Abstract
The most common hereditary disorder in adults, α1-antitrypsin deficiency (AATD), is characterized by reduced plasma levels or the abnormal functioning of α1-antitrypsin (AAT), a major human blood serine protease inhibitor, which is encoded by the SERine Protein INhibitor-A1 (SERPINA1) gene and produced in the liver. Recently, it has been hypothesized that the geographic differences in COVID-19 infection and fatality rates may be partially explained by ethnic differences in SERPINA1 allele frequencies. In our review, we examined epidemiological data on the correlation between the distribution of AATD, SARS-CoV-2 infection, and COVID-19 mortality rates. Moreover, we described shared pathogenetic pathways that may provide a theoretical basis for our epidemiological findings. We also considered the potential use of AAT augmentation therapy in patients with COVID-19.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which originated in Wuhan, China, has resulted in a pandemic, with more than 183 million confirmed cases and approximately 4 million deaths (as of 30 June 2021) [1]. According to the worldwide epidemiologic data, there are remarkably different infection and mortality rates for SARS-CoV-2 between different countries; the numbers of deaths per million of the population in Europe and the Americas were approximately 43 and 21 times higher than those in East Asia, respectively (as of 25 February 2021) [2]. Moreover, some concerns remain regarding the great interindividual differences in the clinical severity of coronavirus disease 2019 (COVID-19), which cannot be completely explained by environmental factors, comorbidities, and age-related fragility [3].
On the basis of these simple observations and the susceptibility of hosts as the main infection component, it could be argued that genetic differences among populations, ethnicities, and individuals may contribute to the different epidemiological and clinical manifestations of COVID-19 [2,4].
Recent studies have investigated genetic susceptibility to SARS-CoV-2 and reported that genetic errors and gene loci are associated with approximately 20% of life-threatening COVID-19 cases, most of which are involved in two immune signaling pathways—i.e., the interferon-mediated antiviral signaling and chemokine-mediated inflammatory signaling pathways [5]. Interestingly, Souilmi et al. showed a strong genomic signature in the East Asian population, likely due to ancient coronavirus epidemics that may have emerged more than 20,000 years ago, involving gene variants encoding virus-interacting proteins (VIPs), which may imply a difference in genetic susceptibility to SARS-CoV-2 infection [6].
In patients with COVID-19, disease severity and high mortality are associated with respiratory failure due to acute respiratory distress syndrome (ARDS) and multiple organ failure due to an uncontrolled cytokine storm, with significantly increased levels of several inflammatory mediators, including IL-6, interferon gamma (INF-γ), Tumor Necrosis Factor alpha (TNF-α), interleukin 17 (IL-17), and IL-8 [3].
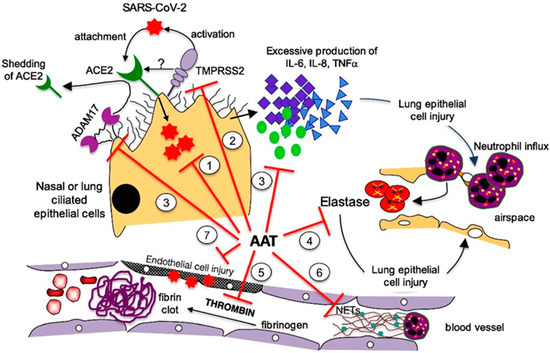
Therefore, we focus on the possible role of α1-antitrypsin deficiency (AATD), an inherited disorder known for nearly 60 years [7], in the vulnerability of some individuals to SARS-CoV-2 infection and severe COVID-19 progression. Indeed, α1-antitrypsin (AAT) inhibits neutrophil elastase (NE), which, when unopposed, can cleave many of the structural proteins of the lung as well as innate immune proteins. The anti inflammatory and tissue-protective properties of AAT are underscored by several studies suggesting its usefulness in transplantation medicine [8].
Moreover, we report epidemiological data on the distribution of both conditions and describe common pathogenic pathways that may support the importance of AAT in antagonizing the SARS-CoV-2 infection.
2. Genetics, Epidemiology, and Clinical Relevance of AATD
AAT, encoded by the SERPINA1 gene, is an acute-phase glycoprotein that is mainly synthesized and secreted by hepatocytes and is the main serum protease inhibitor. However, other cell types can also express this protein. A study by Van’t Wout et al. demonstrated that AAT is produced by differentiated monocyte-derived macrophages, monocyte-derived dendritic cells, and alveolar macrophages [9]. NE, an enzyme responsible for the proteolytic degradation of lung elastase and alveolar and interstitial lung tissue, and AAT, which primarily acts as an inhibitor of the serine protease, act in concert to maintain protease–anti-protease homeostasis, thus protecting the lung tissue from proteolytic digestion [10]. AAT also has anti-coagulation effects and can protect against inflammation, cell death, and the formation of neutrophil extracellular traps (NETs) [11]. Notably, AAT has the capacity to bind and inhibit the SARS-CoV-2-priming protease, transmembrane serine protease 2 (TMPRSS2), on host cells, thus inhibiting the first step in the SARS-CoV-2 cell cycle [12].
