Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Materials
2.2. Cloning of a Full-Length cDNA Fragment Encoding the Precursor Polypeptide of Mastoparan-AF
2.3. Peptide Synthesis
2.4. Membrane Permeabilization Assay
2.5. Hemolytic Activity Assay
2.6. Antimicrobial (Antibacterial) Activity Assay of Mastoparan-AF
2.7. Antibiotic Susceptibility Assay
2.8. Scanning Electron Microscopy (SEM)
2.9. Atomic Force Microscopy (AFM)
2.10. Sequence Alignment
2.11. Physicochemical Property Analysis
3. Results
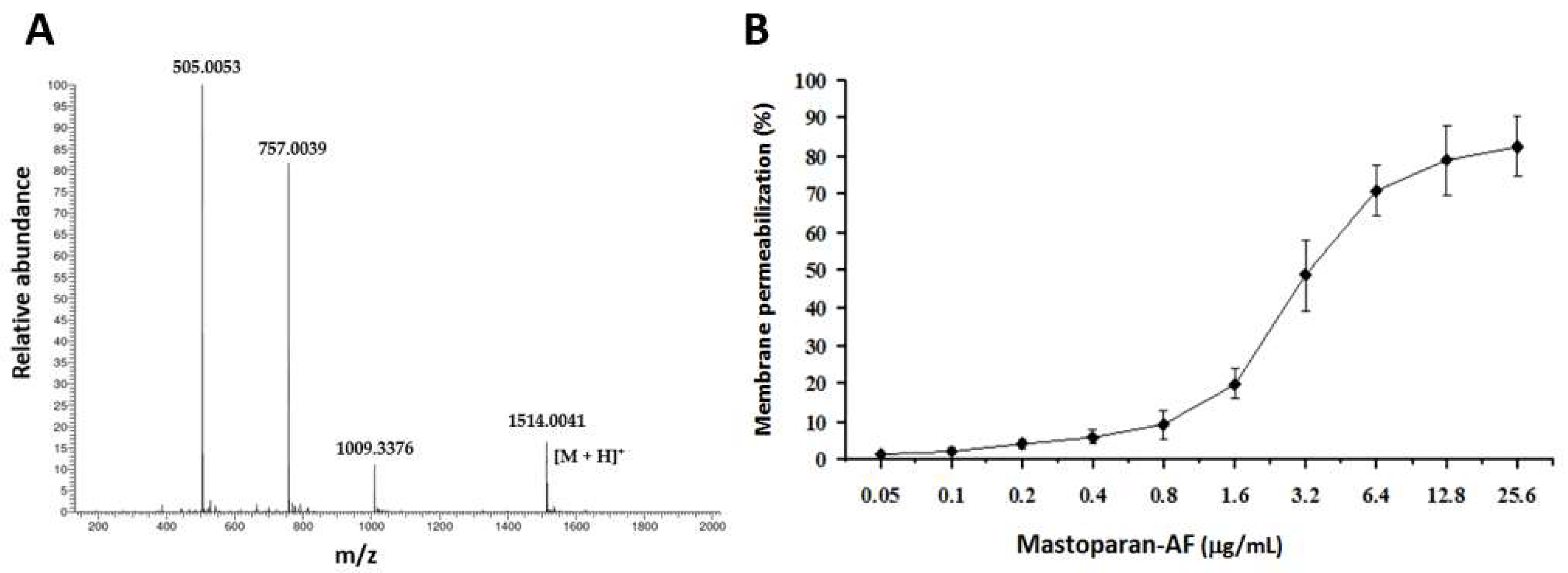
3.1. Cloning of the Full-Length cDNA Fragment Encoding Prepromastoparan-AF
3.2. Using a Synthetic Mastoparan-AF to Measure Its Membrane Permeabilization Activity
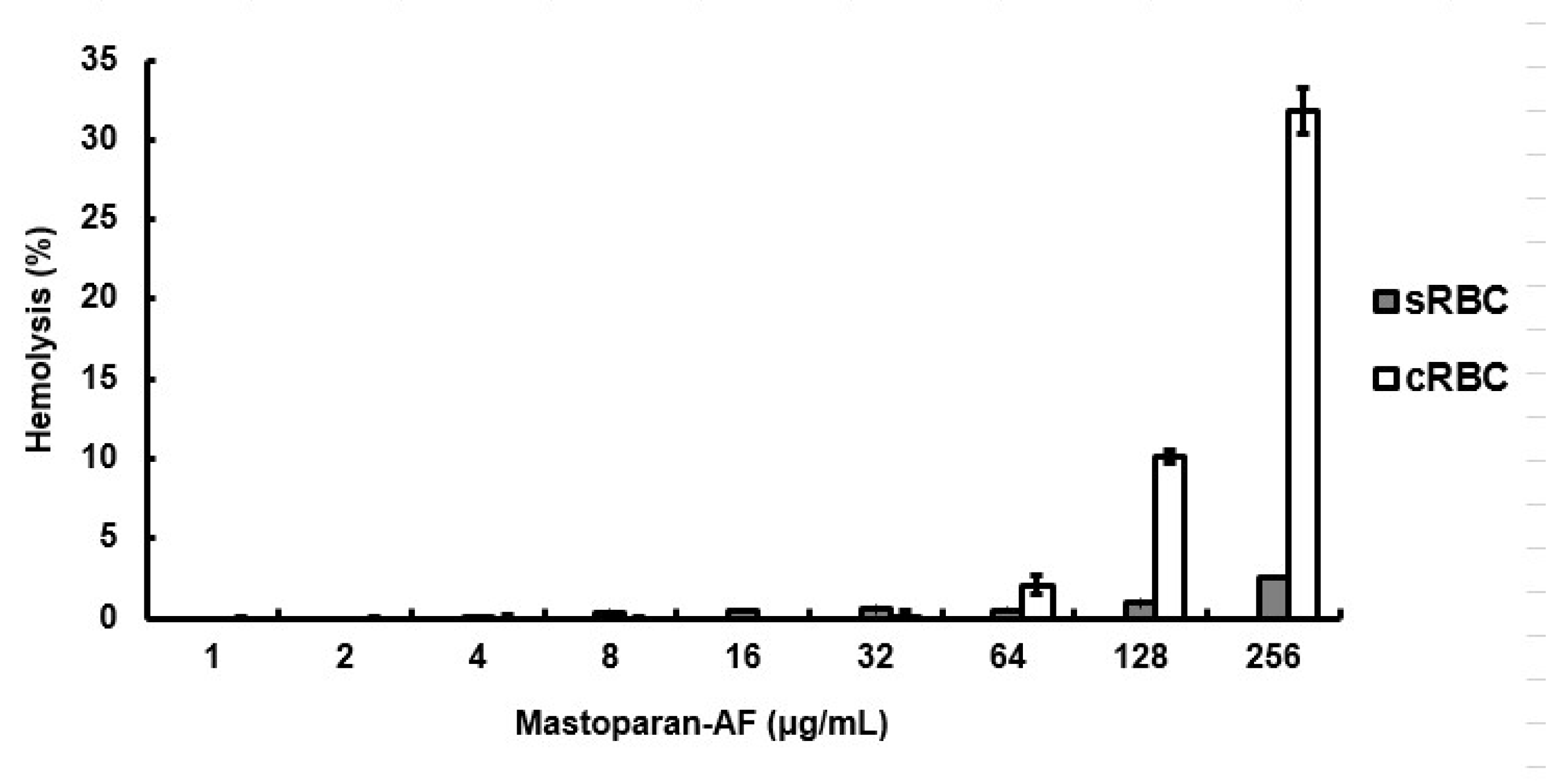
3.3. Limited Hemolytic Activity on RBCs
3.4. Antibacterial Activities of Mastoparan-AF
3.5. Antibiotics Susceptibility of Bacteria
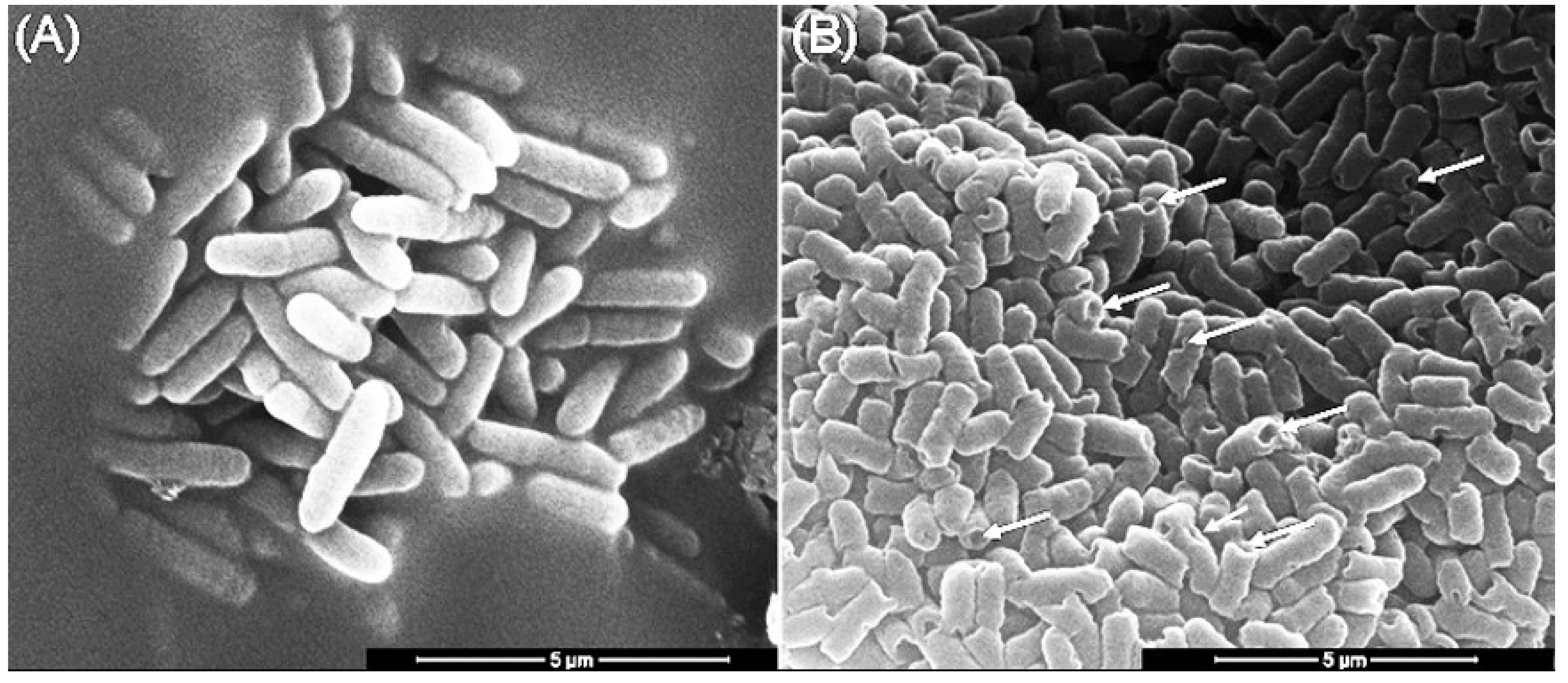
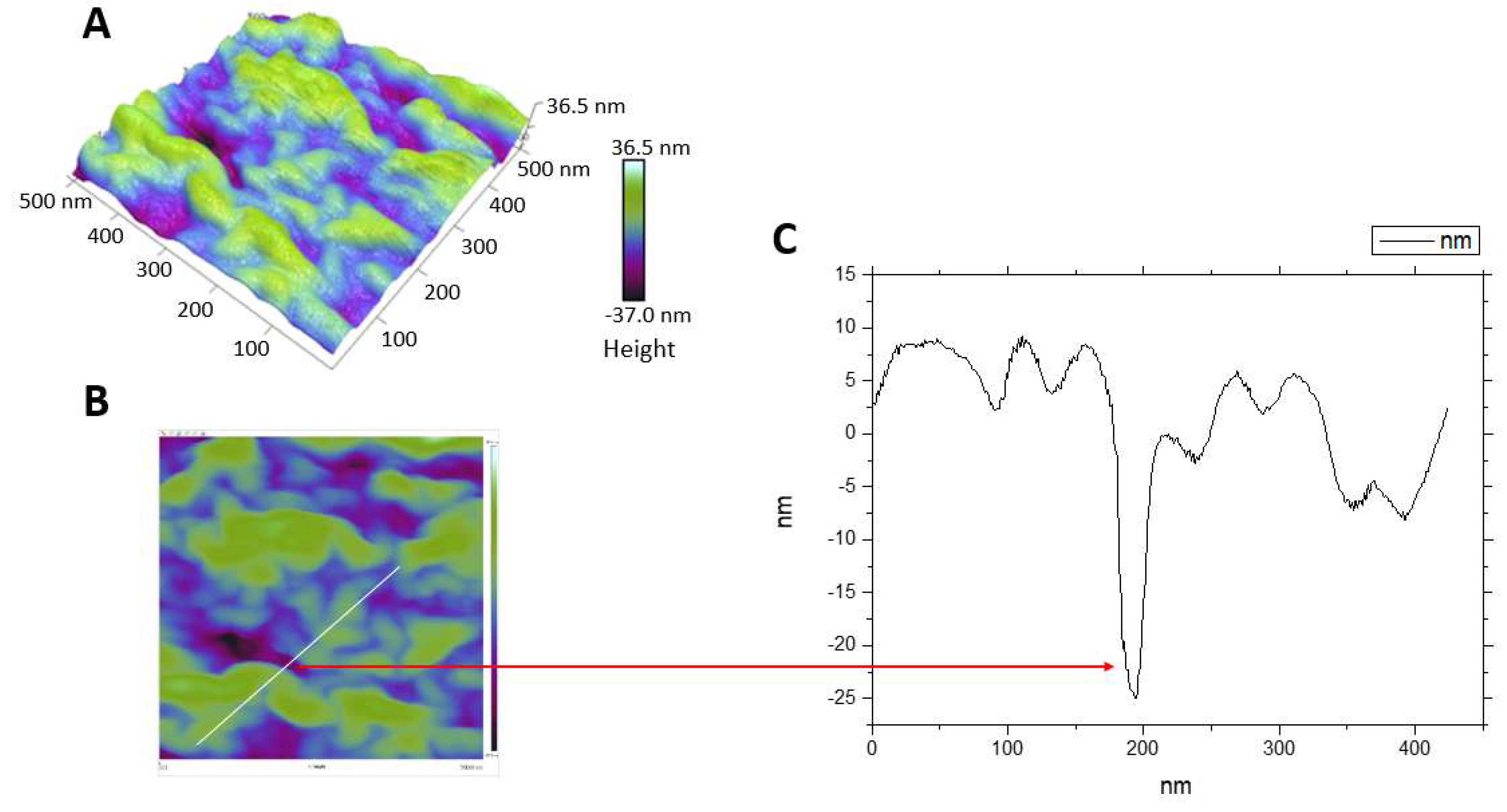
3.6. The Effect of Mastoparan-AF on the Morphology of Hemolytic E. coli O157:H7
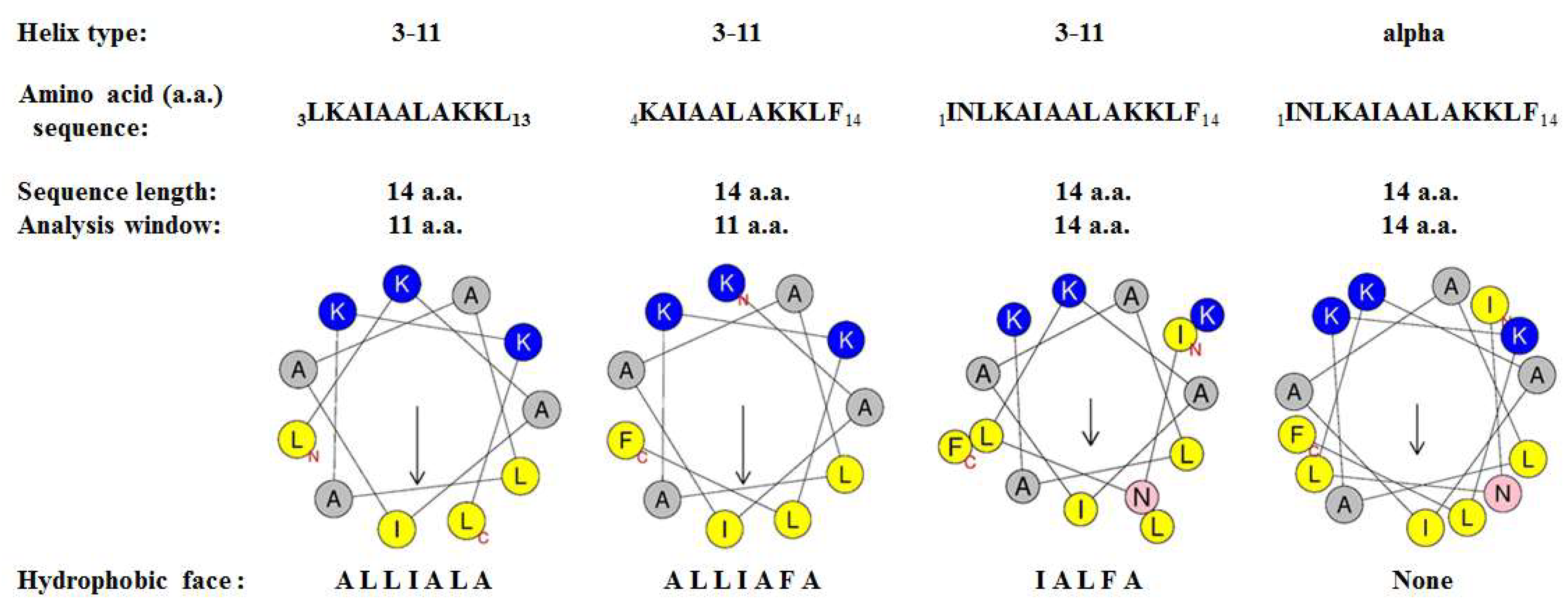
3.7. Physicochemical Properties of Mastoparan-AF and Other Mastoparans
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ghai, I.; Ghai, S. Understanding antibiotic resistance via outer membrane permeability. Infec. Drug Resist. 2018, 11, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Amézquita-López, B.A.; Soto-Beltrán, M.; Lee, B.G.; Yambao, J.C.; Quiñones, B. Isolation, genotyping and antimicrobial resistance of Shiga toxin-producing Escherichia coli. J. Microbiol. Immunol. Infect. 2018, 51, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Slutsker, L.; Ries, A.A.; Greene, K.D.; Wells, J.G.; Hutwagner, L.; Griffin, P.M. Escherichia coli O157:H7 diarrhea in the United States: Clinical and epidemiologic features. Ann. Intern. Med. 1997, 126, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhao, S.; Doyle, M.P.; Joseph, S.W. Antibiotic resistance of Escherichia coli O157:H7 and O157:NM isolated from animals, food, and humans. J. Food Prot. 1998, 61, 1511–1514. [Google Scholar] [CrossRef]
