Learning from Embryogenesis—A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation of Melanocytes and De-Differentiation into Melanoblast-Related Cells
2.2. Melanoma Cell Culture
2.3. microRNA-Sequencing and Bioinformatic Sequence Data Analysis
2.4. Gene Expression Microarray and Gene Set Enrichment Analysis (GSEA) of miRNA Target Genes
2.5. Isolation and Reverse Transcription of miRNAs from Mammalian Cells
2.6. Quantitative RT-PCR with miRNA
3. Results
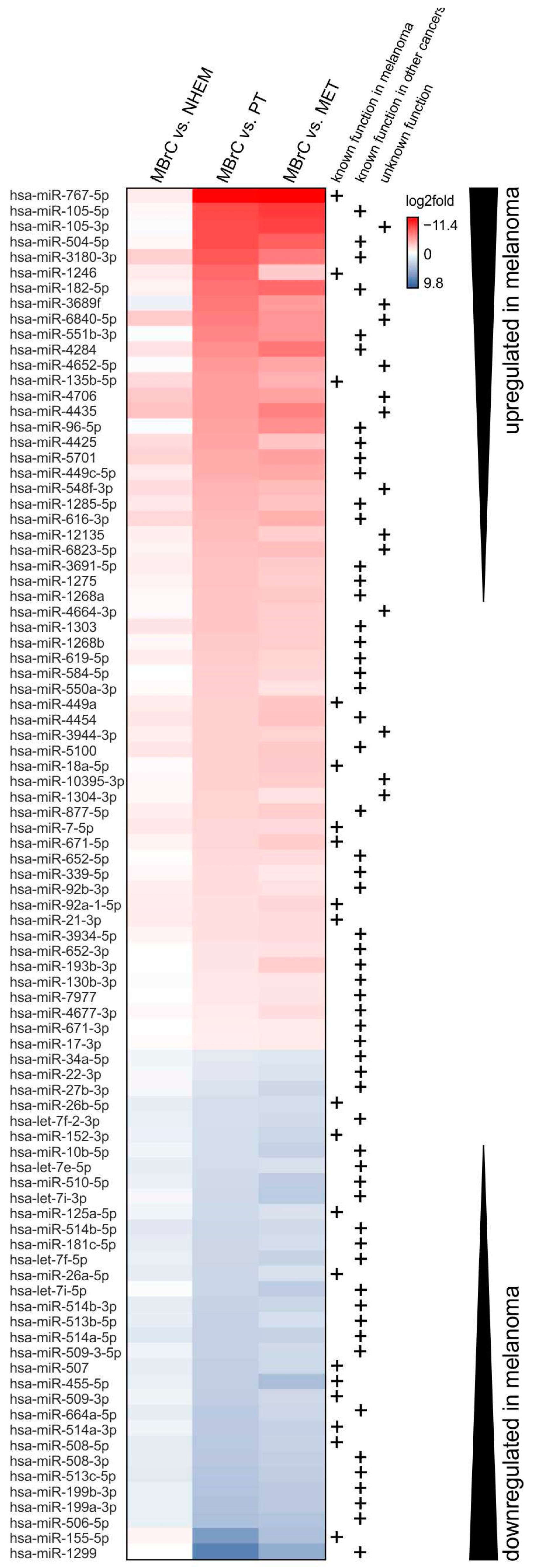
3.1. miRNAs Differentially Expressed Only in Melanoma Cells but Not in NHEMs and Melanoblasts Drive Tumor Development
- Genes and miRNAs, which are equally expressed in melanoma and melanoblasts but differently in melanocytes, are the relevant decisive factors for melanoma development; or
- genes and miRNAs differentially expressed in melanoma compared to melanocytes and melanoblasts are key drivers of melanoma development and progression because these are stabilizing the tumor phenotype. This second hypothesis implicates that hypothesis 1 “only” focuses on genes, which are involved in differentiation/dedifferentiation processes but not in forcing tumor progression.
