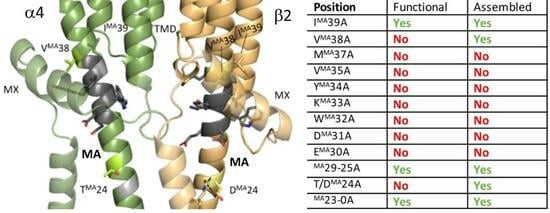
The MA Helix Is Important for Receptor Assembly and Function in the α4β2 nACh Receptor
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
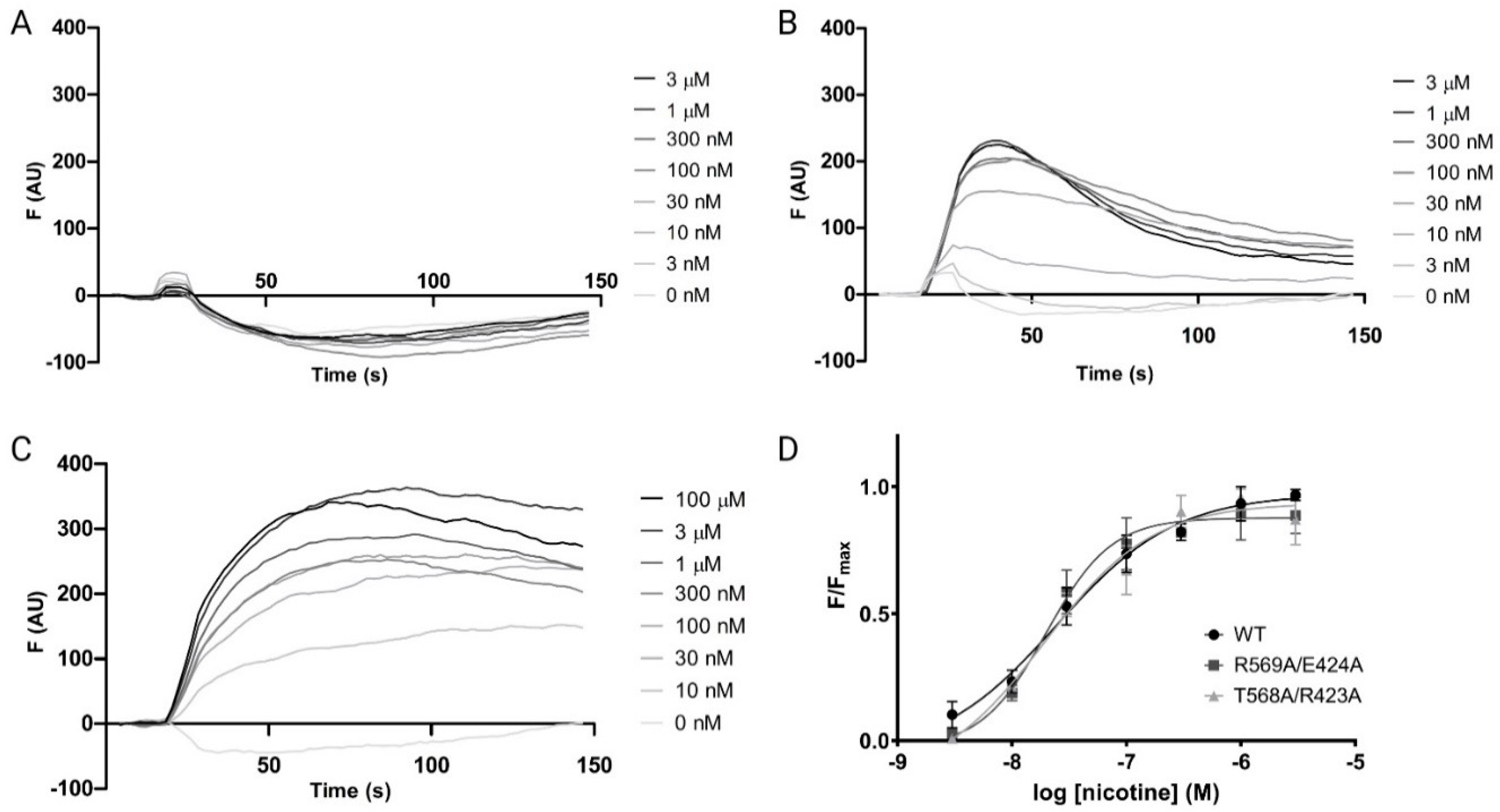
2.2. FlexStation Analysis
2.3. Radioligand Binding
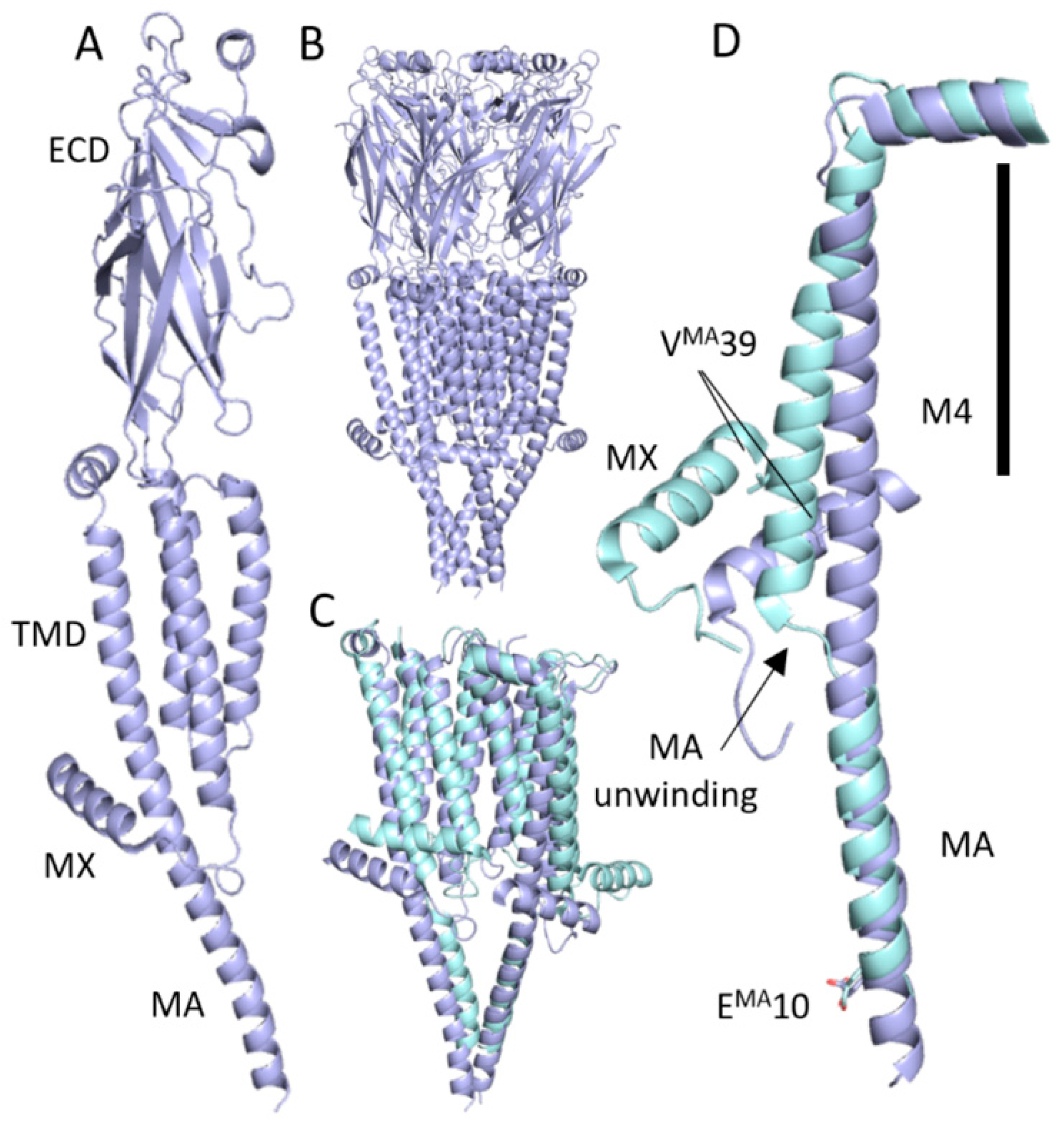
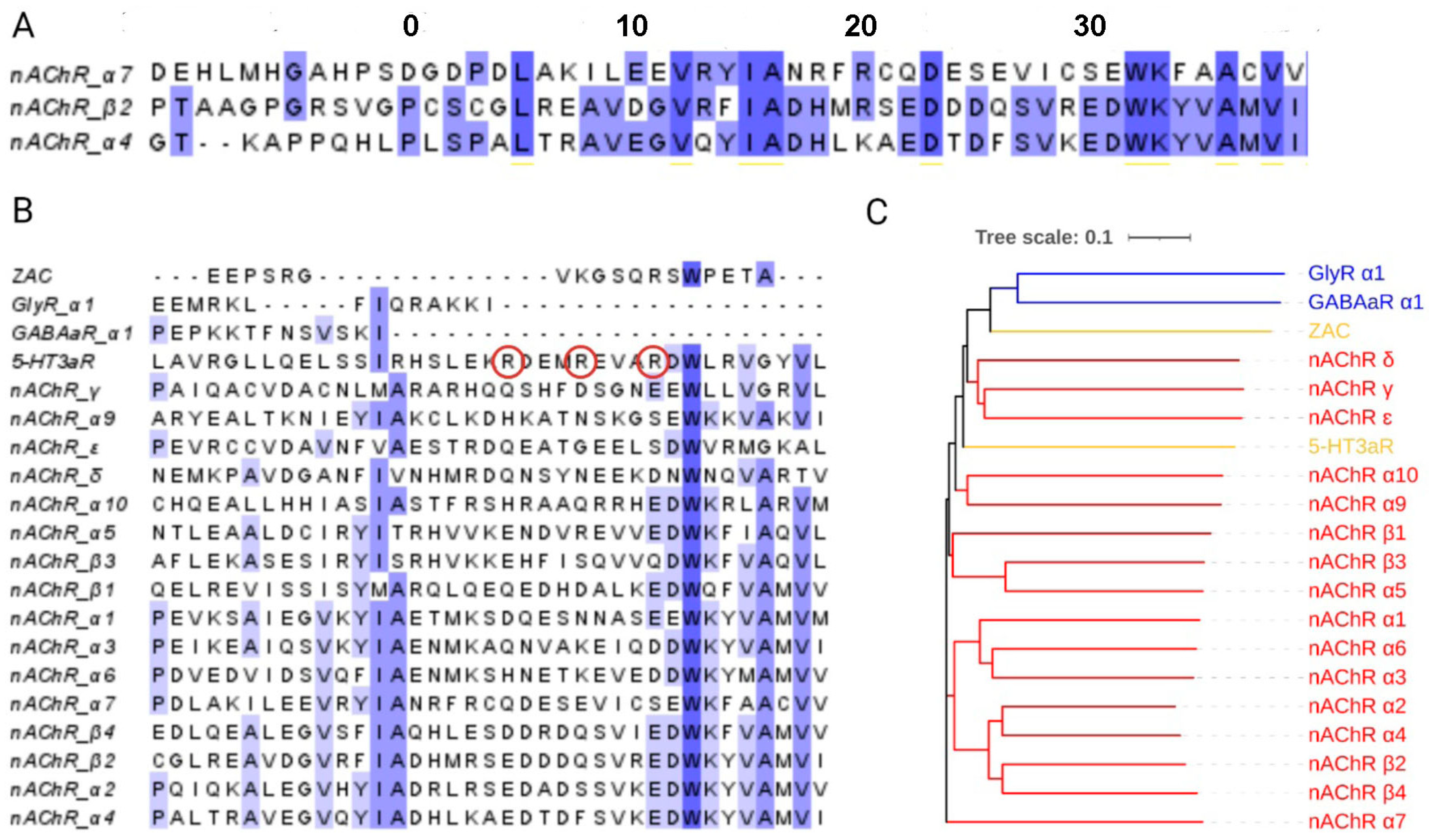
2.4. Protein Structure Prediction
3. Results
3.1. Nine Double-Alanine Mutations in the MA Helix Abolish Function
3.2. Alanine Mutations Are Less Disruptive in the α4 Than in the β2 MA Helix
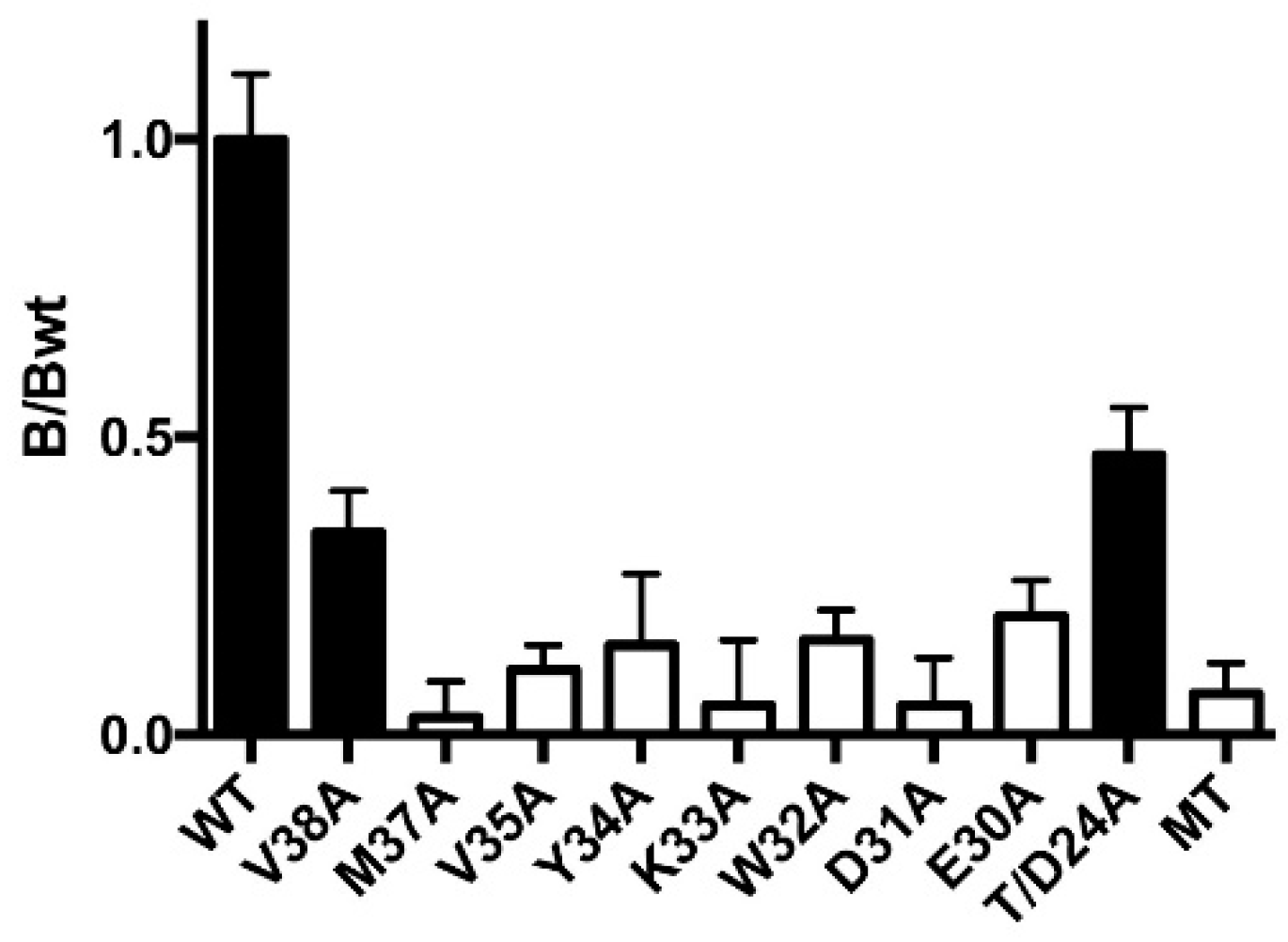
3.3. Two of the Nine Nonfunctional Mutant Receptors Are Expressed
3.4. Non-Alanine Mutations Reveal Required Characteristics of Key MA Helix Residues
4. Discussion
4.1. Two Substitutions Abolished Detectable Ion Channel Function but Not Ligand Binding
4.2. Other Alanine Substitutions in the MA Helix
4.3. Other Non-Alanine Substitutions in the MA Helix
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Bondarenko, V.; Wells, M.M.; Chen, Q.; Tillman, T.S.; Singewald, K.; Lawless, M.J.; Caporoso, J.; Brandon, N.; Coleman, J.A.; Saxena, S.; et al. Structures of highly flexible intracellular domain of human α7 nicotinic acetylcholine receptor. Nat. Commun. 