the standard values for age, while specific IgE levels were considered positive if higher or equal to 0.35 kU/L for inhalants and 0.70 kU/L for food allergens. Blood samples from patients were collected for the genetic analysis.
2.4. FLG Genotyping
Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Mini kit (Qiagen, Hilden, Germany). The loci of interest of the FLG gene were selected on the basis of the allelic frequency of the loss-of-function mutation reported on the ExAc database [22] and from literature. The analyzed loci were: R501X and 2282del4 (in the first repeat, RPT1), 3321delA (RPT2), R1798X (RPT5), S2554X and R2447X (RPT7), S3247X (RPT9), E3429X, E3603X and R3638X (RPT10) (Figure 1). Target regions were amplified by PCR by means of the AmpliTaq Gold (Thermo Fisher, Waltham, MA, USA). Primers were designed with the Primer Express software v 3.0 (Thermo Fisher, Waltham, MA, USA) in the regions with the highest number of mismatches among the different RPTs of the third exon of the gene and the specificity was verified using Primer Blast [23]. The amplified FLG fragments were sequenced by Sanger sequencing procedure with the BigDye Terminator v1.1 Cycle Sequencing kit (Thermo Fisher, Waltham, MA, USA) and loaded on the ABI 3730 DNA Analyzer (Thermo Fisher, Waltham, MA, USA). Primer sequences are available in Supplementary Table S1.
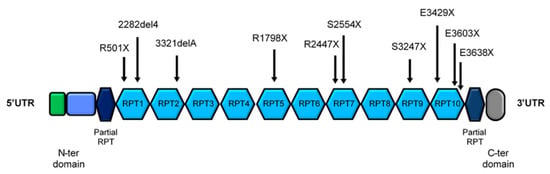
Figure 1.
Structure of the coding portion of FLG gene. The main part of the protein is encoded by the third and larger exon, having a highly complex and repetitive structure with 10–12 near-identical tandem repeats (RPT). The analyzed loss-of-function variants of the FLG gene are reported.
2.5. Diagnostic Criteria and Outcome Variables
An indicator variable was built to classify FLG mutation carriers and wild-type children, with no distinction between the different kinds of detected mutations since they all produce the premature stop of protein translation. Demographic and clinical data included: sex, presence/absence of an atopic condition in at least one of the patients’ parents, age at AD onset, age at the last clinical evaluation, presence/absence of superinfection at diagnosis time, AD subtype, total IgE serum level, presence/absence of asthma, RC and food allergy.
The presence of a family history of atopy was evaluated considering the diagnosis performed by a physician of at least one of the following conditions: RC, allergic asthma, food allergy, IgE-mediated atopic dermatitis.
Regarding AD, disease severity was defined according to the SCORAD (Scoring Atopic Dermatitis) value, with children with a score <15 classified as affected with a mild form, between 15 and 40 with a medium intensity, and with a score >40 with a severe AD manifestation. Moreover, children with a total serum IgE level ≥100 kU/L or specific IgE value ≥0.35 kU/L were classified as affected with an IgE-associated form.
Concerning food allergy, the presence of FA was clinically evaluated and assessed through open-label oral food challenge (OFC), following reported guidelines [21]. Clinically proven severe food allergy reactions or anaphylaxis episodes were assessed through EAACI (European Academy of Allergy and Clinical Immunology) guidelines, and food-allergic children were stratified into two groups considering symptom severity: mild/moderate and severe FA.22 the last category included children who previously exhibited anaphylaxis episodes and/or required the prescription of an adrenaline auto-injector.
Data on the presence of clinically proven episodes of allergy against specific foods (hazelnut, peanut, egg, cow’s milk, kiwi, fish) and on specific serum IgE level for the tested food allergens (wheat, peanut, hazelnut, albumen, kiwi, milk, cod fish) were also recorded for the food allergy-positive children.