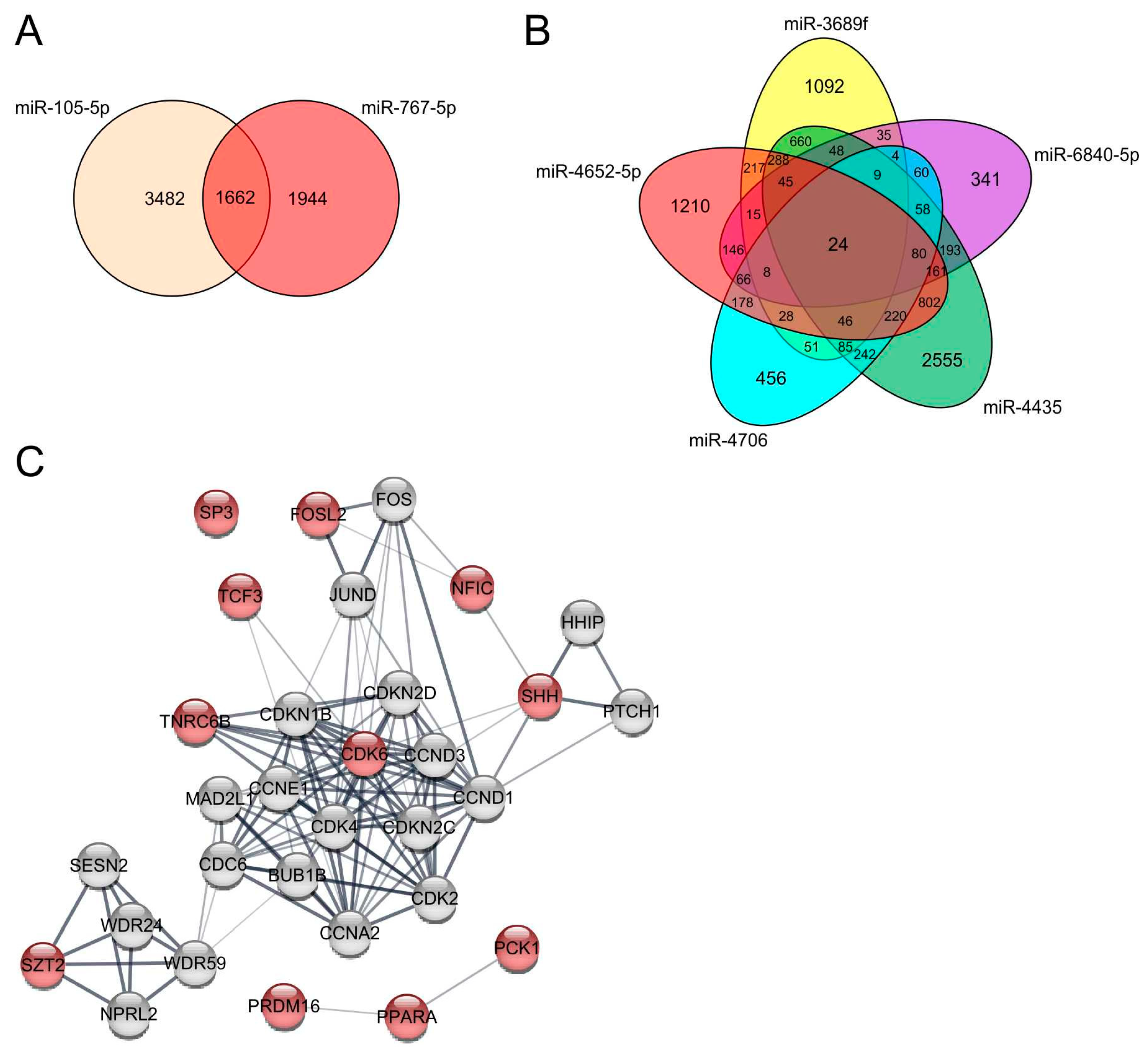
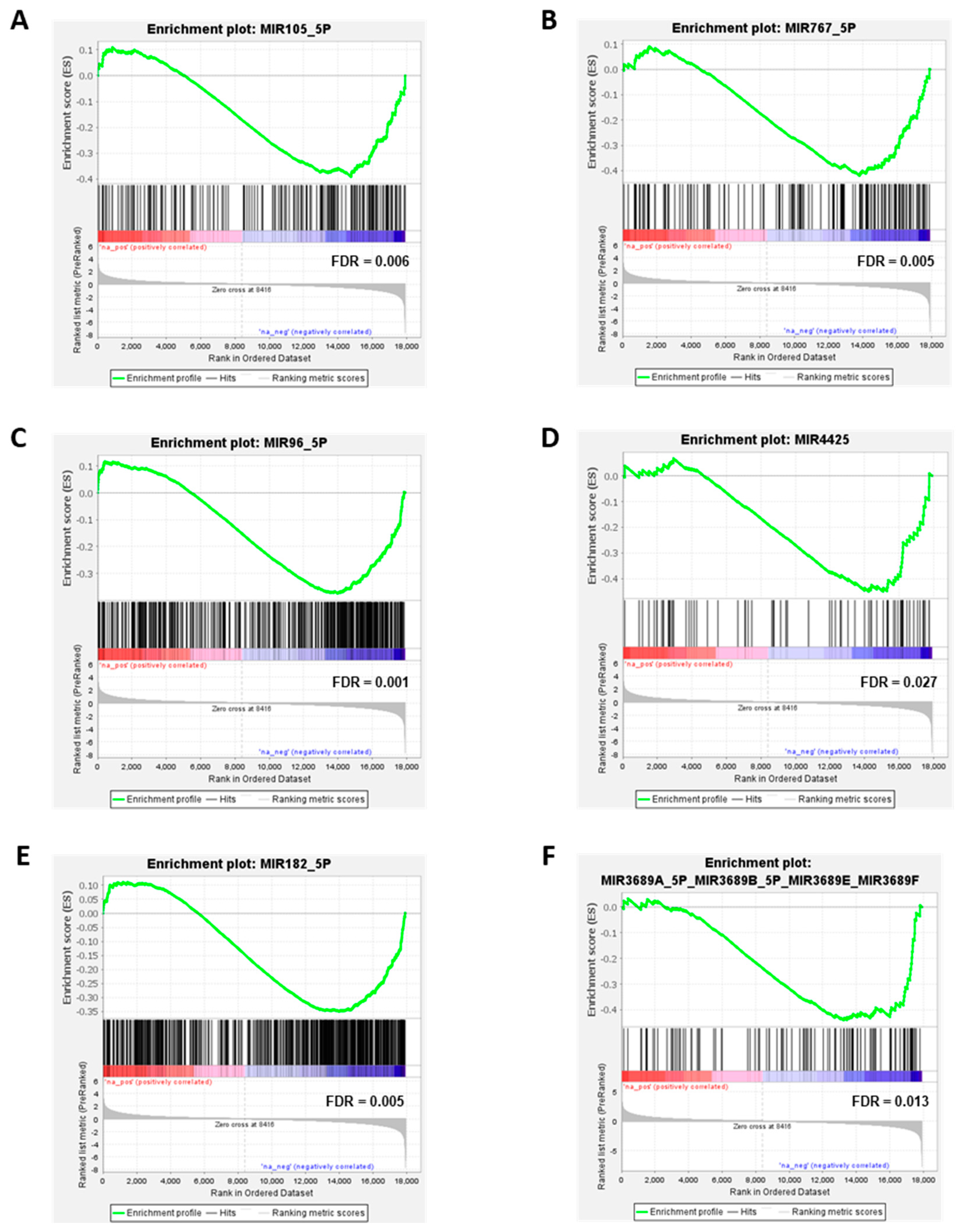
3.2. miRNAs Significantly Upregulated in Melanoma Cells Compared to MBrCs and NHEMs Regulate Important Target Genes Driving Tumorigenesis
3.3. miRNAs Significantly Downregulated in Melanoma Cells Compared to MBrCs and NHEMs Are Implicated in the Regulation of Tumorigenic Pathways
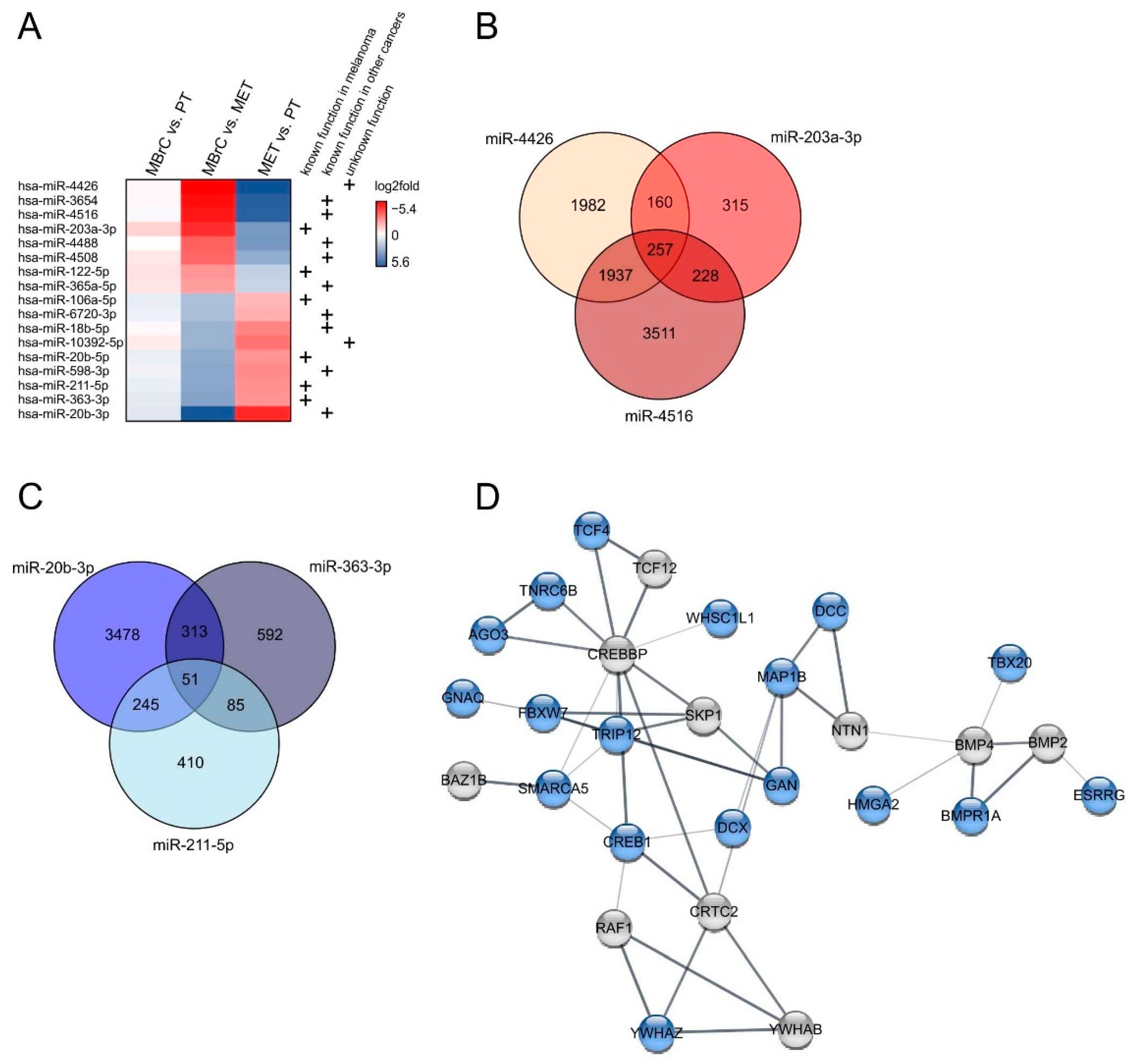
3.4. miRNAs Differentially Expressed in Only Metastasis-Derived Melanoma Cell Lines Compared to Melanoblasts Provide Information about Metastasis Processes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leiter, U.; Eigentler, T.; Garbe, C. Epidemiology of skin cancer. In Sunlight, Vitamin D and Skin Cancer; Springer: Berlin/Heidelberg, Germany, 2014; pp. 120–140. [Google Scholar]
- Garbe, C.; Peris, K.; Hauschild, A.; Saiag, P.; Middleton, M.; Bastholt, L.; Grob, J.-J.; Malvehy, J.; Newton-Bishop, J.; Stratigos, A.J. Diagnosis and treatment of melanoma. European consensus-based interdisciplinary guideline—Update 2016. Eur. J. Cancer 2016, 63, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Bastian, B.C. From melanocytes to melanomas. Nat. Rev. Cancer 2016, 16, 345. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.J.; Erickson, C.A. The making of a melanocyte: The specification of melanoblasts from the neural crest. Pigment Cell Melanoma Res. 2008, 21, 598–610. [Google Scholar] [CrossRef] [PubMed]
- Bosserhoff, A.K.; Ellmann, L.; Kuphal, S. Melanoblasts in culture as an in vitro system to determine molecular changes in melanoma. Exp. Dermatol. 2011, 20, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef] [PubMed]
- Brocker, E.B.; Magiera, H.; Herlyn, M. Nerve growth and expression of receptors for nerve growth factor in tumors of melanocyte origin. J. Investig. Dermatol. 1991, 96, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, M.J.; Seftor, E.A.; Hess, A.R.; Seftor, R.E. Vasculogenic mimicry and tumour-cell plasticity: Lessons from melanoma. Nat. Rev. Cancer 2003, 3, 411–421. [Google Scholar] [CrossRef]
- Vandamme, N.; Berx, G. From neural crest cells to melanocytes: Cellular plasticity during development and beyond. Cell. Mol. Life Sci. 2019, 76, 1919–1934. [Google Scholar] [CrossRef]
- Le Douarin, N.; LeDouarin, N.M.; Kalcheim, C. The Neural Crest; Cambridge University Press: Cambridge, UK, 1999. [Google Scholar]