Over 120 mutations in the SERPINA1 gene encoding AAT have been identified, of which the most common are the Z and S mutations, which are characterized by the substitution of glutamic acid with lysine or valine at positions 342 and 264 of the polypeptide, respectively, as well as by reduced circulating protein levels [13]. Although AATD has long been considered a rare disease, it is now identified as a prevalent hereditary disease among Caucasian individuals in Northern Europe and immigrants from these countries to the New World. Nevertheless, it remains highly underdiagnosed due to its clinical overlap with other pulmonary disorders, including chronic obstructive pulmonary disease (COPD) and severe asthma [14,15]. The highest prevalence of the PI*Z variant was recorded in Northern and Western European countries (mean gene frequency: 0.0140), peaking in southern Scandinavia, Denmark, the Netherlands, the UK, and northern France (gene frequency > 0.0200) [16]. The PI*SZ genotype typically exhibits an AAT level less than half that of a normal (PI*MM) individual and shows a similar lung disease risk between PI*MM and PI*ZZ patients, given the same level of smoke exposure. Epidemiological studies have estimated that there are over 500,000 PI*SZ individuals in Europe [17].
In a worldwide analysis of data from 514 cohorts (69 countries across 11 geographical areas), De Serres estimated the prevalence of PI*ZZ, PI*SZ, and PI*SS individuals to be 3.4 million [16]. Extrapolating this to the world population in 2015, the prevalence of AATD individuals was estimated to be 5.64 million [18].
Clinically, patients with AATD with a homozygous PI*ZZ genotype have an increased risk of developing COPD (including chronic bronchitis, emphysema, and bronchiectasis) and chronic liver disease, an increased susceptibility to viral infection, and a prolonged course of infection [19,20,21,22]. Several multifactorial elements may contribute to the onset and severity of clinical manifestations, including smoking, alcohol intake, air pollutants, and/or passive smoke exposure [19].
Once COPD has been diagnosed, patients with AATD should receive lifestyle suggestions and pharmacological treatment according to standard recommendations [22]. Moreover, patients should be referred to an expert center, where indications for augmentation therapy should be evaluated. AAT augmentation therapy is a disease-specific treatment aimed at restoring protective levels of the deficient enzyme [10]. In line with the COPD treatment guidelines, pulmonary rehabilitation, lung volume reduction, and lung transplantation should also be considered for the management of patients with AATD at the advanced stage of the disease [23,24].
3. AATD and COVID-19: Geographical Overlap and Data from Clinical Registries
During the COVID-19 pandemic, several epidemiologic studies have reported that the geographical distribution of patients with AATD is similar to that of confirmed cases of COVID-19. Of importance, most studies have methodological limitations, including non-official data sources, unknown sample sizes, and different techniques for ascertaining AAT variants, leading to the potential underestimation of the prevalence of AATD [16].
In April 2020, a geographical overlap between the prevalence of allelic variants of SERPINA1 and severe cases of COVID-19 was reported in Italy [25]. In parallel with the distribution of AAT mutations following a north-to-south gradient, with 47% of the total number registered in the Lombardia region (237 out of 508) [26], an inhomogeneous distribution of COVID-19 cases was reported, with 85% of the total fatal cases registered in northern Italy (51.9% in the Lombardia region in particular) and an abrupt decrease from north to south. The AATD population size is much smaller than the COVID-19 patient population, but it should be pointed out that AATD is largely under-recognized in Italy, with a relationship between diagnosed and expected cases of 1 to 460. In particular, one carrier or deficiency-allele combination phenotype is expected for every 13.6 individuals, with greater frequencies in PI*MS and PI*MZ genotypes [16]. Recent data have confirmed a north-to-south gradient of COVID-19 mortality in Italy, with a decrease from 236–378 deaths/100,000 inhabitants in the northern regions to 65 deaths/100,000 inhabitants in southern Italy (Calabria region) [1].
Two later studies confirmed a correlation between the COVID-19 pandemic and the prevalence of AATD on a global scale. In September 2020, Yoshikura reported that the prevalence of AATD was correlated with the epidemiologic distribution of COVID-19, particularly in countries with a high AATD prevalence, showing an epidemic curve characterized by an initial rise, followed by a prolonged duration of high infection and mortality rates, whereas countries with a low AATD prevalence showed a more rapid decrease in mortality rate after the initial rise [27]. Shapira et al. observed that Pi*Z alleles were 8-fold less frequent in Middle East/Far East countries (2:1000 individuals) than in South European countries (17:1000 individuals). This difference was more remarkable (17-fold) when the Pi*S allele was considered, as the prevalence in Far East countries was 5:1000 compared to 86:1000 in South European countries [28]. In addition, a large difference in mortality rates was found between Middle East/Far East and South European countries, such as Thailand, Vietnam, and Cambodia, accounting for 5, 0.34, and 6.9 deaths per 100,000 individuals, respectively, compared with a mortality rate of 216, 174, and 123 per 100,000 individuals in Italy, Spain, and Greece, respectively. The authors also found a significant correlation (R = 0.54; p = 0.00000198) between the frequency of Pi*Z and Pi*S alleles and mortality rates due to COVID-19 in 67 countries. The correlation was still significant after correcting for confounding factors, including urbanization and age distribution. Although these data are suggestive, the authors underline that additional confounding factors should be considered, including the different control measures established by governments, socioeconomic status, and population health [28].
