Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study
Abstract
:1. Introduction
2. Methods
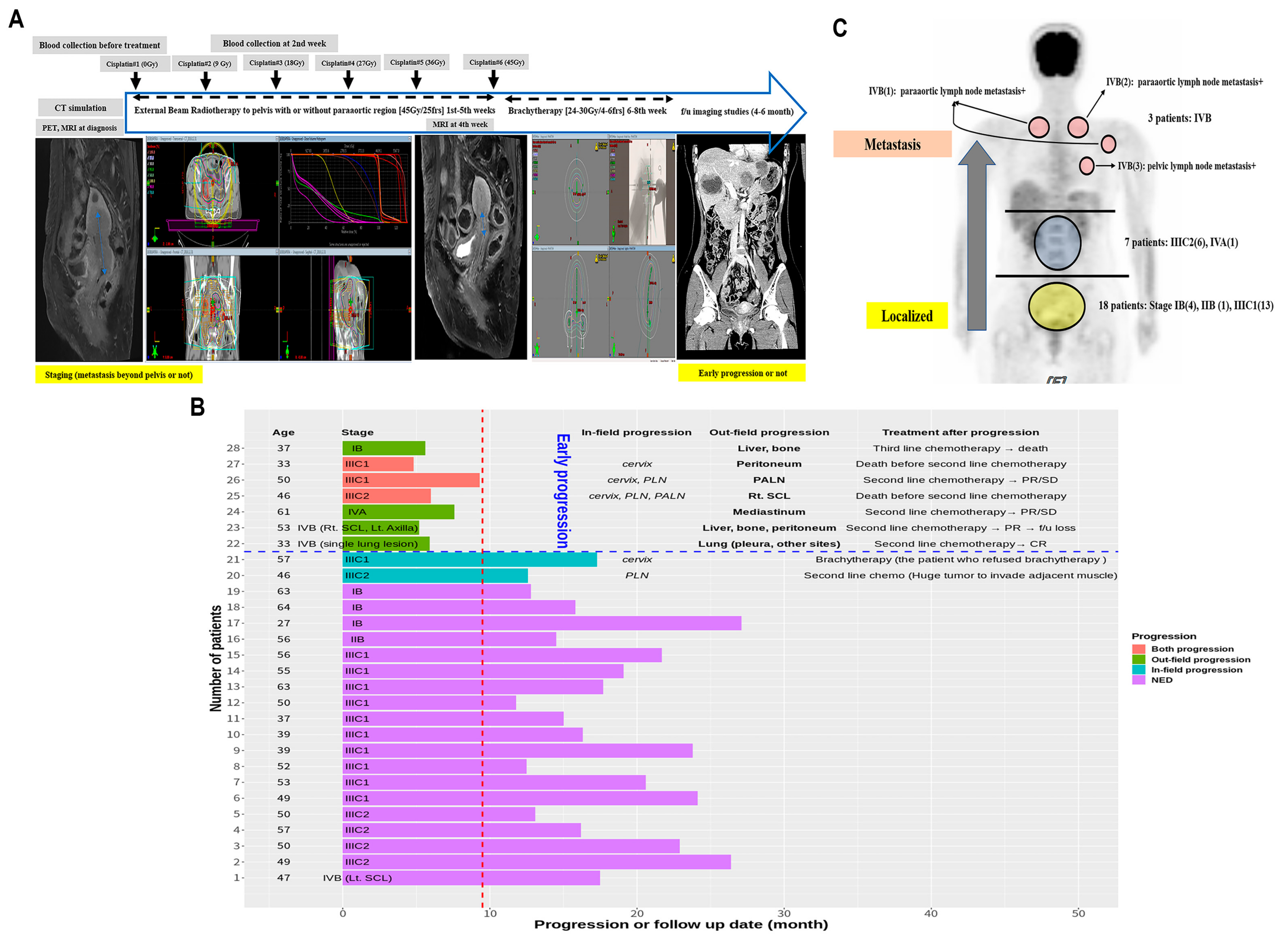
2.1. Patients
2.2. Log2FC and Power Transformation for miRNA and mRNA
2.3. Selection of RNAs to Predict Clinical End Points
2.4. Network Analysis
2.5. Ingenuity Pathway Analysis
3. Results
3.1. Early Progression and Tumor Stage
3.2. Selection of miRNAs That Predicted Early Progression and Tumor Stage Better
3.3. Selection of RNAs According to Disease and Biological Functions Using IPA
3.4. Association between Unresolved Inflammation and Early Progression in miRNA-mRNA Simplified Network Analysis
3.5. Association between Systemic Tumor Microenvironment and Metastasis in miRNA-mRNA Simplified Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tohme, S.; Simmons, R.L.; Tsung, A. Surgery for cancer: A trigger for metastases. Cancer Res. 2017, 77, 1548–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagiannis, G.S.; Condeelis, J.S.; Oktay, M.H. Chemotherapy-Induced Metastasis: Molecular Mechanisms, Clinical Manifestations, Therapeutic Interventions. Cancer Res. 2019, 79, 4567–4576. [Google Scholar] [CrossRef] [Green Version]
- Vilalta, M.; Rafat, M.; Graves, E.E. Effects of radiation on metastasis and tumor cell migration. Cell. Mol. Life Sci. 2016, 73, 2999–3007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Heo, J.; Chun, M.; Oh, Y.T.; Noh, O.K.; Kim, L. Psychiatric comorbidities among ovarian cancer survivors in South Korea: A na-tionwide population-based, longitudinal study. Psychooncology 2018, 27, 1021–1026. [Google Scholar] [CrossRef] [PubMed]
- Fleshner, M.; Crane, C.R. Exosomes, DAMPs and miRNA: Features of Stress Physiology and Immune Homeostasis. Trends Immunol. 2017, 38, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Grigsby, P.W.; Massad, L.S.; Mutch, D.G.; Powell, M.A.; Thaker, P.H.; McCourt, C.; Hagemann, A.; Fuh, K.; Kuroki, L.; Schwarz, J.K.; et al. FIGO 2018 staging criteria for cervical cancer: Impact on stage migration and survival. Gynecol. Oncol. 2020, 157, 639–643. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2008, 9, 239–252. [Google Scholar] [CrossRef]
- Wang, M.; Yu, F.; Ding, H.; Wang, Y.; Li, P.; Wang, K. Emerging Function and Clinical Values of Exosomal MicroRNAs in Cancer. Mol. Ther. Nucleic Acids 2019, 16, 791–804. [Google Scholar] [CrossRef] [Green Version]
- Cho, O.; Noh, O.K.; Oh, Y.-T.; Chang, S.-J.; Ryu, H.-S.; Lee, E.J.; Chun, M. Hematological parameters during concurrent chemoradiotherapy as potential prognosticators in patients with stage IIB cervical cancer. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Cho, O.; Chun, M.; Oh, Y.-T.; Noh, O.K.; Chang, S.-J.; Ryu, H.-S.; Lee, E.J. Prognostic implication of simultaneous anemia and lymphopenia during concurrent chemoradiotherapy in cervical squamous cell carcinoma. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef] [Green Version]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Aqeilan, R.I.; Calin, G.A.; Croce, C.M. miR-15a and miR-16-1 in cancer: Discovery, function and future perspectives. Cell Death Differ. 2009, 17, 215–220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jozic, I.; Stojadinovic, O.; Kirsner, R.S.; Tomic-Canic, M. Skin under the (Spot)-Light: Cross-Talk with the Central Hypothalamic–Pituitary–Adrenal (HPA) Axis. J. Investig. Dermatol. 2015, 135, 1469–1471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maurice, D.H.; Ke, H.; Ahmad, F.; Wang, Y.; Chung, J.; Manganiello, V.C. Advances in targeting cyclic nucleotide phosphodiesterases. Nat. Rev. Drug Discov. 2014, 13, 290–314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, J.; Liu, C.-J.; Li, Z. ADAMTS-18: A metalloproteinase with multiple functions. Front. Biosci. 2014, 19, 1456–1467. [Google Scholar] [CrossRef] [Green Version]
- Van der Spek, A.H.; Fliers, E.; Boelen, A. Thyroid hormone metabolism in innate immune cells. J. Endocrinol. 2017, 232, R67–R81. [Google Scholar] [CrossRef]
- Krashin, E.; Piekiełko-Witkowska, A.; Ellis, M.; Ashur-Fabian, O. Thyroid Hormones and Cancer: A Comprehensive Review of Preclinical and Clinical Studies. Front. Endocrinol. 2019, 10, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergink, S.; Salomons, F.A.; Hoogstraten, D.; Groothuis, T.A.; de Waard, H.; Wu, J.; Li, Y.; Citterio, E.; Houtsmuller, A.B.; Neefjes, J.; et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006, 20, 1343–1352. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Bao, Y.; Ma, M.; Zhang, S.; Zhang, Y.; Yuan, M.; Liu, B.; Yang, Y.; Cui, W.; Ansong, E.; et al. Clinical significance of the correlation between PLCE 1 and PRKCA in esophageal inflammation and esophageal carcinoma. Oncotarget 2017, 8, 33285–33299. [Google Scholar] [CrossRef]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, C.V.; Janakiram, N.B.; Mohammed, A. Molecular Pathways: Mucins and Drug Delivery in Cancer. Clin. Cancer Res. 2017, 23, 1373–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cara, F.; Andreoletti, P.; Trompier, D.; Vejux, A.; Bülow, M.H.; Sellin, J.; Lizard, G.; Cherkaoui-Malki, M.; Savary, S. Peroxisomes in Immune Response and Inflammation. Int. J. Mol. Sci. 2019, 20, 3877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catrysse, L.; Vereecke, L.; Beyaert, R.; van Loo, G. A20 in inflammation and autoimmunity. Trends Immunol. 2014, 35, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Jeanson, L.; Copin, B.; Papon, J.-F.; Moal, F.D.-L.; Duquesnoy, P.; Montantin, G.; Cadranel, J.; Corvol, H.; Coste, A.; Désir, J.; et al. RSPH3 Mutations Cause Primary Ciliary Dyskinesia with Central-Complex Defects and a Near Absence of Radial Spokes. Am. J. Hum. Genet. 2015, 97, 153–162. [Google Scholar] [CrossRef] [Green Version]