- Watson, I.R.; Wu, C.-J.; Zou, L.; Gershenwald, J.E.; Chin, L. Genomic classification of cutaneous melanoma. In Proceedings of the AACR 106th Annual Meeting 2015, Philadelphia, PA, USA, 18–22 April 2015. [Google Scholar]
- Tímár, J.; Vizkeleti, L.; Doma, V.; Barbai, T.; Rásó, E. Genetic progression of malignant melanoma. Cancer Metastasis Rev. 2016, 35, 93–107. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Starega-Roslan, J.; Koscianska, E.; Kozlowski, P.; Krzyzosiak, W.J. The role of the precursor structure in the biogenesis of microRNA. Cell. Mol. Life Sci. 2011, 68, 2859–2871. [Google Scholar] [CrossRef] [PubMed]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Lorio, M.; Croce, C. microRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar]
- Linck, L.; Liebig, J.; Voeller, D.; Eichner, N.; Lehmann, G.; Meister, G.; Bosserhoff, A. microRNA-sequencing data analyzing melanoma development and progression. Exp. Mol. Pathol. 2018, 105, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Mueller, D.W.; Rehli, M.; Bosserhoff, A.K. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Investig. Dermatol. 2009, 129, 1740–1751. [Google Scholar] [CrossRef]
- Ding, N.; Wang, S.; Yang, Q.; Li, Y.; Cheng, H.; Wang, J.; Wang, D.; Deng, Y.; Yang, Y.; Hu, S. Deep sequencing analysis of microRNA expression in human melanocyte and melanoma cell lines. Gene 2015, 572, 135–145. [Google Scholar] [CrossRef]
- Stark, M.S.; Tyagi, S.; Nancarrow, D.J.; Boyle, G.M.; Cook, A.L.; Whiteman, D.C.; Parsons, P.G.; Schmidt, C.; Sturm, R.A.; Hayward, N.K. Characterization of the melanoma miRNAome by deep sequencing. PLoS ONE 2010, 5, e9685. [Google Scholar] [CrossRef] [PubMed]
- Cook, A.L.; Donatien, P.D.; Smith, A.G.; Murphy, M.; Jones, M.K.; Herlyn, M.; Bennett, D.C.; Leonard, J.H.; Sturm, R.A. Human melanoblasts in culture: Expression of BRN2 and synergistic regulation by fibroblast growth factor-2, stem cell factor, and endothelin-3. J. Investig. Dermatol. 2003, 121, 1150–1159. [Google Scholar] [CrossRef]
- Johnson, J.P.; Demmer-Dieckmann, M.; Meo, T.; Hadam, M.R.; Riethmüller, G. Surface antigens of human melanoma cells defined by monoclonal antibodies. I. Biochemical characterization of two antigens found on cell lines and fresh tumors of diverse tissue origin. Eur. J. Immunol. 1981, 11, 825–831. [Google Scholar] [CrossRef]
- Siracký, J.; Blasko, M.; Borovanský, J.; Kovarik, J.; Svec, J.; Vrba, M. Human melanoma cell lines: Morphology, growth, and alpha-mannosidase characteristics. Neoplasma 1982, 29, 661–668. [Google Scholar]
