An Artificial Intelligence Approach to Bloodstream Infections Prediction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Definition of Bloodstream Infection
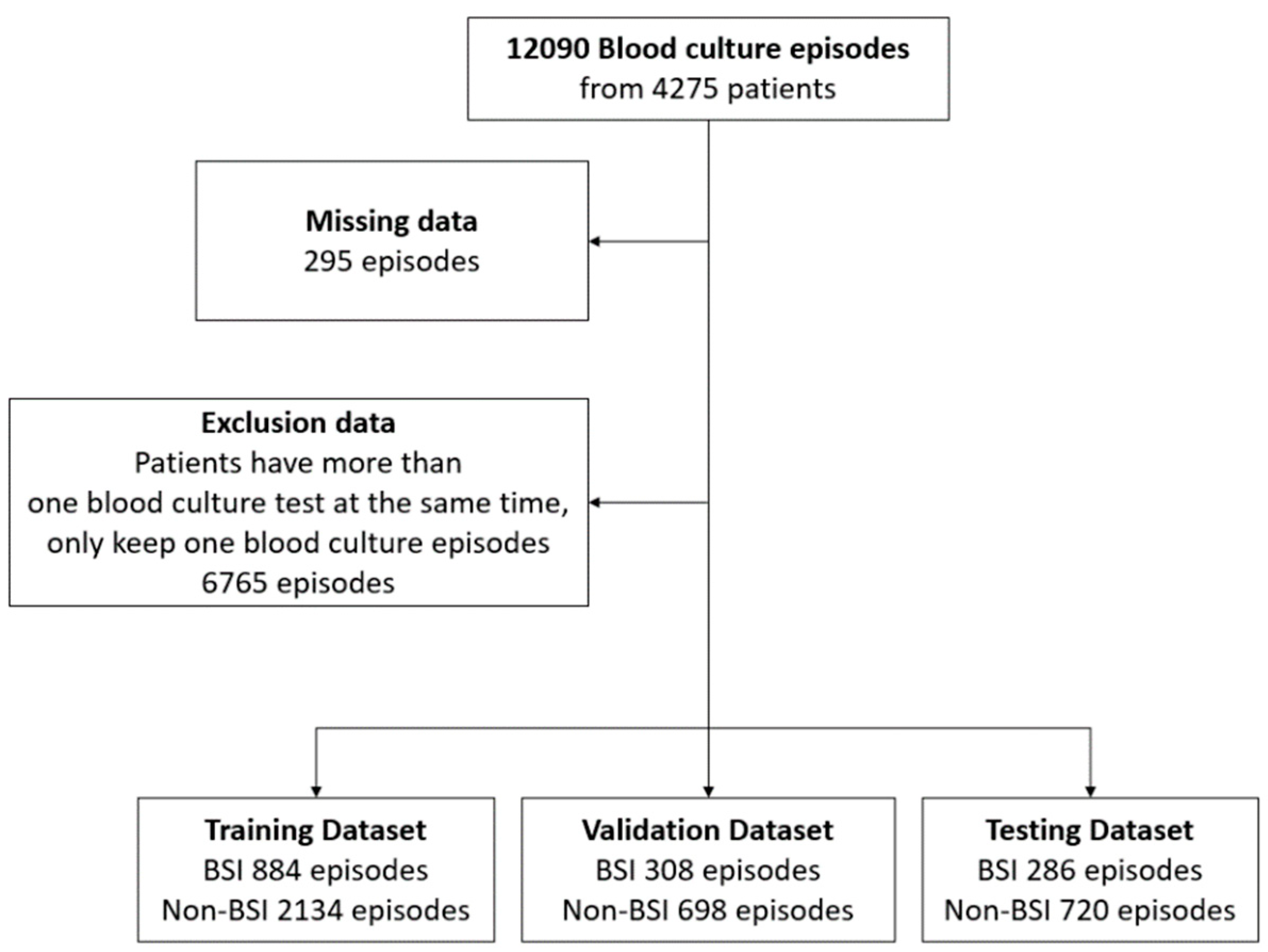
2.2. Data Acquisition
2.3. Data Outcome and Prediction Window
2.4. Clinical Features Selection
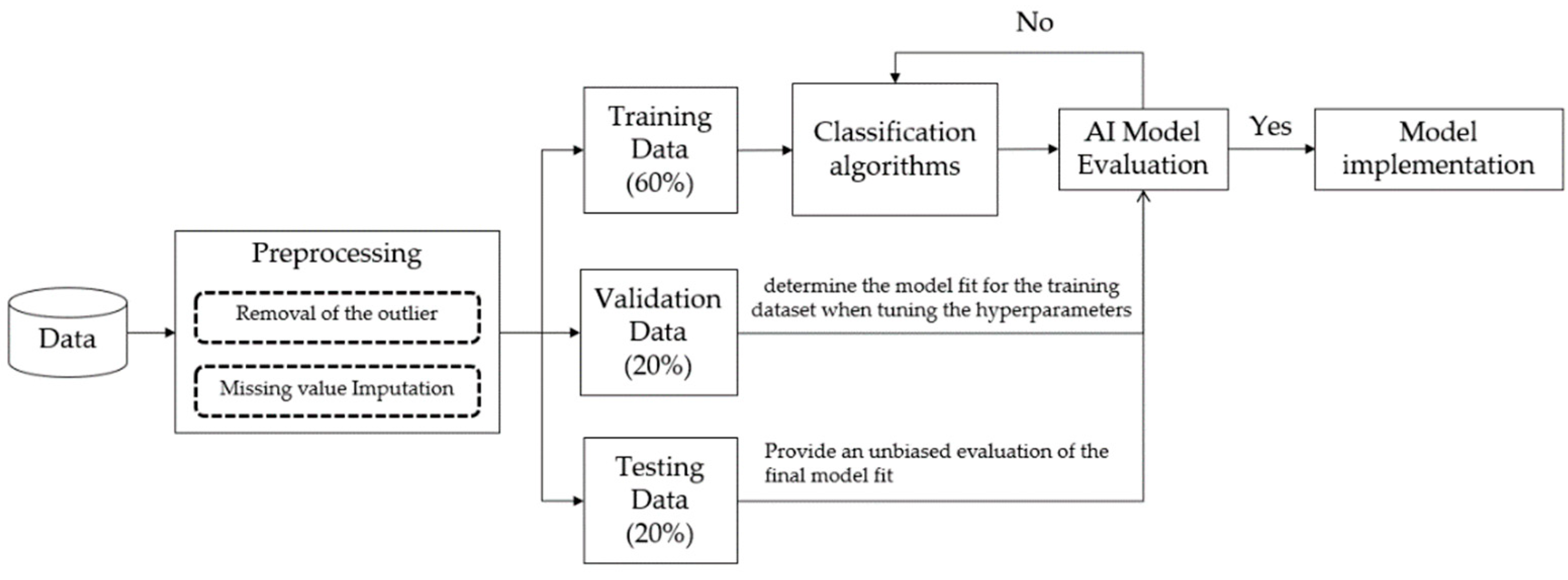
2.5. Study Design and Model Training
2.6. Data Analysis
3. Results
3.1. Evaluation of Different Models
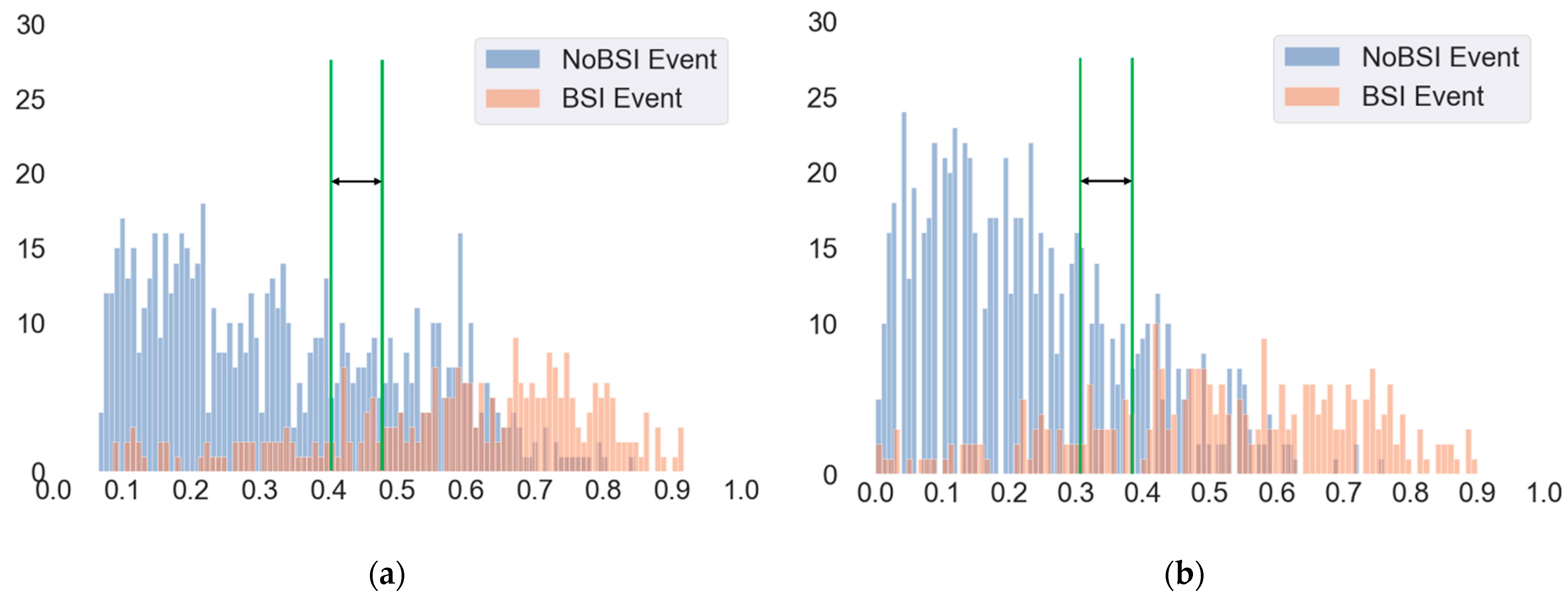
3.2. Evaluation of Different Cut-Off Thresholds
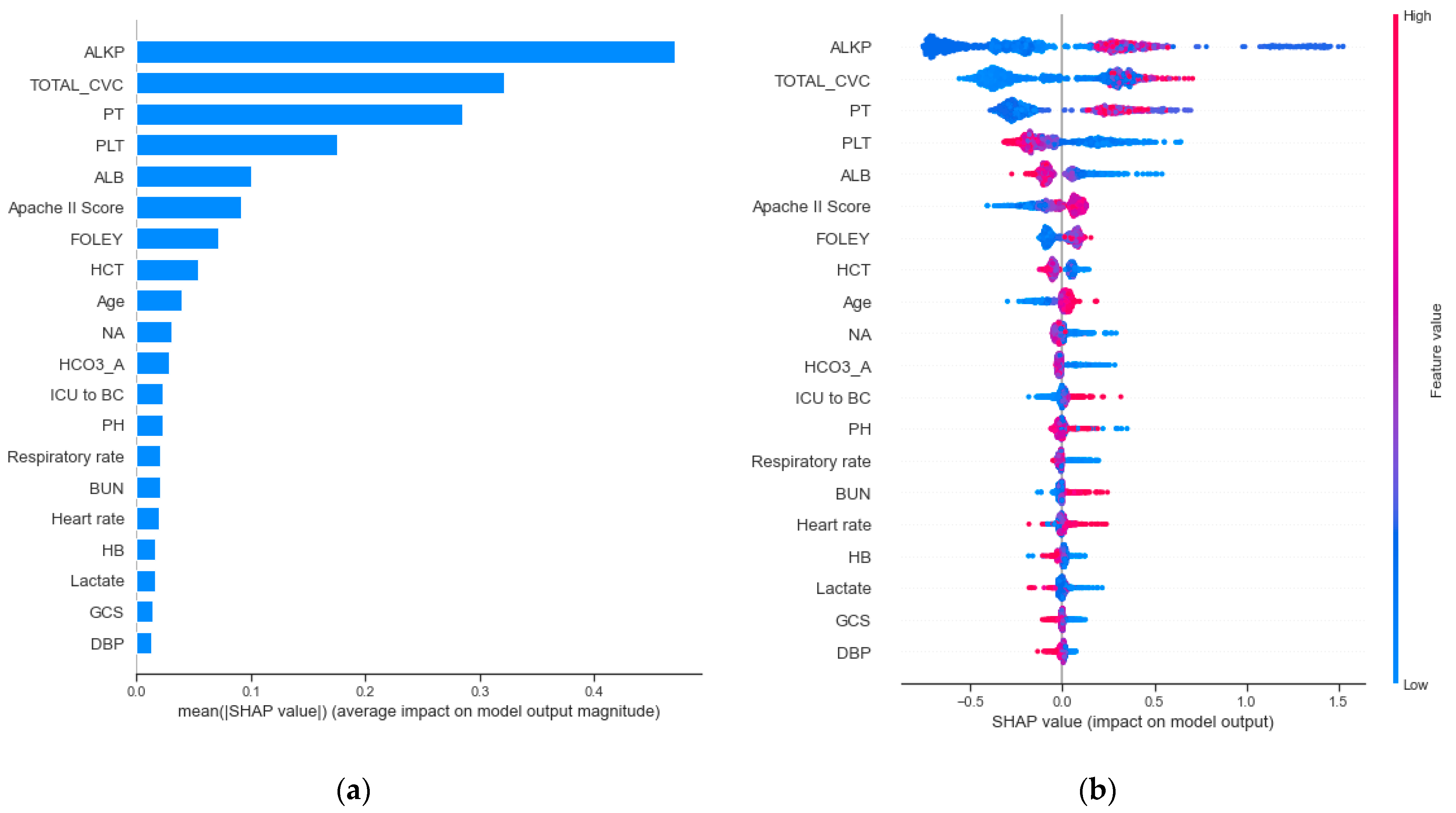
3.3. Clinical Features Importance and Visualization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kang, C.M.; Chen, X.J.; Chih, C.C.; Hsu, C.C.; Chen, P.H.; Lee, T.F.; Teng, L.J.; Hsueh, P.R. Rapid identification of bloodstream bacterial and fungal pathogens and their antibiotic resistance determinants from positively flagged blood cultures using the BioFire FilmArray blood culture identification panel. J. Microbiol. Immunol. Infect. 2020, 53, 882–891. [Google Scholar] [CrossRef]
- Miller, M.A.; Fish, D.N.; Barber, G.R.; Barron, M.A.; Goolsby, T.A.; Moine, P.; Mueller, S.W. A comparison of safety and outcomes with cefazolin versus nafcillin for methicillin-susceptible Staphylococcus aureus bloodstream infections. J. Microbiol. Immunol. Infect. 2020, 53, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.L.; Hsueh, S.C.; Ding, G.S.; Chuang, P.C.; Chen, J.M.; Lu, C.Y.; Chang, L.Y.; Huang, L.M.; Lee, P.I.; Hsueh, P.R. Applicability of an in-House Saponin-Based Extraction Method in Bruker Biotyper Matrix-Assisted Laser Desorption/ionization Time-of-Flight Mass Spectrometry System for Identifying Bacterial and Fungal Species in Positively Flagged Pediatric VersaTREK Blood Cultures. J. Microbiol. Immunol. Infect. 2020, 53, 916–924. [Google Scholar]