- Takagi, H.; Yamane, K.; Matsui, M.; Suzuki, S.; Ito, K. Pathotypes and drug susceptibility of Escherichia coli isolated from companion dogs in Japan. Jpn. J. Infect. Dis. 2020, 73, 253–255. [Google Scholar] [CrossRef]
- Lin, C.H.; Tzen, J.T.; Shyu, C.L.; Yang, M.J.; Tu, W.C. Structural and biological characterization of mastoparans in the venom of Vespa species in Taiwan. Peptides 2011, 32, 2027–2036. [Google Scholar] [CrossRef]
- da Silva, A.V.; De Souza, B.M.; Dos Santos Cabrera, M.P.; Dias, N.B.; Gomes, P.C.; Neto, J.R.; Stabeli, R.G.; Palma, M.S. The effects of the C-terminal amidation of mastoparans on their biological actions and interactions with membrane-mimetic systems. Biochim. Biophys. Acta 2014, 1838, 2357–2368. [Google Scholar] [CrossRef]
- Nakajima, T. Biochemistry of vespid venoms. In Handbook of Natural Toxins, Vol. 2.—Insect Poisons, Allergens, and Other Invertebrate Venoms; Tu, A.T., Ed.; Marcel Dekker: New York, NY, USA, 1984; pp. 109–133. [Google Scholar]
- Hirai, Y.; Yasuhara, T.; Yoshida, H.; Nakajima, T.; Fujino, M.; Kitada, C. A new mast cell degranulating peptide “mastoparan” in the venom of Vespula lewisii. Chem. Pharm. Bull. 1979, 27, 1942–1944. [Google Scholar] [CrossRef]
- Hirai, Y.; Yasuhara, T.; Yoshida, H.; Nakajima, T. A new mast cell degranulating peptide, mastoparan-M, in the venom of the hornet Vespa mandarinia. Biomed. Res. 1981, 2, 447–449. [Google Scholar] [CrossRef]
- Argiolas, A.; Pisano, J.J. Facilitation of phospholipase A2 activity by mastoparans, a new class of mast cell degranulating peptides from wasp venom. J. Biol. Chem. 1983, 258, 13697–13702. [Google Scholar] [CrossRef]
- Ho, C.L.; Hwang, L.L. Structure and biological activities of a new mastoparan isolated from the venom of the hornet Vespa basalis. Biochem. J. 1991, 274, 453–456. [Google Scholar] [CrossRef] [PubMed]