According to epidemiological data, Bhattacharyya et al. observed that within 3 months of the onset of the COVID-19 pandemic, SARS-CoV-2 evolved into 10 additional subtypes, and it was found that the mutant subtype (614G) outcompeted the pre-existing type (614D) significantly faster in Europe and North America than in East Asia. The G mutant is characterized by a novel NE cleavage site near the S1-S2 junction of the spike protein. The authors hypothesized that the elevation of the NE level at the site of infection might have enhanced the activation of the spike protein, thus facilitating host cell entry for 614G. Therefore, AATD could promote the host cell entry of the 614G virus by retarding the inhibition of NE and consequently enhancing the activation of the spike protein. Provided that AATD is more prevalent in European and North American populations, this could explain why the SARS-CoV-2 subtype 614G spread more easily in these geographical regions than in East Asia [29].
Ferrarotti et al. recently investigated whether patients with severe AATD had an increased risk of severe COVID-19 infection. Therefore, they collected data on COVID-19 symptoms, laboratory-confirmed infection, hospitalization, and treatment directly administered to Italian severe AATD subjects using a telephone survey and compared their findings with data collected by the Istituto Superiore di Sanità on the total population in Italy. The authors found a higher frequency of SARS-CoV-2 infection in the AATD cohort (3.8%) compared to the national data regarding infection, and patients with severe AATD had a relative risk of 8.8 (95% confidence interval (CI): 5.1–20.0; p < 0.0001) for symptomatic SARS-CoV-2 infection. Moreover, the relative risk (RR) was higher in patients with AATD and pre-existing lung diseases (RR 13.9; 95% CI: 8.0–33.6; p < 0.001) [30].
Faria et al. collected data from all patients with AATD at the Pulmonology Department in a tertiary hospital in Porto (Portugal) and compared patients with AATD who were diagnosed with COVID-19 with the remaining AATD cohort based on patients’ pre-infection AAT serum levels, clinical, and functional status. The Pi*ZZ genotype was significantly associated with greater COVID-19 incidence (33.3%, p = 0.012), followed by Pi*MS (14.3%) and Pi*SZ (10.0%). Moreover, the baseline AAT levels were significantly lower in patients with COVID-19 (42.9 ± 12.1 vs. 65.5 ± 2.8 mg/dL; p = 0.012) [31].
Unlike previous findings, Schneider and Strnad recently assessed the association between AATD and COVID-19 in the United Kingdom Biobank, a community-based cohort with >500,000 participants, and showed that the most common and mild AATD genotypes were not associated with increased SARS-CoV-2 infection rates or increased SARS-CoV-2 fatalities, whereas the number of severe AATD cases was too low to allow definitive conclusions to be drawn [32].
In conclusion, most epidemiological observations agree that there is a correlation between the SARS-CoV-2 epidemic, the clinical severity of COVID-19, and the prevalence of AATD.
Results from epidemiologic studies are summarized in Table 1.

Table 1.
Main results from epidemiologic studies on the overlap between α1-antitrypsin deficiency and SARS-CoV-2 infection (NA = not available).
5. Augmentation Therapy for Patients with COVID-19
To date, AAT supplement therapy has been largely used to slow disease progression and reduce exacerbations in patients with AATD and COPD. The multiple AAT activities discussed above have suggested its potential therapeutic use for patients with COVID-19. The biological functions of AAT that may antagonize SARS-CoV-2 infection are outlined in Table 2.

Table 2.
Protective effects of augmentation therapy in SARS-CoV-2 infection.
Three clinical trials have been undertaken to examine the potential benefits of α1-proteinase inhibitors in patients with COVID-19 [75,76,77]; however, results were not available at the time of this review. To date, only sporadic case reports and short case series have been published concerning the possible role of AAT as a treatment option for COVID-19.
AAT was successfully used in a 43-year-old patient with COVID-19 who was affected with cystic fibrosis and placed on a waiting list for lung transplantation. She developed severe ARDS and required mechanical ventilation (MV). Poor prognosis and severe inflammatory phenotype prompted physicians to administer AAT as an off-label therapy at 120 mg/kg/week for four consecutive weeks. Rapid clinical, laboratory, and radiographic improvement was detected, and the patient was liberated from MV and discharged from the hospital [78].
Ritzmann et al. administered inhaled or combined inhaled/intravenous AAT in nine patients with mild-to-moderate COVID-19. After the first AAT administration, the C-reactive protein (CRP) levels abruptly decreased. All patients were discharged from the hospital approximately 20 days after admission. Three out of nine patients in a matched control group died and CRP decrease was delayed [79].
Finally, Martini et al. indicated that AAT treatment via aerosol by humid heat vaporization (40–41 °C) should be considered in the early phase of COVID-19 to hamper SARS-CoV-2 entry into the host cells [80].
When advocating for their use in patients with COVID-19, it should be emphasized that AAT products have been marketed for many decades and found to be safe. Moreover, treatment protocols and schedules are largely available.