2022, 13, 793. [Google Scholar] [CrossRef] [PubMed]
- Carland, J.E.; Cooper, M.A.; Livesey, M.R.; Hales, T.G.; Peters, J.A.; Lambert, J.J. Mutagenic Analysis of the Intracellular Portals of the Human 5-HT3A Receptor. J. Biol. Chem. 2013, 288, 31592–31601. [Google Scholar] [CrossRef] [PubMed]
- Cecchini, M.; Changeux, J.P. The nicotinic acetylcholine receptor and its prokaryotic homologues: Structure, conformational transitions & allosteric modulation. Neuropharmacology 2015, 96, e137–e149. [Google Scholar]
- Changeux, J.-P.; Galzi, J.-L.; Devillers-Thiéry, A.; Bertrand, D. The functional architecture of the acetylcholine nicotinic receptor explored by affinity labelling and site-directed mutagenesis. Q. Rev. Biophys. 1992, 25, 395–432. [Google Scholar] [CrossRef] [PubMed]
- Criado, M.; Mulet, J.; Gerber, S.; Sala, S.; Sala, F. A small cytoplasmic region adjacent to the fourth transmembrane segment of the α7 nicotinic receptor is essential for its biogenesis. FEBS Lett. 2011, 585, 2477–2480. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, D.A. Cys-loop neuroreceptors: Structure to the rescue? Chem. Rev. 2008, 108, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Fonck, C.; Cohen, B.N.; Nashmi, R.; Whiteaker, P.; Wagenaar, D.A.; Rodrigues-Pinguet, N.; Deshpande, P.; McKinney, S.; Kwoh, S.; Munoz, J.; et al. Novel Seizure Phenotype and Sleep Disruptions in Knock-In Mice with Hypersensitive α4* Nicotinic Receptors. J. Neurosci. 2005, 25, 11396–11411. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.; Matta, J.A.; Lord, B.; Harrington, A.W.; Sutton, S.W.; Davini, W.B.; Bredt, D.S. Brain α7 Nicotinic Acetylcholine Receptor Assembly Requires NACHO. Neuron 2016, 89, 948–955. [Google Scholar] [CrossRef]
- Hales, T.G.; Dunlop, J.I.; Deeb, T.Z.; Carland, J.E.; Kelley, S.P.; Lambert, J.J.; Peters, J.A. Common Determinants of Single Channel Conductance within the Large Cytoplasmic Loop of 5-Hydroxytryptamine Type 3 and α4β2 Nicotinic Acetylcholine Receptors. J. Biol. Chem. 2006, 281, 8062–8071. [Google Scholar] [CrossRef]