- Mendes, M.A.; de Souza, B.M.; Marques, M.R.; Palma, M.S. Structural and biological characterization of two novel peptides from the venom of the neotropical social wasp Agelaia pallipes. Toxicon 2004, 44, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yang, H.; Yu, H.; Li, J.; Lai, R. The mastoparanogen from wasp. Peptides 2006, 27, 3053–3057. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, X.; Yang, X.; Zhai, L.; Lu, Z.; Liu, J.; Yu, H. Antimicrobial peptides from the venoms of Vespa bicolor Fabricius. Peptides 2008, 29, 1887–1892. [Google Scholar] [CrossRef]
- Nakao, S.; Komagoe, K.; Inoue, T.; Katsu, T. Comparative study of the membrane-permeabilizing activities of mastoparans and related histamine-releasing agents in bacteria, erythrocytes, and mast cells. Biochim. Biophys. Acta 2011, 1808, 490–497. [Google Scholar] [CrossRef]
- Howl, J.; Howl, L.; Jones, S. The cationic tetradecapeptide mastoparan as a privileged structure for drug discovery: Enhanced antimicrobial properties of mitoparan analogues modified at position-14. Peptides 2018, 101, 95–105. [Google Scholar] [CrossRef]
- Hancock, R.E. Cationic peptides: Effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 2001, 1, 156–164. [Google Scholar] [CrossRef]
- Park, N.G.; Yamato, Y.; Lee, S.; Sugihara, G. Interaction of mastoparan-B from venom of a hornet in Taiwan with phospholipid bilayers and its antimicrobial activity. Biopolymers 1995, 36, 793–801. [Google Scholar] [CrossRef]
- Giménez-Andrés, M.; Čopič, A.; Antonny, B. The many faces of amphipathic helices. Biomolecules 2018, 8, 45. [Google Scholar] [CrossRef]
- Vieira-Pires, R.S.; Morais-Cabral, J.H. 310 helices in channels and other membrane proteins. J. Gen. Physiol. 2010, 136, 585–592. [Google Scholar] [CrossRef]
- Zubcevic, L.; Lee, S.Y. The role of π-helices in TRP channel gating. Curr. Opin. Struct. Biol. 2019, 58, 314–323. [Google Scholar] [CrossRef]
- Shi, Y.; Wan, M.; Fu, L.; Zhang, S.; Wang, S.; Gao, L.; Fang, W. Peptide-lipid interaction sites affect vesicles’ responses to antimicrobial peptides. Biophys J. 2018, 115, 1518–1529. [Google Scholar] [CrossRef]
- Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1999, 1462, 55–70. [Google Scholar] [CrossRef]
- Matsuzaki, K. Why and how are peptide–lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim. Biophys. Acta. 1999, 1462, 1–10. [Google Scholar] [CrossRef]
- Hammond, K.; Ryadnov, M.G.; Hoogenboom, B.W. Atomic force microscopy to elucidate how peptides disrupt membranes. Biochim. Biophys. Acta Biomembr. 2021, 1863, 183447. [Google Scholar] [CrossRef]
- Lee, V.S.Y.; Tu, W.C.; Jinn, T.R.; Peng, C.C.; Lin, L.J.; Tzen, J.T.C. Molecular cloning of the precursor polypeptide of mastoparan B and its putative processing enzyme, dipeptidyl peptidase IV, from the black-bellied hornet, Vespa basalis. Insect Mol. Biol. 2007, 16, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Moore, A.J.; Beazley, W.D.; Bibby, M.C.; Devine, D.A. Antimicrobial activity of cecropins. J. Antimicrob. Chemother. 1996, 37, 1077–1089. [Google Scholar] [CrossRef]
- Hunfeld, K.P.; Kraiczy, P.; Wichelhaus, T.A.; Schäfer, V.; Brade, V. New colorimetric microdilution method for in vitro susceptibility testing of Borrelia burgdorferi against antimicrobial substances. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Cavener, D.R. Comparison of the consensus sequence flanking translational start sites in Drosophila and vertebrates. Nucleic Acids Res. 1987, 15, 1353–1361. [Google Scholar] [CrossRef]
- Kozak, M. An analysis of 5’-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987, 15, 8125–8148. [Google Scholar] [CrossRef]
- Eipper, B.A.; Milgram, S.L.; Husten, E.J.; Yun, H.Y.; Mains, R.E. Peptidylglycine α-amidating monooxygenase: A multifunctional protein with catalytic, processing, and routing domains. Protein Sci. 1993, 2, 489–497. [Google Scholar] [CrossRef] [PubMed]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, K.; Sasaki, G.; Ogawa, J.; Miyata, T.; Su, Z.H. Phylogenetic relationships among insect orders based on three nuclear protein-coding gene sequences. Mol. Phylogenet. Evol. 2011, 58, 169–180. [Google Scholar] [CrossRef]