- Cockx, M.; Gouwy, M.; Ruytinx, P.; Lodewijckx, I.; Van Hout, A.; Knoops, S.; Pörtner, N.; Ronsse, I.; Vanbrabant, L.; Godding, V.; et al. Monocytes from patients with Primary Ciliary Dyskinesia show enhanced inflammatory properties and produce higher levels of pro-inflammatory cytokines. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.K.; Strickland, S. A critical role for plasminogen in inflammation. J. Exp. Med. 2020, 217, e20191865. [Google Scholar] [CrossRef] [PubMed]
- Ricciotti, E.; FitzGerald, G.A. Prostaglandins and inflammation. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 986–1000. [Google Scholar] [CrossRef]
- Spann, N.J.; Garmire, L.X.; McDonald, J.G.; Myers, D.S.; Milne, S.B.; Shibata, N.; Reichart, D.; Fox, J.N.; Shaked, I.; Heudobler, D.; et al. Regulated accumulation of desmosterol inte-grates macrophage lipid metabolism and inflammatory responses. Cell 2012, 151, 138–152. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Zhang, X.; Li, Z.; Wei, L.; Peng, Q.; Liu, C.; Wu, Y.; Yan, Q.; Ma, J. A significant role of transcription factors E2F in inflammation and tumor-igenesis of nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2020, 524, 816–824. [Google Scholar] [CrossRef]
- Luyer, M.D.; Greve, J.W.M.; Hadfoune, M.; Jacobs, J.A.; Dejong, C.H.; Buurman, W.A. Nutritional stimulation of cholecystokinin re-ceptors inhibits inflammation via the vagus nerve. J. Exp. Med. 2005, 202, 1023–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yurchenko, M.; Skjesol, A.; Ryan, L.; Richard, G.M.; Kandasamy, R.K.; Wang, N.; Terhorst, C.; Husebye, H.; Espevik, T. SLAMF1 is required for TLR4-mediated TRAM-TRIF–dependent signaling in human macrophages. J. Cell Biol. 2018, 217, 1411–1429. [Google Scholar] [CrossRef] [PubMed]
- Van der Poel, C.E.; Spaapen, R.M.; van de Winkel, J.G.; Leusen, J.H. Functional characteristics of the high affinity IgG receptor, FcγRI. J. Immunol. 2011, 186, 2699–2704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Killick, J.; Morisse, G.; Sieger, D.; Astier, A.L. Complement as a regulator of adaptive immunity. Semin. Immunopathol. 2018, 40, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Hou, W.L.; Yin, J.; Alimujiang, M.; Yu, X.Y.; Ai, L.G.; Bao, Y.Q.; Liu, F.; Jia, W. Inhibition of mitochondrial complex I improves glucose metabo-lism independently of AMPK activation. J. Cell. Mol. Med. 2018, 22, 1316–1328. [Google Scholar] [PubMed] [Green Version]
- E Calvo, S.; Tucker, E.J.; Compton, A.G.; Kirby, D.M.; Crawford, G.; Burtt, N.P.; Rivas, M.; Guiducci, C.; Bruno, D.L.; Goldberger, O.A.; et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nat. Genet. 2010, 42, 851–858. [Google Scholar] [CrossRef]
- Gómez-Valadés, A.G.; Méndez-Lucas, A.; Vidal-Alabró, A.; Blasco, F.X.; Chillon, M.; Bartrons, R.; Bermudez, J.; Perales, J.C. Pck1 Gene Silencing in the Liver Improves Glycemia Control, Insulin Sensitivity, and Dyslipidemia in db/db Mice. Diabetes 2008, 57, 2199–2210. [Google Scholar] [CrossRef] [Green Version]