- Giard, D.J.; Aaronson, S.A.; Todaro, G.J.; Arnstein, P.; Kersey, J.H.; Dosik, H.; Parks, W.P. In vitro cultivation of human tumors: Establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst. 1973, 51, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Cornil, I.; Theodorescu, D.; Man, S.; Herlyn, M.; Jambrosic, J.; Kerbel, R. Fibroblast cell interactions with human melanoma cells affect tumor cell growth as a function of tumor progression. Proc. Natl. Acad. Sci. USA 1991, 88, 6028–6032. [Google Scholar] [CrossRef] [PubMed]
- Marincola, F.M.; Shamamian, P.; Alexander, R.B.; Gnarra, J.R.; Turetskaya, R.L.; Nedospasov, S.A.; Simonis, T.B.; Taubenberger, J.K.; Yannelli, J.; Mixon, A. Loss of HLA haplotype and B locus down-regulation in melanoma cell lines. J. Immunol. 1994, 153, 1225–1237. [Google Scholar]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; Van Den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Love, M.; Anders, S.; Huber, W. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Torre, D.; Lachmann, A.; Ma’ayan, A. BioJupies: Automated generation of interactive notebooks for RNA-Seq data analysis in the cloud. Cell Syst. 2018, 7, 556–561.e3. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.; Johnson, W.; Parker, H.; Jaffe, A.; Storey, J. SVA detailed instruction. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- De la Nava, J.G.; van Hijum, S.; Trelles, O. Saturation and quantization reduction in microarray experiments using two scans at different sensitivities. Stat. Appl. Genet. Mol. Biol. 2004, 3, 11. [Google Scholar] [CrossRef][Green Version]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, Q.; Zhu, X.-H. miR-135b is a novel oncogenic factor in cutaneous melanoma by targeting LATS2. Melanoma Res. 2019, 29, 119–125. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, F.; Sun, P. microRNA-1246 Promotes Melanoma Progression through Targeting FOXA2. Oncotargets Ther. 2020, 13, 1245. [Google Scholar] [CrossRef]
- Shang, J.; Yu, G.; Ji, Z.; Wang, X.; Xia, L. miR-105 inhibits gastric cancer cells metastasis, epithelial-mesenchymal transition by targeting SOX9. Eur. Rev. Med. Pharm. Sci. 2019, 23, 6160–6169. [Google Scholar]
- Li, Y.; Shen, Z.; Jiang, H.; Lai, Z.; Wang, Z.; Jiang, K.; Ye, Y.; Wang, S. microRNA-4284 promotes gastric cancer tumorigenicity by targeting ten-eleven translocation 1. Mol. Med. Rep. 2018, 17, 6569–6575. [Google Scholar] [CrossRef] [PubMed]