- Noderer, W.L.; Flockhart, R.J.; Bhaduri, A.; Diaz de Arce, A.J.; Zhang, J.; Khavari, P.A.; Wang, C.L. Quantitative analysis of mammalian translation initiation sites by FACS-seq. Mol. Syst. Biol. 2014, 10, 748. [Google Scholar] [CrossRef]
- Shirokikh, N.E.; Spirin, A.S. Poly(A) leader of eukaryotic mRNA bypasses the dependence of translation on initiation factors. Proc. Natl. Acad. Sci. USA. 2014, 105, 10738–10743. [Google Scholar] [CrossRef]
- Zhou, Z.; Ergene, C.; Lee, J.Y.; Shirley, D.J.; Carone, B.R.; Caputo, G.A.; Palermo, E.F. Sequence and dispersity are determinants of photodynamic antibacterial activity exerted by peptidomimetic oligo(thiophene)s. ACS Appl. Mater. Interfaces 2019, 11, 1896–1906. [Google Scholar] [CrossRef]
- Čierna, M.; Naumowicz, M.; Bírošová, L.; Krahulec, J.; Sokolová, R.; Kolivoška, V.; Sebechlebská, T.; Kielar, F.; Gál, M. Study of permeabilization of bacterial membrane by electrochemical methods J. Electroanal. Chem. 2020, 857, 113761. [Google Scholar] [CrossRef]
- Chen, A.; Karanastasis, A.; Casey, K.R.; Necelis, M.; Carone, B.R.; Caputo, G.A.; Palermo, E.F. Cationic molecular umbrellas as antibacterial agents with remarkable cell-type selectivity. ACS Appl. Mater. Interfaces 2020, 12, 21270–21282. [Google Scholar] [CrossRef]
- Ingraham, L.M.; Burns, C.P.; Boxer, L.A.; Baehner, R.L.; Haak, R.A. Fluidity properties and liquid composition of erythrocyte membranes in Chediak-Higashi syndrome. J. Cell Biol. 1987, 89, 510–516. [Google Scholar] [CrossRef]
- Al-Qarawi, A.A.; Mousa, H.M. Lipid concentrations in erythrocyte membranes in normal, starved, dehyrated and rehydrated camels (Camelus dromedarius), and in normal sheep (Ovis aries) and goats (Capra hircus). J. Arid. Environ. 2004, 59, 675–683. [Google Scholar] [CrossRef]
- Meincken, M.; Holroyd, D.L.; Rautenbach, M. Atomic force microscopy study of the effect of antimicrobial peptides on the cell envelope of Escherichia coli. Antimicrob. Agents Chemother. 2005, 49, 4085–4092. [Google Scholar] [CrossRef] [PubMed]