Based on multiple early reports of patients admitted to the hospital with COVID-19 showing that subjects with chronic obstructive respiratory disease were significantly under-represented in these cohorts, therapeutic options commonly used in patients with AATD, including bronchodilators (long-acting muscarinic antagonist, long-acting beta-2 agonists, etc.) and inhaled corticosteroids, have been tested aiming to establish whether they would be an effective treatment for early COVID-19. In this regard, a study by Ramakrishnan et al. showed that the early administration of inhaled budesonide can reduce the likelihood of needing urgent medical care and decrease time to recovery after early COVID-19 [81].
6. Conclusions
As AATD represents a large population in geographic areas with a high incidence of COVID-19 and high mortality associated with the disease, understanding the interrelation between these conditions is essential in order to reduce COVID-19-related morbidity and mortality. The potential beneficial effects of AAT make it an ideal candidate for COVID-19 treatment; however, in vivo studies with animal models and clinical trials are needed to validate its large-scale utilization.
Funding
Publishing support and journal styling services were provided by SEEd Medical Publishers and funded by CSL Behring, Italy. APC was funded by the CSL Behring, Italy. CSL Behring had no role in the conduction of the research; the preparation of the article; the study design; the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
All authors declare that they do not have any potential or actual conflicts of interest, including financial or personal relationships with people or organizations that could have inappropriately influenced (biased) the study, except for the publishing support and journal styling services, which were provided by SEEd Medical Publishers and funded by CSL Behring, Italy. No study sponsor was involved in designing or conceptualizing the study; collecting, analyzing, and interpreting the data; drafting the manuscript; or in the decision to submit the manuscript for publication.
References
- World Health Organization. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 21 September 2021).
- Yamamoto, N.; Yamamoto, R.; Ariumi, Y.; Mizokami, M.; Shimotohno, K.; Yoshikura, H. Does genetic predisposition contribute to the exacerbation of COVID-19 symptoms in individuals with comorbidities and explain the huge mortality disparity between the East and the West? Int. J. Mol. Sci. 2021, 22, 5000. [Google Scholar] [CrossRef] [PubMed]
- McElvaney, O.J.; McEvoy, N.L.; McElvaney, O.F.; Carroll, T.P.; Murphy, M.P.; Dunlea, D.M.; Ni Choileain, O.; Clarke, J.; O’Connor, E.; Hogan, G.; et al. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020, 202, 812–821. [Google Scholar] [CrossRef] [PubMed]
- Van Der Made, C.I.; Simons, A.; Schuurs-Hoeijmakers, J.; Heuvel, G.V.D.; Mantere, T.; Kersten, S.; Van Deuren, R.C.; Steehouwer, M.; Van Reijmersdal, S.V.; Jaeger, M.; et al. Presence of Genetic Variants Among Young Men with Severe COVID-19. JAMA 2020, 324, 663. [Google Scholar] [CrossRef]
- McCoy, K.; Peterson, A.; Tian, Y.; Sang, Y. Immunogenetic Association Underlying Severe COVID-19. Vaccines 2020, 8, 700. [Google Scholar] [CrossRef] [PubMed]
- Souilmi, Y.; Lauterbur, M.E.; Tobler, R.; Huber, C.D.; Johar, A.S.; Moradi, S.V.; Johnston, W.A.; Krogan, N.J.; Alexandrov, K.; Enard, D. An ancient viral epidemic involving host coronavirus interacting genes more than 20,000 years ago in East Asia. Curr. Biol. 2021, 31, 3504–3514.e9. [Google Scholar] [CrossRef]
- Laurell, C.-B.; Eriksson, S. The Electrophoretic α1-Globulin Pattern of Serum in α1-Antitrypsin Deficiency. COPD J. Chronic Obstr. Pulm. Dis. 2013, 10, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Strnad, P.; McElvaney, N.G.; Lomas, D.A. Alpha1-Antitrypsin Deficiency. N. Engl. J. Med. 2020, 382, 1443–1455. [Google Scholar] [CrossRef]
- Wout, E.F.A.V.; Van Schadewijk, A.; Savage, N.D.L.; Stolk, J.; Hiemstra, P.S. α1-Antitrypsin Production by Proinflammatory and Antiinflammatory Macrophages and Dendritic Cells. Am. J. Respir. Cell Mol. Biol. 2012, 46, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Ferrarotti, I.; Ottaviani, S.; De Silvestri, A.; Corsico, A.G. Update on α1-antitrypsin deficiency. Breathe 2018, 14, e17–e24. [Google Scholar] [CrossRef]