2.6. Power Analysis
In Europe, the overall prevalence of clinical symptoms of food allergy confirmed by the detection of positive specific IgE is 3.6% among children [24]. Sample dimension was estimated assuming a frequency of the predisposing allele of 0.12, a food allergy prevalence in childhood of 3%, an allele relative risk = 3 and a type I error rate of 0.05. Under these conditions, we obtained an 80% power to detect an association with 27 cases and 211 controls. The estimates were calculated using the Genetic Power Calculator tool [25].
2.7. Statistical Analysis
Statistical analysis was performed through the R software (version 3.1.2).
Correlation between FLG mutations and categorical variables was assessed through the chi-square test of independence or the Fisher’s exact test (when dealing with expected frequencies <5). Considering continuous variables, the association with the presence/absence of the FLG mutation was analyzed using the non-parametric Wilcoxon–Mann–Whitney test. For each variable, a multivariate logistic regression model was estimated, where the stepwise procedure was used for variable selection. P-values ≤ 0.05 were considered statistically significant. When multiple comparisons were performed, the risk was expressed as an odds ratio (OR) and Bonferroni or Benjamini and Hochberg methods were adopted in order to correct the p-values for multiple tests.
3. Results
3.1. Population Description
The database consisted of 238 patients affected with AD, 135 males and 103 females, with a M/F ratio of 1.31. At the enrollment time, the population mean age was 8.2 years (SD: 5.1 years), while at the last clinical evaluation, the mean age was 13.2 years (SD: 4.9 years). The average age at AD onset was equal to 11.6 months (SD: 19.1 months). Clinical features of the population are reported in Table 1.

Table 1.
Clinical features of the analyzed population (n = 238). Data are expressed as number of patients and percentage. The total IgE level variable is reported as the geometric mean with 95% CI.
3.2. FLG Mutation Frequency
Filaggrin sequencing was performed on 10 recurrent loss-of-function mutation sites: R501X, 2282del4, 3321delA, R1798X, S2554X, R2447X, S3247X, E3429X, E3603X and R3638X (Figure 1), including the most common FLG variants analyzed in all the European studies. In the examined cohort of patients, as expected from previous studies, loss-of-function variants were found only in the first repeat (RPT) of the FLG gene (R501X and 2282del4). The R501X mutation was present in 16 children, all in heterozygosis except from one carrier of a homozygous mutation. The 2282del4 heterozygous variant was identified in 15 individuals. One patient carried both the loss-of-function FLG variants in the RPT1 (Table 2). As a consequence, the total number of FLG mutation carriers was 30, representing 12.2% of the studied population. The combined allelic frequency was equal to 6.5% (3.6% for R501X and 3.2% for 2282del4).

Table 2.
Genotype of the RPT1 FLG loci in the 238 patients of the study. One patient was a carrier of both identified RPT1 FLG variants in heterozygosis.
3.3. FLG Mutation Is not Associated with Asthma and RC
Concerning AD general characteristics, no association with the presence of the FLG mutation was detected for sex, age of AD onset, symptoms intensity and family history of atopy (Supplementary Table S2). Since the age of children in the two groups could lead to a selection bias, we confirmed that the age at the last clinical follow-up was not significantly different between the FLG mutant and FLG wt (12.8 ± 5.3 vs. 13.4 ± 4.9 months; p-value = 0.54).
Regarding the distribution of FLG variant carriers and AD subtype, 13.4% of the IgE-associated AD patients were carriers of FLG variants, while 9.7% of the children with the non-IgE-associated AD form were FLG mutated, a difference which is not statistically significant (Supplementary Table S2).
The association between the presence of FLG mutation and allergic diseases showed no correlation with asthma and RC. By contrast, FA was found associated with the FLG loss-of-function variants (p-value= 0.007). Children with the FLG variant suffering from FA were 17 (56.6% of the mutated population), seven of them with mild symptoms and 10 (59%) with a severe manifestation. In the wild-type population, children exhibiting allergic reactions to foods were 78 (32.8% of the non-mutated group), 71% with mild symptoms and 29% with a more serious form (Supplementary Table S2).