- Fantner, G.E.; Barbero, R.J.; Gray, D.S.; Belcher, A.M. Kinetics of antimicrobial peptide activity measured on individual bacterial cells using high-speed atomic force microscopy. Nat. Nanotechnol. 2010, 5, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.M.; Silva, P.M.; Franquelim, H.G.; Carvalho, F.A.; Castanho, M.A.; Santos, N.C. Antimicrobial protein rBPI21-induced surface changes on Gram-negative and Gram-positive bacteria. Nanomed. J. 2014, 10, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Zdybicka-Barabas, A.; Stączek, S.; Pawlikowska-Pawlęga, B.; Mak, P.; Luchowski, R.; Skrzypiec, K.; Mendyk, E.; Wydrych, J.; Gruszecki, W.I.; Cytryńska, M. Studies on the interactions of neutral Galleria mellonella cecropin D with living bacterial cells. Amino Acids 2019, 51, 175–191. [Google Scholar] [CrossRef]
- Dos Santos Cabrera, M.P.; Alvares, D.S.; Leite, N.B.; De Souza, B.M.; Palma, M.S.; Riske, K.A.; Neto, J.R. New insight into the mechanism of action of wasp mastoparan peptides: Lytic activity and clustering observed with giant vesicles. Langmuir 2011, 27, 10805–10813. [Google Scholar] [CrossRef]
- Mileykovskaya, E.; Dowhan, W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J. Bacteriol. 2000, 182, 1172–1175. [Google Scholar] [CrossRef]
- Strandberg, E.; Killian, J.A. Snorkeling of lysine side chains in transmembrane helices: How easy can it get? FEBS Lett. 2003, 544, 69–73. [Google Scholar] [CrossRef]
- Takechi-Haraya, Y.; Ohgita, T.; Kotani, M.; Kono, H.; Saito, C.; Tamagaki-Asahina, H.; Nishitsuji, K.; Uchimura, K.; Sato, T.; Kawano, R.; et al. Effect of hydrophobic moment on membrane interaction and cell penetration of apolipoprotein E-derived arginine-rich amphipathic α-helical peptides. Sci. Rep. 2002, 12, 4959. [Google Scholar] [CrossRef]


| Microorganisms | Mastoparan-AF (μg mL−1) | |||
|---|---|---|---|---|
| MIC a (μg mL−1) | MBC b (μg mL−1) | |||
| Range | M c | Range | M | |
| Gram-positive bacteria | ||||
| S. aureus subsp. aureus | 16–32 | 32 | 32 | 32 |
| Gram-negative bacteria | ||||