- Proctor, K.M.; Miller, S.C.M.; Bryant, N.J.; Gould, G.W. Syntaxin 16 controls the intracellular sequestration of GLUT4 in 3T3-L1 adi-pocytes. Biochem. Biophys. Res. Commun. 2006, 347, 433–438. [Google Scholar] [CrossRef]
- Abdel-Halim, S.M.; Al Madhoun, A.; Nizam, R.; Melhem, M.; Cherian, P.; Al-Khairi, I.; Haddad, D.; Abu-Farha, M.; Abubaker, J.; Bitar, M.S.; et al. Increased Plasma Levels of Adenylate Cyclase 8 and cAMP Are Associated with Obesity and Type 2 Diabetes: Results from a Cross-Sectional Study. Biology 2020, 9, 244. [Google Scholar] [CrossRef]
- Moritoh, Y.; Oka, M.; Yasuhara, Y.; Hozumi, H.; Iwachidow, K.; Fuse, H.; Tozawa, R. Inositol Hexakisphosphate Kinase 3 Regulates Me-tabolism and Lifespan in Mice. Sci. Rep. 2016, 6, 32072. [Google Scholar] [CrossRef]
- Bernhard, F.; Landgraf, K.; Klöting, N.; Berthold, A.; Büttner, P.; Friebe, D.; Kiess, W.; Kovacs, P.; Blüher, M.; Körner, A. Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia 2012, 56, 311–322. [Google Scholar] [CrossRef] [Green Version]
- Uboveja, A.; Satija, Y.K.; Siraj, F.; Sharma, I.; Saluja, D. p73—NAV3 axis plays a critical role in suppression of colon cancer metas-tasis. Oncogenesis 2020, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Z.; Xia, X.Y.; Zhu, F.; Shen, H.; Song, K.; Shang, Z.J. Correlation of deregulated like-acetylglucosaminyl transferase and ab-errant α-dystroglycan expression with human tongue cancer metastasis. J. Oral Maxillofac. Surg. 2014, 72, 1106–1118. [Google Scholar] [CrossRef] [PubMed]
- Van den Boom, J.; Wolter, M.; Blaschke, B.; Knobbe, C.B.; Reifenberger, G. Identification of novel genes associated with astrocytoma progression using suppression subtractive hybridization and real-time reverse transcription-polymerase chain reaction. Int. J. Cancer 2006, 119, 2330–2338. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Cao, Y.; Zhou, H.; Li, Y.; He, B.; Zhou, X.; Nie, Z.; Liang, L.; Liu, Y.; Ye, L. Knockdown of CCNO decreases the tumorigenicity of gastric cancer by induc-ing apoptosis. Onco Targets Ther. 2018, 11, 7471–7481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, M.; O’Brien, J.A.; Bernaudo, S.; Shawer, H.; Ye, G.; Brkić, J.; Amleh, A.; Vanderhyden, B.C.; Refky, B.; Yang, B.B.; et al. miR-590-3p Promotes Ovarian Cancer Growth and Metasta-sis via a Novel FOXA2–Versican Pathway. Cancer Res. 2018, 78, 4175–4190. [Google Scholar] [CrossRef] [Green Version]
- Valer, J.A.; Sánchez-De-Diego, C.; Pimenta-Lopes, C.; Rosa, J.L.; Ventura, F. ACVR1 Function in Health and Disease. Cells 2019, 8, 1366. [Google Scholar] [CrossRef] [Green Version]
- Elamin, Y.Y.; Rafee, S.; Osman, N.; O’byrne, K.J.; Gately, K. Thymidine Phosphorylase in Cancer; Enemy or Friend? Cancer Microenviron. 2016, 9, 33–43. [Google Scholar] [CrossRef] [Green Version]