- Yang, C.; Keshavjee, S.; Liu, M. Alpha-1 Antitrypsin for COVID-19 Treatment: Dual Role in Antiviral Infection and Anti-Inflammation. Front. Pharmacol. 2020, 11, 615398. [Google Scholar] [CrossRef]
- Azouz, N.P.; Klingler, A.M.; Callahan, V.; Akhrymuk, I.V.; Elez, K.; Raich, L.; Henry, B.M.; Benoit, J.L.; Benoit, S.W.; Noé, F.; et al. Alpha 1 Antitrypsin Is an Inhibitor of the SARS-CoV-2–Priming Protease TMPRSS2. Pathog. Immun. 2021, 6, 55–74. [Google Scholar]
- Hatipoğlu, U.; Stoller, J.K. α1-Antitrypsin Deficiency. Clin. Chest Med. 2016, 37, 487–504. [Google Scholar] [CrossRef]
- Miravitlles, M.; Dirksen, A.; Ferrarotti, I.; Koblizek, V.; Lange, P.; Mahadeva, R.; McElvaney, N.G.; Parr, D.; Piitulainen, E.; Roche, N.; et al. European Respiratory Society statement: Diagnosis and treatment of pulmonary disease in α1-antitrypsin deficiency. Eur. Respir. J. 2017, 50, 1700610. [Google Scholar] [CrossRef] [PubMed]
- Vianello, A.; Caminati, M.; Senna, G.; Arcolaci, A.; Chieco-Bianchi, F.; Ferrarotti, I.; Guarnieri, G.; Molena, B.; Crisafulli, E. Effect of α1 antitrypsin deficiency on lung volume decline in severe asthmatic patients undergoing biologic therapy. J. Allergy Clin. Immunol. Pr. 2021, 9, 1414–1416. [Google Scholar] [CrossRef] [PubMed]
- Luisetti, M. 1-Antitrypsin deficiency 1: Epidemiology of 1-antitrypsin deficiency. Thorax 2004, 59, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Green, C.E.; Vayalapra, S.; Hampson, J.A.; Mukherjee, D.; Stockley, R.A.; Turner, A. PiSZ alpha-1 antitrypsin deficiency (AATD): Pulmonary phenotype and prognosis relative to PiZZ AATD and PiMM COPD. Thorax 2015, 70, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Blanco, I.; Bueno, P.; Diego, I.; Pérez-Holanda, S.; Lara, B.; Casas, F.; Esquinas, C.; Miravitlles, M. Alpha-1 antitrypsin Pi*SZ genotype: Estimated prevalence and number of SZ subjects worldwide. Int. J. Chronic Obstr. Pulm. Dis. 2017, 12, 1683–1694. [Google Scholar] [CrossRef] [PubMed]
- De Serres, F.; Blanco, I.; Fernández-Bustillo, E. Genetic epidemiology of alpha-1 antitrypsin deficiency in southern Europe: France, Italy, Portugal and Spain. Clin. Genet. 2003, 63, 490–509. [Google Scholar] [CrossRef]
- O’Brien, M.E.; Fee, L.; Browne, N.; Carroll, T.P.; Meleady, P.; Henry, M.; McQuillan, K.; Murphy, M.P.; Logan, M.; McCarthy, C.; et al. Activation of complement component 3 is associated with airways disease and pulmonary emphysema in alpha-1 antitrypsin deficiency. Thorax 2020, 75, 321–330. [Google Scholar] [CrossRef]
- Fischer, H.-P.; Ortiz-Pallardó, M.; Ko, Y.; Esch, C.; Zhou, H. Chronic liver disease in heterozygous α1-antitrypsin deficiency PiZ. J. Hepatol. 2000, 33, 883–892. [Google Scholar] [CrossRef]
- Hill, A.T.; Campbell, E.J.; Bayley, D.L.; Hill, S.L.; Stockley, R.A. Evidence for Excessive Bronchial Inflammation during an Acute Exacerbation of Chronic Obstructive Pulmonary Disease in Patients with α1-Antitrypsin Deficiency (PiZ). Am. J. Respir. Crit. Care Med. 1999, 160, 1968–1975. [Google Scholar] [CrossRef]
- Vogelmeier, C.F.; Criner, G.J.; Martinez, F.J.; Anzueto, A.; Barnes, P.J.; Bourbeau, J.; Celli, B.R.; Chen, R.; Decramer, M.; Fabbri, L.M.; et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease 2017 Report: GOLD Executive Summary. Eur. Respir. J. 2017, 49, 1700214. [Google Scholar] [CrossRef] [PubMed]
- Gulack, B.C.; Mulvihill, M.S.; Ganapathi, A.; Speicher, P.J.; Chery, G.; Snyder, L.; Davis, R.D.; Hartwig, M.G. Survival after lung transplantation in recipients with alpha-1-antitrypsin deficiency compared to other forms of chronic obstructive pulmonary disease: A national cohort study. Transpl. Int. 2017, 31, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Vianello, A.; Braccioni, F. Geographical Overlap Between Alpha-1 Antitrypsin Deficiency and COVID-19 Infection in Italy: Casual or Causal? Arch. Bronconeumol. 2020, 56, 609–610. [Google Scholar] [CrossRef]
- Italian Registry of Patients with Alpha-1 Antitrypsin Deficiency. Available online: http://alfa1antitripsina.it/it (accessed on 31 August 2021).