- Koizumi, Y.; Sakanashi, D.; Ohno, T.; Nakamura, A.; Yamada, A.; Shibata, Y.; Shiota, A.; Hideo, K.; Mao, H.; Nobuhiro, A.; et al. Plasma procalcitonin levels remain low at the onset of gram-positive bacteremia regardless of severity or the presence of shock: A retrospective analysis of patients with detailed clinical characteristics. J. Microbiol. Immunol. Infect. 2020. [Google Scholar] [CrossRef]
- Pan, Y.P.; Fang, Y.P.; Xu, Y.H.; Wang, Z.X.; Shen, J.L. The Diagnostic Value of Procalcitonin versus Other Biomarkers in Prediction of Bloodstream Infection. Clin. Lab. 2017, 63, 277–285. [Google Scholar] [CrossRef]
- Lee, K.H.; Dong, J.J.; Jeong, S.J.; Chae, M.-H.; Lee, B.S.; Kim, H.J.; Ko, S.H.; Song, Y.G. Early Detection of Bacteraemia Using Ten Clinical Variables with an Artificial Neural Network Approach. J. Clin. Med. 2019, 8, 1592. [Google Scholar] [CrossRef] [Green Version]
- Roimi, M.; Neuberger, A.; Shrot, A.; Paul, M.; Geffen, Y.; Bar-Lavie, Y. Early diagnosis of bloodstream infections in the intensive care unit using machine-learning algorithms. Intensive Care Med. 2020, 46, 454–462. [Google Scholar] [CrossRef]
- Park, H.J.; Jung, D.Y.; Ji, W.; Choi, C.M. Detection of Bacteremia in Surgical In-Patients Using Recurrent Neural Network Based on Time Series Records: Development and Validation Study. J. Med. Internet Res. 2020, 46, e19512. [Google Scholar] [CrossRef]
- Mahmoud, E.; Dhoayan, M.; Bosaeed, M.; Johani, S.; Arabi, Y.M. Developing Machine-Learning Prediction Algorithm for Bacteremia in Admitted Patients. Infect. Drug Resist. 2021, 14, 757–765. [Google Scholar] [CrossRef]