- Bissey, P.-A.; Mathot, P.; Guix, C.; Jasmin, M.; Goddard, I.; Costechareyre, C.; Gadot, N.; Delcros, J.-G.; Mali, S.M.; Fasan, R. Blocking SHH/Patched interaction triggers tumor growth inhibition through Patched-induced apoptosis. Cancer Res. 2020, 80, 1970–1980. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.-Y.; Feng, S.-C.; Sun, Y.-Q.; Jiang, G.-Q. miR-96-5p promotes breast cancer migration by activating MEK/ERK signaling. J. Gene Med. 2020, 22, e3188. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, Y.; Zheng, X.; Chen, L.; Tuo, X.; Chen, H.; Xue, M.; Chen, Q.; Chen, W.; Li, X. 20(S)-Rg3 upregulates FDFT1 via reducing miR-4425 to inhibit ovarian cancer progression. Arch. Biochem. Biophys. 2020, 693, 108569. [Google Scholar] [CrossRef]
- Cao, M.-Q.; You, A.-B.; Zhu, X.-D.; Zhang, W.; Zhang, Y.-Y.; Zhang, S.-Z.; Zhang, K.-w.; Cai, H.; Shi, W.-K.; Li, X.-L. miR-182-5p promotes hepatocellular carcinoma progression by repressing FOXO3a. J. Hematol. Oncol. 2018, 11, 1–12. [Google Scholar]
- Gao, J.; Zeng, K.; Liu, Y.; Gao, L.; Liu, L. LncRNA SNHG5 promotes growth and invasion in melanoma by regulating the miR-26a-5p/TRPC3 pathway. OncoTargets Ther. 2019, 12, 169. [Google Scholar] [CrossRef]
- Li, Y.; Wang, P.; Wu, L.-L.; Yan, J.; Pang, X.-Y.; Liu, S.-J. miR-26a-5p inhibit gastric cancer cell proliferation and invasion through mediated Wnt5a. OncoTargets Ther. 2020, 13, 2537. [Google Scholar] [CrossRef]
- Zhou, X.; Yan, T.; Huang, C.; Xu, Z.; Wang, L.; Jiang, E.; Wang, H.; Chen, Y.; Liu, K.; Shao, Z. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 1–15. [Google Scholar] [CrossRef]
- Dabbah, M.; Attar-Schneider, O.; Zismanov, V.; Tartakover Matalon, S.; Lishner, M.; Drucker, L. Letter to the Editor: miR-199b-3p and miR-199a-3p are isoforms with identical sequence and established function as tumor and metastasis suppressors. J. Leukoc. Biol. 2017, 101, 1069. [Google Scholar] [CrossRef] [PubMed]
- Streicher, K.L.; Zhu, W.; Lehmann, K.P.; Georgantas, R.W.; Morehouse, C.A.; Brohawn, P.; Carrasco, R.A.; Xiao, Z.; Tice, D.A.; Higgs, B.W.; et al. A novel oncogenic role for the miRNA-506-514 cluster in initiating melanocyte transformation and promoting melanoma growth. Oncogene 2012, 31, 1558–1570. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Li, L.; Li, J.; Zhao, H. miR-1299 Impedes the Progression of Non-Small-Cell Lung Cancer Through EGFR/PI3K/AKT Signaling Pathway. OncoTargets Ther. 2020, 13, 7493. [Google Scholar] [CrossRef] [PubMed]