3.4. FLG Mutation Is Associated with Severe FA
In order to identify the variables independently associated with FLG mutations, a multivariate logistic regression model was estimated, which showed that the only variables significantly associated with the FLG mutation were severe FA and total IgE levels (Table 3 and Table 4).

Table 3.
Multivariate model results (n = 228). Multivariate model including all the considered covariates obtained from the first step of the selection model procedure.

Table 4.
Multivariate model results, final model (n = 228). Only the significant variables emerging from the previous step of the model were considered. “Severe FA” and “Total IgE” are the only variables associated with FLG mutations.
Only the association between the subgroup of severe food-allergic patients and the presence of FLG mutations proved to be significant (OR = 8.94, CI: 3.02–28.27), suggesting that people with a RPT1 FLG mutation have a nine-times increased risk of developing severe FA. Conversely, no statistical evidence of an increased risk of developing mild/moderate FA was identified for FLG-mutated patients (p-value = 0.10). If the analysis was carried out without FA stratification into mild and severe categories, the multivariate analysis showed a smaller but significant OR (3.74, CI: 1.56–9.71), implying that the increased risk observed for the entire FA category was driven by the severe FA subgroup.
Regarding the total IgE variable, there was a small association with FLG mutations with a weakly positive OR (1.02, CI: 1.01–1.03), implying that carrying FLG mutation has a slightly protective effect on the total IgE level increase among children with DA.
3.5. FLG Mutation Carriers Have an Increased Risk of Peanut and Hazelnut Allergy
Since FA seemed to be the most relevant variable associated with the FLG mutation, a deeper analysis on specific food sensitization and on allergic reactions was performed in children with and without FLG variants. Since specific IgE antibodies are very sensitive to cross-reaction with other kind of allergens, often not belonging to the food category, only the dataset of clinically proven FA patients (n = 95) was used for this analysis, thus increasing the confidence that the considered food IgE antibodies were specific for the tested allergens.
The levels of specific IgE antibodies to selected food allergens proved to be higher in the FLG-mutated compared to the wild-type population both in the case of peanut and hazelnut (Figure 2), even if the difference reached statistical significance only for hazelnut (p-value = 0.03).
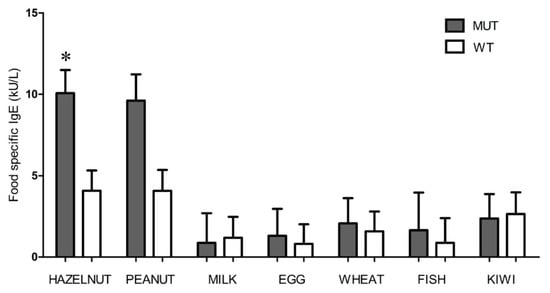
Figure 2.
Food specific IgE levels in the FLG mutated (grey) and wild-type (white) groups. Geometric IgE mean values with standard error are reported. Hazelnut (*, p-value = 0.03) is the only variable with a statistically significant difference between the FLG mutated and wild-type groups according to the multivariate model.
The association analysis between the FLG genotype and food allergy reactions to specific food allergens was performed again only on clinically proven food-allergic patients (Table 5). The association between each allergen and FLG mutations was firstly evaluated using the Chi square test, showing that a significant association was present only in the case of peanut (p-value = 0.05) and hazelnut (p-value = 0.04). No significant association with FLG mutations was found for the other considered food allergens (cow’s milk, egg, fish and kiwi). To prevent multicollinearity, since in the model estimation hazelnut and peanut resulted strongly correlated (p-value = 1.7 × 10 − 6, Fisher’s exact test), the regression model was performed once considering all the variables excluding hazelnut and a second time excluding peanut instead of hazelnut. In the first model, only the peanut coefficient emerged as statistically significant (OR = 3.2, CI: 1.1–9.3 and p-value = 0.05), while in the second one, a significant association was found for hazelnut (OR = 4.1, CI: 1.5–13.0 and (p-value = 0.03), proving that carriers of a RPT1 FLG mutation have a four-times increased risk of developing hazelnut allergy and a three-times increased risk of showing peanut allergy.