- Yoshikura, H. Epidemiological correlation between COVID-19 epidemic and prevalence of α-1 antitrypsin deficiency in the world. Glob. Health Med. 2021, 3, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Shapira, G.; Shomron, N.; Gurwitz, D. Ethnic differences in alpha-1 antitrypsin deficiency allele frequencies may partially explain national differences in COVID-19 fatality rates. FASEB J. 2020, 34, 14160–14165. [Google Scholar] [CrossRef]
- Bhattacharyya, C.; Das, C.; Ghosh, A.; Singh, A.K.; Mukherjee, S.; Majumder, P.P.; Basu, A.; Biswas, N.K. SARS-CoV-2 mutation 614G creates an elastase cleavage site enhancing its spread in high AAT-deficient regions. Infect. Genet. Evol. 2021, 90, 104760. [Google Scholar] [CrossRef]
- Ferrarotti, I.; Ottaviani, S.; Balderacchi, A.; Barzon, V.; De Silvestri, A.; Piloni, D.; Mariani, F.; Corsico, A. COVID-19 infection in severe Alpha 1-antitrypsin deficiency: Looking for a rationale. Respir. Med. 2021, 183, 106440. [Google Scholar] [CrossRef]
- Faria, N.; Costa, M.I.; Gomes, J.; Sucena, M. Alpha-1 antitrypsin deficiency severity and the risk of COVID-19: A Portuguese cohort. Respir. Med. 2021, 181, 106387. [Google Scholar] [CrossRef]
- Schneider, C.V.; Strnad, P. SARS-CoV-2 Infection in Alpha1-Antitrypsin Deficiency. Respir. Med. 2021, 184, 106466. [Google Scholar] [CrossRef]
- Bai, X.; Hippensteel, J.; Leavitt, A.; Maloney, J.P.; Beckham, D.; Garcia, C.; Li, Q.; Freed, B.M.; Ordway, D.; Sandhaus, R.A.; et al. Hypothesis: Alpha-1-antitrypsin is a promising treatment option for COVID-19. Med. Hypotheses 2020, 146, 110394. [Google Scholar] [CrossRef]
- Harbig, A.; Mernberger, M.; Bittel, L.; Pleschka, S.; Schughart, K.; Steinmetzer, T.; Stiewe, T.; Nist, A.; Böttcher-Friebertshäuser, E. Transcriptome profiling and protease inhibition experiments identify proteases that activate H3N2 influenza A and influenza B viruses in murine airways. J. Biol. Chem. 2020, 295, 11388–11407. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, Z.; Zhang, J.; Adelsberger, J.W.; Yang, J.; Burton, G.F. Alpha-1-antitrypsin interacts with gp41 to block HIV-1 entry into CD4+ T lymphocytes. BMC Microbiol. 2016, 16, 172. [Google Scholar] [CrossRef]
- Bryan, C.L.; Beard, K.S.; Pott, G.B.; Rahkola, J.; Gardner, E.M.; Janoff, E.N.; Shapiro, L. HIV infection is associated with reduced serum alpha-1-antitrypsin concentrations. Clin. Investig. Med. 2010, 33, E384–E389. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Shapiro, L.; Fellingham, G.; Willardson, B.M.; Burton, G.F. HIV Replication in CD4+ T Lymphocytes in the Presence and Absence of Follicular Dendritic Cells: Inhibition of Replication Mediated by α-1-Antitrypsin through Altered IκBα Ubiquitination. J. Immunol. 2011, 186, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Whitney, J.B.; Asmal, M.; Geiben-Lynn, R. Serpin Induced Antiviral Activity of Prostaglandin Synthetase-2 against HIV-1 Replication. PLoS ONE 2011, 6, e18589. [Google Scholar] [CrossRef]
- Shapiro, L.; Pott, G.B.; Ralston, A.H. Alpha-1-antitrypsin inhibits human immunodeficiency virus type 1. FASEB J. 2000, 15, 115–122. [Google Scholar] [CrossRef]
- Pott, G.B.; Beard, K.S.; Bryan, C.L.; Merrick, D.T.; Shapiro, L. Alpha-1 Antitrypsin Reduces Severity of Pseudomonas Pneumonia in Mice and Inhibits Epithelial Barrier Disruption and Pseudomonas Invasion of Respiratory Epithelial Cells. Front. Public Health 2013, 1, 19. [Google Scholar] [CrossRef]
- Bai, X.; Bai, A.; Honda, J.R.; Eichstaedt, C.; Musheyev, A.; Feng, Z.; Huitt, G.; Harbeck, R.; Kosmider, B.; Sandhaus, R.A.; et al. Alpha-1-Antitrypsin Enhances Primary Human Macrophage Immunity Against Non-tuberculous Mycobacteria. Front. Immunol. 2019, 10, 1417. [Google Scholar] [CrossRef]
- Gassen, N.C.; Niemeyer, D.; Muth, D.; Corman, V.M.; Martinelli, S.; Gassen, A.; Hafner, K.; Papies, J.; Mösbauer, K.; Zellner, A.; et al. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef]
- Peng, R.; Wu, L.-A.; Wang, Q.; Qi, J.; Gao, G.F. Cell Entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Prior, P. Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. J. Biol. Chem. 2021, 296, 100135. [Google Scholar] [CrossRef] [PubMed]