- Tsai, C.M.; Lin, C.R.; Zhang, H.; Chiu, I.M.; Cheng, C.Y.; Yu, H.R.; Huang, Y.H. Using Machine Learning to Predict Bacteremia in Febrile Children Presented to the Emergency Department. Diagnostics 2020, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- Ratzinger, F.; Haslacher, H.; Perkmann, T.; Pinzan, M.; Anner, P.; Makristathis, H.; Burgmann, H.; Heinze, G.; Dorffner, G. Machine learning for fast identification of bacteraemia in SIRS patients treated on standard care wards: A cohort study. Sci. Rep. 2018, 8, 12233. [Google Scholar] [CrossRef] [Green Version]
- Levy, M.M.; Fink, M.P.; Marshall, J.C.; Abraham, E.; Angus, D.; Cook, D.; Cohen, J.; Opal, S.M.; Vincent, J.L.; Ramsay, G. SCCM/ESICM/ACCP/ATS/SIS. 2001 SCCM/ESICM/ACCP/ATS/SIS International Sepsis Definitions Conference. Crit. Care Med. 2003, 31, 1250–1256. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Righi, E.; Carnelutti, A. Bloodstream infections in the Intensive Care Unit. Virulence 2016, 7, 267–279. [Google Scholar] [CrossRef]
- Davis, P.K. Generalizing Concepts of Verification, Validation and Accreditation (VV&A) for Military Simulation; RAND Corporation: Santa Monica, CA, USA, 1992. [Google Scholar]
- Obuchowski, N.A. Receiver operating characteristic curves and their use in radiology. Radiology 2003, 229, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Orbach, J. Principles of Neurodynamics. Perceptrons and the Theory of Brain Mechanisms. Arch. Gen. Psychiatry 1962, 7, 218–219. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Štrumbelj, E.; Kononenko, I. Explaining prediction models and individual predictions with feature contributions. Knowl. Inf. Syst. 2014, 41, 647–665. [Google Scholar] [CrossRef]