- Hanek, A.P.; Lester, H.A.; Dougherty, D.A. A Stereochemical Test of a Proposed Structural Feature of the Nicotinic Acetylcholine Receptor. J. Am. Chem. Soc. 2008, 130, 13216–13218. [Google Scholar] [CrossRef][Green Version]
- Hassaine, G.; Deluz, C.; Grasso, L.; Wyss, R.; Tol, M.B.; Hovius, R.; Graff, A.; Stahlberg, H.; Tomizaki, T.; Desmyter, A.; et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature 2014, 512, 276–281. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Kelley, S.P.; Dunlop, J.I.; Kirkness, E.F.; Lambert, J.J.; Peters, J.A. A cytoplasmic region determines single-channel conductance in 5-HT3 receptors. Nature 2003, 424, 321–324. [Google Scholar] [CrossRef]
- Lansdell, S.J.; Gee, V.J.; Harkness, P.C.; Doward, A.I.; Baker, E.R.; Gibb, A.J.; Millar, N.S. RIC-3 Enhances Functional Expression of Multiple Nicotinic Acetylcholine Receptor Subtypes in Mammalian Cells. Mol. Pharmacol. 2005, 68, 1431–1438. [Google Scholar] [CrossRef]
- Mazzaferro, S.; Whiteman, S.T.; Alcaino, C.; Beyder, A.; Sine, S.M. NACHO and 14-3-3 promote expression of distinct subunit stoichiometries of the α4β2 acetylcholine receptor. Cell Mol. Life Sci. 2020, 78, 1565–1575. [Google Scholar] [CrossRef]