- De Loyola, M.B.; dos Reis, T.T.A.; de Oliveira, G.X.L.M.; Palmeira, J.; Argañaraz, G.A.; Argañaraz, E.R. Alpha-1-antitrypsin: A possible host protective factor against Covid-19. Rev. Med. Virol. 2020, 31, e2157. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Kawase, M.; Shirato, K.; van der Hoek, L.; Taguchi, F.; Matsuyama, S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. J. Virol. 2012, 86, 6537–6545. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. USA 2020, 117, 7001–7003. [Google Scholar] [CrossRef] [PubMed]
- Wettstein, L.; Conzelmann, C.; Müller, J.; Weil, T.; Groß, R.; Hirschenberger, M.; Seidel, A.; Klute, S.; Prelli Bozzo, C.; Zech, F.; et al. Alpha-1 Antitrypsin Inhibits SARS-CoV-2 Infection. bioRxiv 2020. [Google Scholar] [CrossRef]
- Oguntuyo, K.Y.; Stevens, C.S.; Siddiquey, M.N.; Schilke, R.M.; Woolard, M.D.; Zhang, H.; Acklin, J.A.; Ikegame, S.; Hung, C.-T.; Lim, J.K.; et al. In Plain Sight: The Role of Alpha-1-Antitrypsin in COVID-19 Pathogenesis and Therapeutics. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chan, E.D.; Pott, G.B.; Silkoff, P.E.; Ralston, A.H.; Bryan, C.L.; Shapiro, L. Alpha-1-antitrypsin inhibits nitric oxide production. J. Leukoc. Biol. 2012, 92, 1251–1260. [Google Scholar] [CrossRef]
- Ehlers, M.R. Immune-modulating effects of alpha-1 antitrypsin. Biol. Chem. 2014, 395, 1187–1193. [Google Scholar] [CrossRef]
- Bergin, D.A.; Reeves, E.P.; Meleady, P.; Henry, M.; McElvaney, O.J.; Carroll, T.; Condron, C.; Chotirmall, S.H.; Clynes, M.; O’Neill, S.J.; et al. α-1 Antitrypsin regulates human neutrophil chemotaxis induced by soluble immune complexes and IL-8. J. Clin. Investig. 2010, 120, 4236–4250. [Google Scholar] [CrossRef]
- Bai, D.; Ueno, L.; Vogt, P.K. Akt-mediated regulation of NFκB and the essentialness of NFκB for the oncogenicity of PI3K and Akt. Int. J. Cancer 2009, 125, 2863–2870. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Sun, K.; Guo, S.; Wang, J.; Li, A.; Rong, X.; Wang, T.; Shang, Y.; Chang, W.; Wang, S. Early Warning Indicators of Severe COVID-19: A Single-Center Study of Cases from Shanghai, China. Front. Med. 2020, 7, 432. [Google Scholar] [CrossRef] [PubMed]
- Bucurenci, N.; Blake, D.R.; Chidwick, K.; Winyard, P. Inhibition of neutrophil superoxide production by human plasma α1-antitrypsin. FEBS Lett. 1992, 300, 21–24. [Google Scholar] [CrossRef]
- Wang, J.-Z.; Zhang, R.-Y.; Bai, J. An anti-oxidative therapy for ameliorating cardiac injuries of critically ill COVID-19-infected patients. Int. J. Cardiol. 2020, 312, 137–138. [Google Scholar] [CrossRef]
- Heurich, A.; Hofmann-Winkler, H.; Gierer, S.; Liepold, T.; Jahn, O.; Pöhlmann, S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J. Virol. 2013, 88, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Iwai, M.; Horiuchi, M. Devil and angel in the renin–angiotensin system: ACE–angiotensin II–AT1 receptor axis vs. ACE2–angiotensin-(1–7)–Mas receptor axis. Hypertens. Res. 2009, 32, 533–536. [Google Scholar] [CrossRef]
- Belaaouaj, A.; McCarthy, R.; Baumann, M.; Gao, Z.; Ley, T.J.; Abraham, S.N.; Shapiro, S.D. Mice lacking neutrophil elastase reveal impaired host defense against gram negative bacterial sepsis. Nat. Med. 1998, 4, 615–618. [Google Scholar] [CrossRef]
- Ishii, T.; Doi, K.; Okamoto, K.; Imamura, M.; Dohi, M.; Yamamoto, K.; Fujita, T.; Noiri, E. Neutrophil Elastase Contributes to Acute Lung Injury Induced by Bilateral Nephrectomy. Am. J. Pathol. 2010, 177, 1665–1673. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in Covid-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Levi, M.; Hunt, B.J. Thrombosis and coagulopathy in COVID-19: An illustrated review. Res. Pr. Thromb. Haemost. 2020, 4, 744–751. [Google Scholar] [CrossRef]
- Middleton, E.A.; He, X.-Y.; Denorme, F.; Campbell, R.A.; Ng, D.; Salvatore, S.P.; Mostyka, M.; Baxter-Stoltzfus, A.; Borczuk, A.C.; Loda, M.; et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood 2020, 136, 1169–1179. [Google Scholar] [CrossRef]
- Rao, A.N.; Kazzaz, N.M.; Knight, J.S. Do neutrophil extracellular traps contribute to the heightened risk of thrombosis in inflammatory diseases? World J. Cardiol. 2015, 7, 829–842. [Google Scholar] [CrossRef]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.N.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil extracellular traps in COVID-19. JCI Insight 2020, 5, e138999. [Google Scholar] [CrossRef]