- Jerome, H.F. Greedy function approximation: A gradient boosting machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar]
- Eliakim-Raz, N.; Bates, D.W.; Leibovici, L. Predicting bacteraemia in validated models—A systematic review. Clin Microbiol. Infect. 2015, 21, 295–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, M.L.; Apisarnthanarak, A.; Jaggi, N.; Harrington, G.; Morikane, K.; Thule, T.A.; Ching, P.; Villanueva, V.; Zong, Z.; Jeong, J.S.; et al. APSIC guide for prevention of Central Line Associated Bloodstream Infections (CLABSI). Antimicrob. Resist. Infect. Control. 2016, 4, 5–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeon, C.Y.; Neidell, M.; Jia, H.; Sinisi, M.; Larson, E. On the role of length of stay in healthcare-associated bloodstream infection. Infect. Control. Hosp. Epidemiol. 2012, 33, 1213–1218. [Google Scholar] [CrossRef]
- Jin, S.J.; Kim, M.; Yoon, J.H.; Song, Y.G. A new statistical approach to predict bacteremia using electronic medical records. Scand. J. Infect. Dis. 2013, 45, 672–680. [Google Scholar] [CrossRef] [PubMed]



| All (n = 5030) | BSI (n = 1478) | Non-BSI (n = 3552) | p-Value | |
|---|---|---|---|---|
| Basic characteristics | ||||
| Age, years | 66.36 ± 15.83 | 67.50 ± 14.74 | 65.89 ± 16.23 | <0.001 |
| Sex (male) | 3354 (65.15%) | 1040 (70.37%) | 2314 (66.68%) | <0.001 |
| Charlson comorbidity index | 2.23 ± 1.43 | 2.35 ± 1.41 | 2.19 ± 1.43 | <0.001 |
| APACHE II score | 25.73 ± 6.13 | 26.47 ± 6.11 | 25.42 ± 6.12 | <0.001 |
| Divisions | <0.001 | |||
| MICU | 2024 (40.24%) | 615 (41.61%) | 1409 (39.67%) | |
| SICU | 1292 (25.69%) | 489 (33.09%) | 803 (22.61%) | |
| CCU | 727 (14.45%) | 252 (17.05%) | 475 (13.37%) | |
| CV | 404 (8.03%) | 115 (7.78%) | 289 (8.14%) | |