- Behm-Ansmant, I.; Rehwinkel, J.; Doerks, T.; Stark, A.; Bork, P.; Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 2006, 20, 1885–1898. [Google Scholar] [CrossRef]
- Cui, T.; Bell, E.H.; McElroy, J.; Becker, A.P.; Gulati, P.M.; Geurts, M.; Mladkova, N.; Gray, A.; Liu, K.; Yang, L. miR-4516 predicts poor prognosis and functions as a novel oncogene via targeting PTPN14 in human glioblastoma. Oncogene 2019, 38, 2923–2936. [Google Scholar] [CrossRef]
- Hao, T.; Li, C.; Ding, X.; Xing, X. microRNA-363-3p/p21 (Cip1/Waf1) Axis Is Regulated by HIF-2α in Mediating Stemness of Melanoma Cells. Neoplasma 2019, 66, 427–436. [Google Scholar] [CrossRef]
- Xue, M.; Tao, W.; Yu, S.; Yan, Z.; Peng, Q.; Jiang, F.; Gao, X. lncRNA ZFPM2-AS1 promotes proliferation via miR-18b-5p/VMA21 axis in lung adenocarcinoma. J. Cell. Biochem. 2020, 121, 313–321. [Google Scholar] [CrossRef]
- Wang, J.; Tao, Y.; Bian, Q. miRNA and mRNA expression profiling reveals potential biomarkers for metastatic cutaneous melanoma. Expert Rev. Anticancer Ther. 2021, 21, 557–567. [Google Scholar] [CrossRef]
- Chamcheu, J.C.; Roy, T.; Uddin, M.B.; Banang-Mbeumi, S.; Chamcheu, R.-C.N.; Walker, A.L.; Liu, Y.-Y.; Huang, S. Role and therapeutic targeting of the PI3K/Akt/mTOR signaling pathway in skin cancer: A review of current status and future trends on natural and synthetic agents therapy. Cells 2019, 8, 803. [Google Scholar] [CrossRef]
- Busse, A.; Keilholz, U. Role of TGF-β in melanoma. Curr. Pharm. Biotechnol. 2011, 12, 2165–2175. [Google Scholar] [CrossRef]
- Linck-Paulus, L.; Hellerbrand, C.; Bosserhoff, A.K.; Dietrich, P. Dissimilar Appearances Are Deceptive–Common microRNAs and Therapeutic Strategies in Liver Cancer and Melanoma. Cells 2020, 9, 114. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. microRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar] [CrossRef]
- Müller, D.; Bosserhoff, A.-K. Integrin β 3 expression is regulated by let-7a miRNA in malignant melanoma. Oncogene 2008, 27, 6698–6706. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Ahn, J.-H.; Lee, M. Upregulation of microRNA-1246 is associated with BRAF inhibitor resistance in melanoma cells with mutant BRAF. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2017, 49, 947. [Google Scholar] [CrossRef]