Table 5.
Association between FLG mutations and allergic reactions to different foods. Number of patients, OR, 95% CI and p-value of the univariate and multivariate analysis in the subgroup of FLG-mutated and wild-type individuals. n = 95 (for all the variables there are some missing values).
4. Discussion
In the European population, the prevalence of FLG gene alterations, especially of R501X and 2282del4 mutations, varies among different regions, depending on the ethnic groups [3,4,5,26,27,28,29,30,31,32].
In our study, 12.2% of patients (30 out of 238) are carriers of FLG mutations with an allelic frequency of 6.5% (3.6% for the R501X mutation and 3.2% for 2282del4 variant). This result is in line with the European general tendency even if not coherent with another study performed on an Italian population [28] showing only 1.4% of combined allelic frequency. The heterogeneity in the reported FLG mutation frequencies could be explained by the wide spectrum of clinical features of the population under study (i.e., percentage of IgE phenotypes, different grade of severity), thus resulting in a scattered mutation distribution. Moreover, the heterogeneity of inclusion criteria and the type of sequencing approach may influence the percentage of FLG mutation carriers.
Previously published papers [3,4,27] showed that FLG variants represent a risk factor for asthma and RC development. According to our data, asthma and RC are not associated with FLG variants; however, we cannot exclude that in the coming years, given the average age of these patients (13.2 years), some of them will develop asthma and that a significant relationship with asthma occurrence can be uncovered. On the contrary, we can indeed exclude AD as a possible confounding factor, since all the patients included in this study are affected by AD.
The most interesting result of this study is the strong association between severe FA and FLG variants. We show that children carrying the R501X or 2282del4 mutations suffer from severe FA almost nine times more than children with wild-type FLG. Several reports have already described a correlation between FLG variants and FA [10,33,34,35,36], without considering the severity of the allergic reaction. Interestingly, FLG RPT1 mutations were identified as predisposing to food allergy also in a genome-wide association study [37], with an OR similar to the one identified in our study when correlating with FA as a whole, and not specifically with severe FA. Moreover, a recent study confirmed the correlation between low FLG expression, trans-epidermal water loss and FA in AD children, confirming the same percentage of FLG-mutation carriers in AD children with FA (24% vs. 18% in our series) [38]. More importantly, this study confirmed the role of skin barrier dysfunction and food allergy, while our study, beyond confirming association with FA identified in previous studies, shows as a new finding the correlation of FLG mutations with disease severity. Indeed, this is the first work analyzing, in a pediatric population with a predominant IgE-associated phenotype, the link between FLG mutations and FA focusing on the intensity of the allergic conditions, that shows that FLG mutation carriers are a high-risk population for severe allergic reactions as anaphylaxis.
In our study, foods identified as causative of allergic reactions associated with FLG variants are peanut and hazelnut (also confirmed by specific IgE sensitization). Other epidemiological and clinical studies [39,40,41,42] reported the connection between FLG mutation and peanut allergy, while the hazelnut appeared for the first time in this study. Notably, hazelnut and peanut are highly allergenic foods and they are very common allergens among dried fruit in Italy, so it is reasonable that the severity of FA could be at least partly related to their effect and frequency of use.
The linkage between allergic sensitization and FLG mutations is supposed to be represented by the skin barrier defect. An inadequate protection against the external environment allows the contact of environmental allergens, such as food proteins, with the immune system via antigen-presenting cells in the superficial epidermis, leading to sensitization, which could worsen AD symptoms and may also be a precursor condition of food allergies. In these terms, an early identification of the FLG mutations in AD children could be important to direct primary and secondary prevention efforts to these allergy-prone children. Taking into account that the onset age of AD does not seem to be associated with the presence of FLG mutations, the genetic identification of FLG-mutated children could facilitate the implementation of efforts aimed primarily at improving skin barrier function in a high-risk subset of AD children [43]. Nevertheless, the study also shows some limitations, particularly related to the reduced sample size that could limit the power to detect some associations, and to the small number of patients with severe AD included in the cohort. Anyway, these results strongly suggest that the association between severe food allergy and FLG mutations warrants further investigations.