| CVS | 323 (6.42%) | 137 (9.27%) | 186 (5.24%) | |
| NICU | 987 (19.62%) | 122 (8.25%) | 865 (24.35%) | |
| NEURO | 166 (3.30%) | 25 (1.69%) | 141 (3.97%) | |
| NS | 821 (16.32%) | 97 (6.56%) | 724 (20.38%) | |
| The etiology for ICU admission | <0.001 | |||
| Scheduled surgery | 241 (5.09%) | 49 (3.49%) | 192 (5.76%) | |
| Emergency surgery | 113 (2.39%) | 31 (2.20%) | 82 (2.46%) | |
| NS surgery, scheduled | 50 (1.06%) | 4 (0.28%) | 46 (1.38%) | |
| NS surgery, emergency | 318 (6.71%) | 22 (1.56%) | 296 (8.89%) | |
| Acute respiratory failure | 1130 (23.85%) | 325 (23.12%) | 805 (24.17%) | |
| Pneumonia | 327 (6.90%) | 92 (6.54%) | 235 (7.05%) | |
| Sepsis, non-pneumonia | 308 (6.50%) | 125 (8.89%) | 183 (5.49%) | |
| Acute cardiac conditions | 544 (11.48%) | 164 (11.66%) | 380 (11.41%) | |
| Acute neurological conditions | 170 (3.59%) | 29 (2.06%) | 141 (4.23%) | |
| Pulmonary embolism | 0 (0%) | 0 (0%) | 0 (0%) | |
| Acute renal conditions | 58 (1.22%) | 14 (1.00%) | 44 (1.32%) | |
| Acute GI condition | 688 (14.52%) | 276 (19.63%) | 412 (12.37%) | |
| Post-PCI | 22 (0.46%) | 6 (0.43%) | 16 (0.48%) | |
| OHCA/INCA | 39 (0.82%) | 12 (0.85%) | 27 (0.81%) | |
| Others | 729 (15.39%) | 257 (18.28%) | 472 (14.17%) | |
| Outcomes | ||||
| ICU stay, days | 25.65 ± 20.05 | 34.10 ± 25.39 | 22.13 ± 16.09 | <0.001 |
| Hospital stay, days | 49.12 ± 40.57 | 58.37 ± 39.95 | 45.27 ± 40.21 | <0.001 |
| Clinical Variable | BSI (n = 1478) | Non-BSI (n = 3552) | p-Value | Standard Cut-off |
|---|---|---|---|---|
| Vital sign | ||||
| Temperature (°C) | 36.62 ± 0.49 | 36.66 ± 0.48 | 0.007 | |
| SBP (mmHg) | 121.40 ± 14.95 | 123.78 ± 15.31 | <0.001 | |
| DBP (mmHg) | 66.46 ± 10.50 | 67.95 ± 10.79 | <0.001 | |
| GCS | 7.37 ± 3.20 | 7.56 ± 3.53 | 0.065 | |