- Cicchese, J.M.; Evans, S.; Hult, C.; Joslyn, L.R.; Wessler, T.; Millar, J.A.; Marino, S.; Cilfone, N.A.; Mattila, J.T.; Linderman, J.J.; et al. Dynamic balance of pro- and anti-inflammatory signals controls disease and limits pathology. Immunol. Rev. 2018, 285, 147–167. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Cho, O.; Chun, M.; Chang, S.J.; Oh, Y.T.; Noh, O.K. Prognostic value of severe lymphopenia during pelvic concurrent chemo-radiotherapy in cervical cancer. Anticancer Res. 2016, 36, 3541–3547. [Google Scholar] [PubMed]
- Lee, S.; Cho, O.; Chun, M.; Chang, S.J.; Kong, T.W.; Lee, E.J.; Lee, Y. Association Between Radiation Tolerance of Lymphocytes and Clinical Outcomes in Cervical Cancer. In Vivo 2019, 33, 2191–2198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Liu, M.; Wang, Z.; Huang, M.; Xu, N.; Wu, L. Serum microRNAs related with chemoradiotherapy resistance in ad-vanced-stage cervical squamous cell carcinoma. Transl. Oncol. 2017, 10, 378–384. [Google Scholar] [CrossRef] [PubMed]




| All | Early Progression | p | ||
|---|---|---|---|---|
| (N = 28) | No (N = 21) | Yes (N = 7) | ||
| Age (years) (IQR) | 50.0 (42.5;56.0) | 50.0 (47.0;56.0) | 46.0 (35.0;51.5) | 0.184 |
| FIGO staging 2018, n (%) | 0.298 | |||
| - IB | 4 (14.3%) | 3 (14.3%) | 1 (14.3%) | |
| - IIB-IIIC1 | 14 (50.0%) | 12 (57.1%) | 2 (28.6%) | |
| - IIIC2-IVA | 7 (25.0%) | 5 (23.8%) | 2 (28.6%) | |
| - IVB | 3 (10.7%) | 1 (4.8%) | 2 (28.6%) | |
| Pathology, n (%) | 0.017 | |||
| - Adenocarcinoma | 4 (14.3%) | 1 (4.8%) | 3 (42.9%) | |
| - Adenosquamous cell carcinoma | 1 (3.6%) | 0 (0.0%) | 1 (14.3%) | |
| - Unclassified carcinoma | 1 (3.6%) | 1 (4.8%) | 0 (0.0%) | |
| - Squamous cell carcinoma | 22 (78.6%) | 19 (90.5%) | 3 (42.9%) | |
| RT field, n (%) | 0.815 | |||
| Pelvis | 19 (67.9%) | 15 (71.4%) | 4 (57.1%) | |
| Pelvis with para-aortic region | 9 (32.1%) | 6 (28.6%) | 3 (42.9%) | |
| Total dose (EQD2) (IQR) | 76.2 (72.2;84.2) | 75.5 (72.2;84.2) | 84.2 (74.2;84.2) | 0.357 |
| Intracavitary brachytherapy, n (%) | 0.483 | |||
| - No treatment | 2 (7.1%) | 1 (4.8%) Refusal | 1 (14.3%) EBRT | |
| - 24 Gy in four fractions | 10 (35.7%) | 9 (42.9%) | 1 (14.3%) | |
| - 24 Gy in six fractions | 5 (17.9%) | 4 (19.0%) | 1 (14.3%) | |
| - 25 Gy in five fractions | 1 (3.6%) | 1 (4.8%) | 0 (0.0%) | |
| - 30 Gy in six fractions | 10 (35.7%) | 6 (28.6%) | 4 (57.1%) | |
| Dexamethasone during RT, n (%) | 1.000 | |||
| No | 21 (75.0%) | 16 (76.2%) | 5 (71.4%) | |
| Yes | 7 (25.0%) | 5 (23.8%) | 2 (28.6%) | |
| Dexamethasone after RT, n (%) | 0.061 | |||
| No | 24 (85.7%) | 20 (95.2%) | 4 (57.1%) | |
| Yes | 4 (14.3%) | 1 (4.8%) | 3 (42.9%) | |
| Death, n (%) | 0.014 | |||
| No | 25 (89.3%) | 21 (100.0%) | 4 (57.1%) | |
| Yes | 3 (10.7%) | 0 (0.0%) | 3 (42.9%) | |
| Regulatory miRNAs | mRNAs | Related Function | References |
|---|---|---|---|
| Pro-inflammation | |||