| E. coli JM109 pAcUW21 (AmpR) d | 16 | 16 | 16 | 16 |
| E. coli O157:H7 | 16–32 | 16 | 16–32 | 32 |
| E. coli 232 e | 4–8 | 8 | 4–8 | 8 |
| E. coli 237 e | 4 | 4 | 4 | 4 |
| Penicillins | Cephalosporins | Aminoglycosides | TET | Sulfa | CHL | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | AMP b | AMX/ CLA | TIC | TIC/ CLA | CFZ | FOX | AMK | GEN | DOX | TMP/ SXT | CHL |
| E. coli strain | |||||||||||
| JM109 pAcUW21 (AmpR) | >1024 R | 32/16 R | >1024 R | >1024/2 R | 32 R | 16 I | 4 S | 1 S | 2 S | 16 R | 8 S |
| O157:H7 | >1024 R | 64/32 R | >1024 R | >1024/2 R | 8 R | 16 I | 4 S | 1 S | 16 R | >32 R | >1024 R |
| 232 | 16 I | 128/64 R | 8 S | 32/2 I | 256 R | 128 R | 4 S | 1 S | 8 I | 8 R | 8 S |
| 237 | >1024 R | 16/8 I | >1024 R | 64/2 I | 4 I | 8 S | 4 S | 1 S | 64 R | >32 R | 8 S |
| S. aureus subsp. aureus | 256 R | 8/4 R | 32 R | 256/2 R | 4 N | 8 N | 4 S | 2 S | 16 R | 0.25 S | 8 S |
| Peptide Name | Helical Type | Sequence | Analysis Window | Hydrophobicity H | Hydrophobic Moment μH | Net Charge z (+) | Hydrophobic Face |
|---|---|---|---|---|---|---|---|
| Mastoparan-AF | 3–11 | 1INLKAIAALAKKLF14 | 14 aa | 0.583 | 0.418 | 3 | IALFA |
| 3–11 | 3LKAIAALAKKL13 | 11 aa | 0.470 | 0.623 | 3 | ALLIALA | |
| 3–11 | 4KAIAALAKKLF14 | 11 aa | 0.478 | 0.625 | 3 | ALLIAFA | |
| α | 1INLKAIAALAKKLF14 | 14 aa | 0.583 | 0.400 | 3 | none | |
| Mastoparan-A | 3–11 | 1IKWKAILDAVKKVI14 | 14 aa | 0.549 | 0.532 | 3 | IVWIL |
| 3–11 | 3WKAILDAVKKV13 | 11 aa | 0.461 | 0.747 | 2 | AAVIVML | |
| 3–11 | 4KAILDAVKKVI14 | 11 aa | 0.420 | 0.718 | 2 | AAVIVIL | |
| α | 1IKWKAILDAVKKVI14 | 14 aa | 0.549 | 0.544 | 3 | none | |
| Mastoparan-B | 3–11 | 1LKLKSIVSWAKKVL14 | 14 aa | 0.561 | 0.404 | 4 | IALLV |
| 3–11 | 3LKSIVSWAKKV13 | 11 aa | 0.495 | 0.622 | 3 | WVIALV | |
| 3–11 | 4KSIVSWAKKVL14 | 11 aa | 0.495 | 0.622 | 3 | WVIALV | |
| α | 1LKLKSIVSWAKKVL14 | 14 aa | 0.561 | 0.404 | 4 | none | |
| Mastoparan-D | 3–11 | 1INLKAIAAFAKKLL14 | 14 aa | 0.583 | 0.419 | 3 | IALLA |
| 3–11 | 3LKAIAAFAKKL13 | 11 aa | 0.478 | 0.628 | 3 | AFLIALA | |