- Mesoy, S.M.; Lummis, S.C.R. M4, the Outermost Helix, is Extensively Involved in Opening of the α4β2 nACh Receptor. ACS Chem. Neurosci. 2020, 12, 133–139. [Google Scholar] [CrossRef]
- Mocatta, J.; Mesoy, S.M.; Dougherty, D.A.; Lummis, S.C.R. 5-HT3 Receptor MX Helix Contributes to Receptor Function. ACS Chem. Neurosci. 2022, 13, 2338–2345. [Google Scholar] [CrossRef]
- Morales-Perez, C.L.; Noviello, C.M.; Hibbs, R.E. X-ray structure of the human α4β2 nicotinic receptor. Nature 2016, 538, 411–415. [Google Scholar] [CrossRef]
- Nelson, M.E.; Kuryatov, A.; Choi, C.H.; Zhou, Y.; Lindstrom, J.; Kohout, T.A.; Lefkowitz, R.J. Alternate Stoichiometries of α4β2 Nicotinic Acetylcholine Receptors. Mol. Pharmacol. 2003, 63, 332–341. [Google Scholar] [CrossRef]
- Nemecz, Á.; Prevost, M.S.; Menny, A.; Corringer, P.J. Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron 2016, 90, 452–470. [Google Scholar] [CrossRef]
- Nguyen, M.; Alfonso, A.; Johnson, C.D.; Rand, J.B. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics 1995, 140, 527–535. [Google Scholar] [CrossRef]