In conclusion, this study demonstrated that in the Italian population there is a considerable percentage of children affected by AD carrying an FLG mutation (12.2%) and that these loss-of-function mutations are a relevant risk factor for severe FA manifestations. The knowledge of the FLG genotype of children with AD could be useful in order to properly treat the disease and all its related clinical manifestations and provide the best preventive measures for this high-risk population.
Supplementary Materials
The following are available online at https://www.mdpi.com/2077-0383/10/2/233/s1. Table S1: List of PCR primers; Table S2: Univariate analysis results.
Author Contributions
Conceptualization, F.C., A.A., G.R. and A.P. (Andrea Pession); methodology, F.C., A.A., D.M., C.D.C., L.R., and I.N.; validation, F.C., A.A., D.M., V.I. and A.G.; formal analysis: V.I. and M.D.L.; data curation, F.C., A.A., D.M., A.G. and V.I.; writing—original draft preparation, F.C., A.A., D.M., M.D.L. and G.R.; writing—review and editing, G.R., A.P. (Annalisa Patrizi), A.P. (Andrea Pession), and I.N.; supervision, A.P. (Annalisa Patrizi), G.R. and A.P. (Andrea Pession). All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of S.Orsola-Malpighi Hospital of Bologna (protocol code 040/2011/U/Tess).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy issues.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Leung, D.Y.M. Atopic dermatitis: New insights and opportunities for therapeutic intervention. J. Allergy Clin. Immunol. 2000, 105, 860–876. [Google Scholar] [CrossRef]
- Flohr, C.; Mann, J. New insights into the epidemiology of childhood atopic dermatitis. Allergy 2014, 69, 3–16. [Google Scholar] [CrossRef]
- Palmer, C.N.A.; Irvine, A.D.; Terron-Kwiatkowski, A.; Zhao, Y.; Liao, H.; Lee, S.P.; Goudie, D.R.; Sandilands, A.; Campbell, L.E.; Smith, F.J.D.; et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 2006, 38, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, S.; Illig, T.; Baurecht, H.; Irvine, A.D.; Rodríguez, E.; Diaz-Lacava, A.; Klopp, N.; Wagenpfeil, S.; Zhao, Y.; Liao, H. Loss-of-function variations within the filaggrin gene predispose for atopic dermatitis with allergic sensitizations. J. Allergy Clin. Immunol. 2006, 118, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Barker, J.; Palmer, C.N.; Zhao, Y.; Liao, H.; Hull, P.R.; Lee, S.P.; Allen, M.H.; Meggitt, S.J.; Reynolds, N.J.; Trembath, R.C.; et al. Null Mutations in the Filaggrin Gene (FLG) Determine Major Susceptibility to Early-Onset Atopic Dermatitis that Persists into Adulthood. J. Investig. Derm. 2007, 127, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.F.; Lack, G. Food allergy and anaphylaxis in pediatrics: Update 2010–2012. Pediatr. Allergy Immunol. 2012, 23, 698–706. [Google Scholar] [CrossRef]
- Eller, E.; Kjaer, H.F.; Høst, A.; Andersen, K.E.; Bindslev-Jensen, C. Food allergy and food sensitization in early childhood: Results from the DARC cohort. Allergy 2009, 64, 1023–1029. [Google Scholar] [CrossRef]
- Ricci, G.; Patrizi, A.; Giannetti, A.; Dondi, A.; Bendandi, B.; Masi, M. Does improvement management of atopic dermatitis influence the appearance of respiratory allergic diseases? A follow-up study. Clin. Mol. Allergy 2010, 8, 8. [Google Scholar] [CrossRef]