| Heart rate (bpm) | 93.36 ± 15.57 | 90.28 ± 15.58 | <0.001 | |
| Respiratory rate (breath/min) | 19.03 ± 3.53 | 18.65 ± 3.61 | <0.001 | |
| Laboratory | ||||
| Albumin (g/dL) | 2.64 ± 0.55 | 2.84 ± 0.59 | <0.001 | 3.5–5 |
| Alkaline phosphatase (U/L) | 211.00 ± 181.11 | 191.23 ± 220.81 | 0.015 | 50–190 |
| BUN (mg/dL) | 53.81 ± 37.91 | 42.31 ± 33.90 | <0.001 | 5–25 |
| Creatinine (mg/dL) | 2.26 ± 1.90 | 1.94 ± 1.99 | <0.001 | 0.5–1.4 |
| CRP (mg/dL) | 11.09 ± 8.99 | 9.93 ± 9.80 | 0.016 | <0.3 |
| Glucose (mg/dL) | 180.08 ± 91.35 | 182.99 ± 103.07 | 0.574 | 70–200 |
| HCO3-A (mmol/L) | 23.38 ± 5.16 | 24.40 ± 5.24 | <0.001 | 22–26 |
| Hematocrit (%) | 27.42 ± 4.78 | 29.26 ± 5.79 | <0.001 | 37–52 |
| Hemoglobin (g/dL) | 9.13 ± 1.46 | 9.76 ± 1.79 | <0.001 | 12–17.5 |
| Potassium(K) (mEq/L) | 3.90 ± 0.67 | 3.95 ± 0.67 | 0.011 | 3.5–5.3 |
| Na (mEq/L) | 140.26 ± 7.41 | 140.76 ± 7.01 | 0.024 | 137–153 |
| pH (blood gas) | 7.43 ± 0.07 | 7.43 ± 0.07 | 0.83 | 7.35–7.45 |
| Platelet count (/UL) | 140.39 ± 107.39 | 196.45 ± 125.71 | <0.001 | 150–400 |
| PO2-A (mmHg) | 122.80 ± 49.18 | 124.50 ± 62.55 | 0.414 | 80–100 |
| Prothrombin time (PT) (s) | 13.98 ± 6.68 | 12.77 ± 5.48 | <0.001 | 9.5–11.7 |
| WBC (/UL) | 11.81 ± 7.55 | 12.22 ± 7.13 | 0.066 | 3.5–11 |
| Lactate (mg/dL) | 18.32 ± 17.01 | 16.08 ± 14.54 | <0.001 | 3.0–12 |
| Clinical information | ||||
| ICU day to blood culture, days | 19.70 ± 17.53 | 8.99 ± 4.86 | <0.001 | |
| Central venous catheter (h | 1233.57 ± 2965.12 | 698.78 ± 3261.02 | <0.001 | |
| ENDO (h) | 2927.38 ± 5080.96 | 1600.29 ± 3972.20 | <0.001 | |
| FOLEY (h) | 528.04 ± 536.07 | 325.87 ± 632.31 | <0.001 | |
| Dataset | Algorithms 1 | AUROC (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Brier Score |