| 3–11 | 4KAIAAFAKKLL14 | 11 aa | 0.478 | 0.628 | 3 | AFLIALA | |
| α | 1INLKAIAAFAKKLL14 | 14 aa | 0.583 | 0.402 | 3 | none | |
| Mastoparan-M | 3–11 | 1INLKAIAALAKKLL14 | 14 aa | 0.576 | 0.416 | 3 | IALLA |
| 3–11 | 3LKAIAALAKKL13 | 11 aa | 0.470 | 0.623 | 3 | ALLIALA | |
| 3–11 | 4KAIAALAKKLL14 | 11 aa | 0.470 | 0.623 | 3 | ALLIALA | |
| α | 1INLKAIAALAKKLL14 | 14 aa | 0.576 | 0.399 | 3 | none | |
| Mastoparan-V | 3–11 | 1INWKGIAAMAKKLL14 | 14 aa | 0.560 | 0.421 | 3 | IAWLA |
| 3–11 | 3WKGIAAMAKKL13 | 11 aa | 0.449 | 0.622 | 3 | MLIAWA | |
| 3–11 | 4KGIAAMAKKLL14 | 11 aa | 0.399 | 0.602 | 3 | MLIALA | |
| α | 1INWKGIAAMAKKLL14 | 14 aa | 0.560 | 0.419 | 3 | none |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-H.; Shyu, C.-L.; Wu, Z.-Y.; Wang, C.-M.; Chiou, S.-H.; Chen, J.-Y.; Tseng, S.-Y.; Lin, T.-E.; Yuan, Y.-P.; Ho, S.-P.; et al. Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction. Membranes 2023, 13, 251. https://doi.org/10.3390/membranes13020251
Lin C-H, Shyu C-L, Wu Z-Y, Wang C-M, Chiou S-H, Chen J-Y, Tseng S-Y, Lin T-E, Yuan Y-P, Ho S-P, et al. Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction. Membranes. 2023; 13(2):251. https://doi.org/10.3390/membranes13020251
Chicago/Turabian StyleLin, Chun-Hsien, Ching-Lin Shyu, Zong-Yen Wu, Chao-Min Wang, Shiow-Her Chiou, Jiann-Yeu Chen, Shu-Ying Tseng, Ting-Er Lin, Yi-Po Yuan, Shu-Peng Ho, and et al. 2023. "Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction" Membranes 13, no. 2: 251. https://doi.org/10.3390/membranes13020251
APA StyleLin, C.-H., Shyu, C.-L., Wu, Z.-Y., Wang, C.-M., Chiou, S.-H., Chen, J.-Y., Tseng, S.-Y., Lin, T.-E., Yuan, Y.-P., Ho, S.-P., Tung, K.-C., Mao, F. C., Lee, H.-J., & Tu, W.-C. (2023). Antimicrobial Peptide Mastoparan-AF Kills Multi-Antibiotic Resistant Escherichia coli O157:H7 via Multiple Membrane Disruption Patterns and Likely by Adopting 3–11 Amphipathic Helices to Favor Membrane Interaction. Membranes, 13(2), 251. https://doi.org/10.3390/membranes13020251