- Stark, M.S.; Bonazzi, V.F.; Boyle, G.M.; Palmer, J.M.; Symmons, J.; Lanagan, C.M.; Schmidt, C.W.; Herington, A.C.; Ballotti, R.; Pollock, P.M.; et al. miR-514a regulates the tumour suppressor NF1 and modulates BRAFi sensitivity in melanoma. Oncotarget 2015, 6, 17753–17763. [Google Scholar] [CrossRef]
- Dong, X.; Chang, M.; Song, X.; Ding, S.; Xie, L.; Song, X. Plasma miR-1247-5p, miR-301b-3p and miR-105-5p as potential biomarkers for early diagnosis of non-small cell lung cancer. Thorac. Cancer 2021, 12, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zhang, Z. Serum miR-3180-3p and miR-124-3p may Function as Noninvasive Biomarkers of Cisplatin Resistance in Gastric Cancer. Clin. Lab. 2020, 66. [Google Scholar] [CrossRef]
- Valentini, V.; Zelli, V.; Gaggiano, E.; Silvestri, V.; Rizzolo, P.; Bucalo, A.; Calvieri, S.; Grassi, S.; Frascione, P.; Donati, P. miRNAs as potential prognostic biomarkers for metastasis in thin and thick primary cutaneous melanomas. Anticancer Res. 2019, 39, 4085–4093. [Google Scholar] [CrossRef]
- Loriot, A.; Van Tongelen, A.; Blanco, J.; Klaessens, S.; Cannuyer, J.; van Baren, N.; Decottignies, A.; De Smet, C. A novel cancer-germline transcript carrying pro-metastatic miR-105 and TET-targeting miR-767 induced by DNA hypomethylation in tumors. Epigenetics 2014, 9, 1163–1171. [Google Scholar] [CrossRef]
- Jia, M.; Li, Z.; Pan, M.; Tao, M.; Wang, J.; Lu, X. LINC-PINT Suppresses the Aggressiveness of Thyroid Cancer by Downregulating miR-767-5p to Induce TET2 Expression. Mol. Ther. Nucleic Acids 2020, 22, 319–328. [Google Scholar] [CrossRef]
- Zhang, L.; Geng, Z.; Wan, Y.; Meng, F.; Meng, X.; Wang, L. Functional analysis of miR-767-5p during the progression of hepatocellular carcinoma and the clinical relevance of its dysregulation. Histochem. Cell Biol. 2020, 154, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Li, H.; Shi, M.; Zhu, Y.; Ma, Y.; Zhong, Y.; Xiong, C.; Chen, H.; Peng, C. TET1-mediated DNA hydroxymethylation activates inhibitors of the Wnt/β-catenin signaling pathway to suppress EMT in pancreatic tumor cells. J. Exp. Clin. Cancer Res. 2019, 38, 1–17. [Google Scholar] [CrossRef]
- Gajos-Michniewicz, A.; Czyz, M. WNT signaling in melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Webster, M.R.; Marchbank, K.; Behera, R.; Ndoye, A.; Kugel, C.H.; Dang, V.M.; Appleton, J.; O’Connell, M.P.; Cheng, P. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 2016, 532, 250–254. [Google Scholar] [CrossRef]