- Venkataraman, D.; Soto-Ramírez, N.; Kurukulaaratchy, R.J.; Holloway, J.W.; Karmaus, W.; Ewart, S.L.; Arshad, S.H.; Erlewyn-Lajeunesse, M. Filaggrin loss-of-function mutations are associated with food allergy in childhood and adolescence. J. Allergy Clin. Immunol. 2014, 134, 876–882.e4. [Google Scholar] [CrossRef]
- Brough, H.A.; Simpson, A.; Makinson, K.; Hankinson, J.; Brown, S.; Douiri, A.; Belgrave, D.C.; Penagos, M.; Stephens, A.C.; McLean, W.I.; et al. Peanut allergy: Effect of environmental peanut exposure in children with filaggrin loss-of-function mutations. J. Allergy Clin. Immunol. 2014, 134, 867–875.e1. [Google Scholar] [CrossRef]
- Kim, B.E.; Leung, D.Y. Epidermal Barrier in Atopic Dermatitis. Allergy Asthma Immunol. Res. 2012, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Tenn, M.W.; Ellis, A.K. The clinical relevance of filaggrin mutations: Effect on allergic disease. Ann. Allergy Asthma Immunol. 2016, 117, 483–489. [Google Scholar] [CrossRef] [PubMed]
- Skaaby, T.; Husemoen, L.L.N.; Thyssen, J.P.; Meldgaard, M.; Thuesen, B.; Pisinger, C.; Jorgensen, T.; Carlsen, K.; Johansen, J.D.; Menne, T.; et al. Filaggrin loss-of-function mutations and incident cancer: A population-based study. Br. J. Dermatol. 2014, 171, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Andersen, Y.M.; Egeberg, A.; Balslev, E.; Jørgensen, C.; Szecsi, P.B.; Stender, S.; Kaae, J.; Linneberg, A.; Gislason, G.; Skov, L.; et al. Filaggrin loss-of-function mutations, atopic dermatitis and risk of actinic keratosis: Results from two cross-sectional studies. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 1038–1043. [Google Scholar] [CrossRef]
- Lowe, A.; Su, J.C.; Allen, K.; Abramson, M.; Cranswick, N.; Robertson, C.; Forster, D.; Varigos, G.; Hamilton, S.; Kennedy, R.; et al. A randomized trial of a barrier lipid replacement strategy for the prevention of atopic dermatitis and allergic sensitization: The PEBBLES pilot study. Br. J. Dermatol. 2018, 178, e19–e21. [Google Scholar] [CrossRef]
- Ricci, G.; Dondi, A.; Neri, I.; Ricci, L.; Patrizi, A.; Pession, A. Atopic dermatitis phenotypes in childhood. Ital. J. Pediatr. 2014, 40, 46. [Google Scholar] [CrossRef]
- Garmhausen, D.; Hagemann, T.; Bieber, T.; Dimitiou, I.; Fimmers, R.; Diepgen, T.; Novak, N. Characterization of different courses of atopic dermatitis in adolescent and adult patients. Allergy 2013, 68, 498–506. [Google Scholar] [CrossRef]
- Hanifin, J.M.; Rajka, G. Diagnostic Features of Atopic Dermatitis. Acta Derm. Venereol. 1980, 92, 44–47. [Google Scholar]
- Eichenfield, L.F.; Hanifin, J.M.; Luger, T.A.; Stevens, S.R.; Pride, H.B. Consensus conference on pediatric atopic dermatitis. J. Am. Acad. Dermatol. 2003, 49, 1088–1095. [Google Scholar] [CrossRef]
- Stalder, J.-F.; Taïeb, A. Severity Scoring of Atopic Dermatitis: The SCORAD Index. Dermatology 1993, 186, 23–31. [Google Scholar]
- Calvani, M.; Berti, I.; Fiocchi, A.G.; Galli, E.; Giorgio, V.; Martelli, A.; Sopo, S.M.; Panetta, V. Oral food challenge: Safety, adherence to guidelines and predictive value of skin prick testing. Pediatr. Allergy Immunol. 2012, 23, 754–760. [Google Scholar] [CrossRef] [PubMed]
- ExAc Browser. Available online: http://exac.broadinstitute.org/ (accessed on 16 March 2016).