|---|---|---|---|---|---|
| Validation dataset | LR | 0.709 (0.679–0.737) | 0.679 (0.624–0.728) | 0.660 (0.625–0.695) | 0.218 |
| SVM | 0.728 (0.699–0.756) | 0.578 (0.522–0.632) | 0.779 (0.747–0.809) | 0.195 | |
| MLP | 0.735 (0.707–0.761) | 0.494 (0.438–0.549) | 0.832 (0.803–0.858) | 0.231 | |
| XGBoost | 0.825 (0.802–0.849) | 0.724 (0.672–0.771) | 0.777 (0.744–0.806) | 0.165 | |
| RF | 0.855 (0.832–0.877) | 0.565 (0.509–0.619) | 0.927 (0.905–0.944) | 0.139 | |
| Testing dataset | LR | 0.685 (0.653–0.715) | 0.615 (0.558–0.670) | 0.644 (0.609–0.679) | 0.223 |
| SVM | 0.704 (0.673–0.733) | 0.566 (0.508–0.623) | 0.756 (0.723–0.786) | 0.199 | |
| MLP | 0.668 (0.633–0.698) | 0.406 (0.350–0.463) | 0.811 (0.781–0.838) | 0.254 | |
| XGBoost | 0.821 (0.795–0.843) | 0.706 (0.651–0.756) | 0.775 (0.743–0.804) | 0.163 | |
| RF | 0.851 (0.824–0.872) | 0.577 (0.519–0.633) | 0.940 (0.921–0.955) | 0.134 |
| Algorithms | Cut-Off Threshold | Sensitivity | Specificity | Precision | True Positive | True Negative | False Positive | False Negative |
|---|---|---|---|---|---|---|---|---|
| RF | 0.3 | 82.9% | 69.2% | 51.6% | 237 (23.6%) | 498 (49.5%) | 222 (22.1%) | 49 (4.9%) |
| 0.4 | 69.9% | 85.8% | 66.2% | 200 (19.9%) | 618 (61.4%) | 102 (10.1%) | 86 (8.6%) | |
| 0.41 | 68.2% | 86.0% | 65.9% | 195 (19.4%) | 619 (61.5%) | 101 (10.0%) | 91 (9.1%) | |
| 0.5 | 57.7% | 94.0% | 79.3% | 165 (16.4%) | 677 (67.3%) | 43 (4.3%) | 121 (12.0%) | |
| 0.53 | 51.0% | 96.5% | 85.4% | 146 (14.5%) | 695 (69.1%) | 25 (2.5%) | 140 (13.9%) | |
| 0.6 | 38.8% | 98.2% | 89.5% | 111 (11.0%) | 707 (70.3%) | 13 (1.3%) | 175 (17.4%) | |
| 0.7 | 21.3% | 99.6% | 95.3% | 61 (6.1%) | 717 (71.3%) | 3 (0.3%) | 225 (22.3%) | |