- Regad, T. Molecular and cellular pathogenesis of melanoma initiation and progression. Cell. Mol. Life Sci. 2013, 70, 4055–4065. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hu, W.; Fu, W.; Dai, L.; Jiang, Z.; Zhong, S.; Deng, B.; Zhao, J. HGF/MET regulated epithelial-mesenchymal transitions and metastasis By FOSL2 in non-small cell lung cancer. OncoTargets Ther. 2019, 12, 9227. [Google Scholar] [CrossRef] [PubMed]
- Kuphal, S.; Wallner, S.; Bosserhoff, A.K. Impact of LIF (leukemia inhibitory factor) expression in malignant melanoma. Exp. Mol. Pathol. 2013, 95, 156–165. [Google Scholar] [CrossRef]
- Maruta, S.; Takiguchi, S.; Ueyama, M.; Kataoka, Y.; Oda, Y.; Tsuneyoshi, M.; Iguchi, H. A role for leukemia inhibitory factor in melanoma-induced bone metastasis. Clin. Exp. Metastasis 2009, 26, 133. [Google Scholar] [CrossRef] [PubMed]
- Larribère, L.; Kuphal, S.; Sachpekidis, C.; Hüser, L.; Bosserhoff, A.; Utikal, J. Targeted therapy-resistant melanoma cells acquire transcriptomic similarities with human melanoblasts. Cancers 2018, 10, 451. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linck-Paulus, L.; Lämmerhirt, L.; Völler, D.; Meyer, K.; Engelmann, J.C.; Spang, R.; Eichner, N.; Meister, G.; Kuphal, S.; Bosserhoff, A.K. Learning from Embryogenesis—A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development. J. Clin. Med. 2021, 10, 2259. https://doi.org/10.3390/jcm10112259
Linck-Paulus L, Lämmerhirt L, Völler D, Meyer K, Engelmann JC, Spang R, Eichner N, Meister G, Kuphal S, Bosserhoff AK. Learning from Embryogenesis—A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development. Journal of Clinical Medicine. 2021; 10(11):2259. https://doi.org/10.3390/jcm10112259
Chicago/Turabian StyleLinck-Paulus, Lisa, Lisa Lämmerhirt, Daniel Völler, Katharina Meyer, Julia C. Engelmann, Rainer Spang, Norbert Eichner, Gunter Meister, Silke Kuphal, and Anja Katrin Bosserhoff. 2021. "Learning from Embryogenesis—A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development" Journal of Clinical Medicine 10, no. 11: 2259. https://doi.org/10.3390/jcm10112259
APA StyleLinck-Paulus, L., Lämmerhirt, L., Völler, D., Meyer, K., Engelmann, J. C., Spang, R., Eichner, N., Meister, G., Kuphal, S., & Bosserhoff, A. K. (2021). Learning from Embryogenesis—A Comparative Expression Analysis in Melanoblast Differentiation and Tumorigenesis Reveals miRNAs Driving Melanoma Development. Journal of Clinical Medicine, 10(11), 2259. https://doi.org/10.3390/jcm10112259