- Zuo, Y.; Yalavarthi, S.; Shi, H.; Gockman, K.; Zuo, M.; Madison, J.A.; Blair, C.; Weber, A.; Barnes, B.J.; Egeblad, M.; et al. Neutrophil Extracellular Traps (NETs) as Markers of Disease Severity in COVID-19. medRxiv 2020, 2020, 2009. [Google Scholar] [CrossRef]
- Zuo, Y.; Zuo, M.; Yalavarthi, S.; Gockman, K.; Madison, J.A.; Shi, H.; Woodard, W.; Lezak, S.P.; Lugogo, N.L.; Knight, J.S.; et al. Neutrophil extracellular traps and thrombosis in COVID-19. J. Thromb. Thrombolysis 2020, 51, 446–453. [Google Scholar] [CrossRef]
- Barnes, B.J.; Adrover, J.M.; Baxter-Stoltzfus, A.; Borczuk, A.; Cools-Lartigue, J.; Crawford, J.M.; Daßler-Plenker, J.; Guerci, P.; Huynh, C.; Knight, J.S.; et al. Targeting potential drivers of COVID-19: Neutrophil extracellular traps. J. Exp. Med. 2020, 217, e20200652. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, E.; Korenbaum, E.; Hegermann, J.; Ochs, M.; Koepke, J.; Koczulla, A.R.; Welte, T.; Köhnlein, T.; Janciauskiene, S. Does Augmentation with Alpha1-Antitrypsin Affect Neutrophil Extracellular Traps Formation? Int. J. Biol. Sci. 2012, 8, 1023–1025. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Petrache, I.; Fijalkowska, I.; Medler, T.R.; Skirball, J.; Cruz, P.; Zhen, L.; Petrache, H.I.; Flotte, T.R.; Tuder, R.M. α-1 Antitrypsin Inhibits Caspase-3 Activity, Preventing Lung Endothelial Cell Apoptosis. Am. J. Pathol. 2006, 169, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Petrache, I.; Fijalkowska, I.; Zhen, L.; Medler, T.R.; Brown, E.; Cruz, P.; Choe, K.-H.; Taraseviciene-Stewart, L.; Scerbavicius, R.; Shapiro, L.; et al. A Novel Antiapoptotic Role for α1-Antitrypsin in the Prevention of Pulmonary Emphysema. Am. J. Respir. Crit. Care Med. 2006, 173, 1222–1228. [Google Scholar] [CrossRef]
- Yang, C.; Chapman, K.R.; Wong, A.; Liu, M. α1-Antitrypsin deficiency and the risk of COVID-19: An urgent call to action. Lancet Respir. Med. 2021, 9, 337–339. [Google Scholar] [CrossRef]
- Shimi, G.; Zand, H. Association of alpha-1-antitrypsin deficiency with vitamin D status: Who is most at risk of getting severe COVID-19? Inflamm. Res. 2021, 70, 375–377. [Google Scholar] [CrossRef] [PubMed]
- Study to Evaluate the Safety and Efficacy of Liquid Alpha1-Proteinase Inhibitor (Human) in Hospitalized Participants with Coronavirus Disease (COVID-19)—Tabular View—ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/record/NCT04547140 (accessed on 27 November 2020).
- Trial of Alpha One Antitrypsin Inhalation in Treating Patient with Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)—Tabular View—ClinicalTrials.Gov. Available online: https://clinicaltrials.gov/ct2/show/record/NCT04385836 (accessed on 27 November 2020).
- McEvoy, N.L.; Clarke, J.L.; Mc Elvaney, O.J.; Mc Elvaney, O.F.; Boland, F.; Hyland, D.; Geoghegan, P.; Donnelly, K.; Friel, O.; Cullen, A.; et al. A randomised, double-blind, placebo-controlled, pilot trial of intravenous plasma purified alpha-1 antitrypsin for SARS-CoV-2-induced Acute Respiratory Distress Syndrome: A structured summary of a study protocol for a randomised, controlled trial. Trials 2021, 22, 1–3. [Google Scholar] [CrossRef] [PubMed]
- McElvaney, O.J.; O’Connor, E.; McEvoy, N.L.; Fraughan, D.D.; Clarke, J.; McElvaney, O.F.; Gunaratnam, C.; O’Rourke, J.; Curley, G.F.; McElvaney, N.G. Alpha-1 antitrypsin for cystic fibrosis complicated by severe cytokinemic COVID-19. J. Cyst. Fibros. 2020, 20, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Ritzmann, F.; Chitirala, P.; Krüger, N.; Hoffmann, M.; Zuo, W.; Lammert, F.; Smola, S.; Tov, N.; Alagem, N.; Lepper, P.M.; et al. Therapeutic Application of alpha-1-antitrypsin in COVID-19. Am. J. Respir. Crit. Care Med. 2021, 204, 2. [Google Scholar] [CrossRef]
- Martini, F.; De Mattei, M.; Contini, C.; Tognon, M.G. Potential Use of Alpha-1 Anti-trypsin in the Covid-19 Treatment. Front. Cell Dev. Biol. 2020, 8, 577528. [Google Scholar] [CrossRef]
- Ramakrishnan, S.; Nicolau, D.V.; Langford, B.; Mahdi, M.; Jeffers, H.; Mwasuku, C.; Krassowska, K.; Fox, R.; Binnian, I.; Glover, V.; et al. Inhaled budesonide in the treatment of early COVID-19 (STOIC): A phase 2, open-label, randomised controlled trial. Lancet Respir. Med. 2021, 9, 763–772. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).