| XGBoost | 0.3 | 89.9% | 48.8% | 41.1% | 257 (25.5%) | 351 (34.9%) | 369 (36.7%) | 29 (2.9%) |
| 0.4 | 82.2% | 64.3% | 47.8% | 235 (23.4%) | 463 (46.0%) | 257 (25.5%) | 51 (5.1%) | |
| 0.41 | 80.8% | 67.0% | 49.4% | 231 (23.0%) | 483 (48.0%) | 237 (23.6%) | 55 (5.4%) | |
| 0.5 | 70.6% | 77.5% | 55.5% | 202 (20.1%) | 558 (55.5%) | 162 (16.1%) | 84 (8.3%) | |
| 0.53 | 67.1% | 81.0% | 58.4% | 192 (19.1%) | 583 (58.0%) | 137 (13.6%) | 94 (9.3%) | |
| 0.6 | 53.5% | 90.7% | 69.5% | 153 (15.2%) | 653 (64.9%) | 67 (6.7%) | 133 (13.2%) | |
| 0.7 | 33.2% | 97.9% | 86.3% | 60 (5.9%) | 693 (68.9%) | 5 (0.5%) | 248 (24.7%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pai, K.-C.; Wang, M.-S.; Chen, Y.-F.; Tseng, C.-H.; Liu, P.-Y.; Chen, L.-C.; Sheu, R.-K.; Wu, C.-L. An Artificial Intelligence Approach to Bloodstream Infections Prediction. J. Clin. Med. 2021, 10, 2901. https://doi.org/10.3390/jcm10132901
Pai K-C, Wang M-S, Chen Y-F, Tseng C-H, Liu P-Y, Chen L-C, Sheu R-K, Wu C-L. An Artificial Intelligence Approach to Bloodstream Infections Prediction. Journal of Clinical Medicine. 2021; 10(13):2901. https://doi.org/10.3390/jcm10132901
Chicago/Turabian StylePai, Kai-Chih, Min-Shian Wang, Yun-Feng Chen, Chien-Hao Tseng, Po-Yu Liu, Lun-Chi Chen, Ruey-Kai Sheu, and Chieh-Liang Wu. 2021. "An Artificial Intelligence Approach to Bloodstream Infections Prediction" Journal of Clinical Medicine 10, no. 13: 2901. https://doi.org/10.3390/jcm10132901
APA StylePai, K.-C., Wang, M.-S., Chen, Y.-F., Tseng, C.-H., Liu, P.-Y., Chen, L.-C., Sheu, R.-K., & Wu, C.-L. (2021). An Artificial Intelligence Approach to Bloodstream Infections Prediction. Journal of Clinical Medicine, 10(13), 2901. https://doi.org/10.3390/jcm10132901