- Primer Blast. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/ (accessed on 20 March 2016).
- Muraro, A.; Werfel, T.; Hoffmann-Sommergruber, K.; A Roberts, G.; Beyer, K.; Bindslev-Jensen, C.; Cardona, V.; Dubois, A.; Dutoit, G.; Eigenmann, P.; et al. EAACI Food Allergy and Anaphylaxis Guidelines: Diagnosis and management of food allergy. Allergy 2014, 69, 1008–1025. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Cherny, S.S.; Sham, P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics 2003, 19, 149–150. [Google Scholar] [CrossRef] [PubMed]
- Marenholz, I.; Nickel, R.; Rüschendorf, F.; Schulz, F.; Esparza-Gordillo, J.; Kerscher, T.; Grüber, C.; Lau, S.; Worm, M.; Keil, T. Filaggrin loss-of-function mutations predispose to phenotypes involved in the atopic march. J. Allergy Clin. Immunol. 2006, 118, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Morar, N.; Cookson, W.O.C.M.; Harper, J.I.; Moffatt, M.F. Filaggrin mutations in children with severe atopic dermatitis. J. Investig. Dermatol. 2007, 127, 1667–1672. [Google Scholar] [CrossRef] [PubMed]
- Giardina, E.; Paolillo, N.; Sinibaldi, C.; Novelli, G. R501X and 2282del4 Filaggrin Mutations Do Not Confer Susceptibility to Psoriasis and Atopic Dermatitis in Italian Patients. Dermatology 2008, 216, 83–84. [Google Scholar] [CrossRef]
- Müller, S.; Marenholz, I.; Lee, Y.-A.; Sengler, C.; Zitnik, S.E.; Griffionen, R.W.; Meglio, P.; Wahn, U.; Nickel, R. Association of Filaggrin loss-of-function-mutations with atopic dermatitis and asthma in the Early Treatment of the Atopic Child (ETAC) population. Pediatr. Allergy Immunol. 2009, 20, 358–361. [Google Scholar]
- Woźniak, M.; Kaczmarek-Skamira, E.; Romańska-Gocka, K.; Czajkowski, R.; Kałużna, L.; Zegarska, B. The prevalence of mutations in the gene encoding filaggrin in the population of Polish patients with atopic dermatitis. Postep. Dermatol. Alergol. 2016, 33, 128–133. [Google Scholar] [CrossRef]
- Filipowska-Grońska, A.; Weryńska-Kalemba, M.; Bozek, A.; Filipowska, B.; Żebracka-Gala, J.; Rusinek, D.; Kula, D.; Jarząb, J. The frequency of polymorphic variants of filaggrin gene and clinical atopic dermatitis. Adv. Dermatol. Allergol. 2016, 1, 37–41. [Google Scholar] [CrossRef]
- González-Tarancón, R.; Sanmartín, R.; Lorente, F.; Salvador-Rupérez, E.; Hernández-Martín, A.; Rello, L.; Puzo, J.; Gilaberte, Y. Prevalence of FLG loss-of-function mutations R501X, 2282del4, and R2447X in Spanish children with atopic dermatitis. Pediatr. Dermatol. 2020, 37, 98–102. [Google Scholar] [CrossRef]
- Van Ginkel, C.D.; Flokstra-de Blok, B.M.J.; Kollen, B.J.; Kukler, J.; Koppelman, G.H.; Dubois, A.E.J. Loss-of-function variants of the filaggrin gene are associated with clinical reactivity to foods. Allergy 2015, 70, 461–464. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.J.; Asai, Y.; Cordell, H.J.; Campbell, L.E.; Zhao, Y.; Liao, H.; Northstone, K.; Henderson, J.; Alizadehfar, R.; Ben-Shoshan, M.; et al. Loss-of-function variants in the filaggrin gene are a significant risk factor for peanut allergy. J. Allergy Clin. Immunol. 2011, 127, 661–667. [Google Scholar] [CrossRef] [PubMed]