| miR-1228-5p↓, miR-33a-5p↓, miR-146a-3p↑ | PDE3A↓ | Cardiac contractility↑ Vascular contractility↑ | [15] |
| miR-1228-5p↓, miR-146a-3p↑ | ADAMTS-18↓ | Platelet activation↑ | [16] |
| miR-3200-3p↓ | TG↑ | Inflammatory cytokine↑ Cancer proliferation↑ | [17,18] |
| miR-33a-5p↓ | HIST2H2AA3/4↓ | DNA damage↑ | [19] |
| miR-3200-3p↓ | PLCE1↑ | Inflammatory cytokine↑ Cancer proliferation↑ | [20] |
| miR-3200-3p↓ | GCNT3↑ | Inflammatory cytokine↑ | [21,22] |
| miR-146a-3p↑ | PHYH↑ | Peroxisome ↑ | [23] |
| miR-6815-5p↓ | TNIP1↓ | Anti-inflammation↓ | [24] |
| miR-6815-5p↓ | RSPH3↓ | Inflammatory cytokine↑ | [25,26] |
| Anti-inflammation | |||
| miR-1228-5p↓, miR-33a-5p↓, miR-146a-3p↑ | PDE3A↓ | Platelet aggregation↓ | [15] |
| miR-146a-3p↑ | PLAUR↓ | Plasminogen activation↓ | [27] |
| miR-33a-5p↓ | PTGS1↓ | Prostaglandins↓ -> anti-inflammation↑ | [28] |
| miR-1228-5p↓ | DHCR24↓ | Inflammatory gene expression↓ | [29] |
| miR-146a-3p↑ | E2F2↓ | Inflammatory signal↓ | [30] |
| miR-3200-3p↓ | CCKBR↑ | Vagus nerve stimulation -> anti-inflammation↑ | [31] |
| Cell mediated immunity↓ | |||
| miR-1228-5p↓, miR-146a-3p↑ | SLAMF1↑ | Activation of macrophages↓ | [32] |
| miR-1228-5p↓, miR-3200-3p↓ | FCGR1A↓ | Antigen presentation↓ | [33] |
| miR-1228-5p↓ | C1QB↓ | Antigen presentation↓ | [34] |
| Blood glucose↑ | |||
| miR-3200-3p↓ | NUBPL↑ | Mitochondrial complex 1↑ -> Blood glucose↑ | [35,36] |
| miR-1228-5p↓, miR-146a-3p↑ | PCK1↑ | Blood glucose↑ | [37] |
| miR-1228-5p↓, miR-146a-3p↑ | STX16↓ | Intracellular glucose transport↓ | [38] |
| miR-3200-3p↓ | ADCY8↑ | Obese and type 2 diabetes | [39] |
| miR-3200-3p↓ | IP6K3↑ | Blood glucose↑ | [40] |
| miR-3200-3p↓ | NEGR1↑ | Obese and insulin resistance↑ | [41] |
| Cancer progression | |||
| miR-1228-5p↓, miR-146a-3p↑ | NAV3↓ | Cancer metastasis↑ | [42] |
| miR-1228-5p↓ | LARGE1↓ | Cancer metastasis↑ | [43] |
| miR-33a-5p↓ | PSD3↓ | Cancer proliferation↑ | [44] |
| miR-146a-3p↑ | CCNO↑ | Cancer proliferation↑ | [45] |
| miR-6815-5p↓ | miR-590-3p↑ | Cancer progression↑ | [46] |
| Unclassified | |||
| miR-33a-5p↓ | ACVR1↓ | Oncogene vs. tumor suppressor gene | [47] |
| miR-3200-3p↓ | TYMP↓ | Cancer proliferation↓ vs. chemo response↓ | [48] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, O.; Kim, D.-W.; Cheong, J.-Y. Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study. J. Clin. Med. 2021, 10, 2110. https://doi.org/10.3390/jcm10102110
Cho O, Kim D-W, Cheong J-Y. Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study. Journal of Clinical Medicine. 2021; 10(10):2110. https://doi.org/10.3390/jcm10102110
Chicago/Turabian StyleCho, Oyeon, Do-Wan Kim, and Jae-Youn Cheong. 2021. "Plasma Exosomal miRNA Levels after Radiotherapy Are Associated with Early Progression and Metastasis of Cervical Cancer: A Pilot Study" Journal of Clinical Medicine 10, no. 10: 2110. https://doi.org/10.3390/jcm10102110