- Brough, H.A.; Liu, A.H.; Sicherer, S.H.; Makinson, K.; Douiri, A.; Brown, S.J.; Stephens, A.C.; McLean, W.I.; Turcanu, V.; Wood, R.A.; et al. Atopic dermatitis increases the effect of exposure to peanut antigen in dust on peanut sensitization and likely peanut allergy. J. Allergy Clin. Immunol. 2015, 135, 164–170.e4. [Google Scholar] [CrossRef] [PubMed]
- Hirota, T.; Nakayama, T.; Sato, S.; Yanagida, N.; Matsui, T.; Sugiura, S.; Takaoka, Y.; Hizawa, N.; Fujieda, S.; Miyatake, A.; et al. Association study of childhood food allergy with genome-wide association studies–discovered loci of atopic dermatitis and eosinophilic esophagitis. J. Allergy Clin. Immunol. 2017, 140, 1713–1716. [Google Scholar] [CrossRef][Green Version]
- Marenholz, I.; Grosche, S.; Kalb, B.; Rüschendorf, F.; Blümchen, K.; Schlags, R.; Harandi, N.; Price, M.; Hansen, G.; Seidenberg, J.; et al. Genome-wide association study identifies the SERPINB gene cluster as a susceptibility locus for food allergy. Nat. Commun. 2017, 8, 1056. [Google Scholar] [CrossRef]
- Leung, D.Y.; Calatroni, A.; Zaramela, L.S.; Lebeau, P.K.; Dyjack, N.; Brar, K.; David, G.; Johnson, K.; Leung, S.; Ramirez-Gama, M.; et al. The nonlesional skin surface distinguishes atopic dermatitis with food allergy as a unique endotype. Sci. Transl. Med. 2019, 11, eaav2685. [Google Scholar] [CrossRef]
- Du Toit, G.; Roberts, G.; Sayre, P.H.; Marshall, P.H.; Bahnson, H.T.; Mitchell, H.; Radulovic, S.; Chan, S.; Fox, A.; Turcanu, V.; et al. Identifying infants at high risk of peanut allergy: The Learning Early About Peanut Allergy (LEAP) screening study. J. Allergy Clin. Immunol. 2013, 131, 135–143. [Google Scholar] [CrossRef]
- Strid, J.; Hourihane, J.; Kimber, I.; Callard, R.; Strobel, S. Epicutaneous exposure to peanut protein prevents oral tolerance and enhances allergic sensitization. Clin. Exp. Allergy 2005, 35, 757–766. [Google Scholar] [CrossRef]
- Moghaddam, A.E.; Hillson, W.R.; Noti, M.; Gartlan, K.H.; Johnson, S.; Thomas, B.; Artis, D.; Sattentau, Q.J. Dry roasting enhances peanut-induced allergic sensitization across mucosal and cutaneous routes in mice. J. Allergy Clin. Immunol. 2014, 134, 1453–1456. [Google Scholar] [CrossRef]
- Asai, Y.; Greenwood, C.M.T.; Hull, P.R.; Alizadehfar, R.; Ben-Shoshan, M.; Brown, S.J.; Campbell, L.; Michel, D.; Bussières, J.; Rousseau, F.; et al. Filaggrin gene mutation associations with peanut allergy persist despite variations in peanut allergy diagnostic criteria or asthma status. J. Allergy Clin. Immunol. 2013, 132, 239–242.e7. [Google Scholar] [CrossRef]
- Lowe, A.J.; Leung, D.Y.M.; Tang, M.L.K.; Su, J.C.; Allen, K.J. The skin as a target for prevention of the atopic march. Ann. Allergy Asthma Immunol. 2018, 120, 145–151. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).