- Noviello, C.M.; Gharpure, A.; Mukhtasimova, N.; Cabuco, R.; Baxter, L.; Borek, D.; Sine, S.M.; Hibbs, R.E. Structure and gating mechanism of the α7 nicotinic acetylcholine receptor. Cell 2021, 184, 2121–2134.e13. [Google Scholar] [CrossRef]
- Plested, A.J.R. Structural mechanisms of activation and desensitization in neurotransmitter-gated ion channels. Nat. Struct. Mol. Biol. 2016, 23, 494–502. [Google Scholar] [CrossRef]
- Price, K.L.; Lummis, S.C. FlexStation examination of 5-HT3 receptor function using Ca2+- and membrane potential-sensitive dyes: Advantages and potential problems. J. Neurosci. Methods 2005, 149, 172–177. [Google Scholar] [CrossRef]
- Stuebler, A.G.; Jansen, M. Mobility of Lower MA-Helices for Ion Conduction through Lateral Portals in 5-HT3A Receptors. Biophys. J. 2020, 119, 2593–2603. [Google Scholar] [CrossRef]
- Tapper, A.R.; McKinney, S.L.; Nashmi, R.; Schwarz, J.; Deshpande, P.; Labarca, C.; Whiteaker, P.; Marks, M.J.; Collins, A.C.; Lester, H.A. Nicotine Activation of α4* Receptors: Sufficient for Reward, Tolerance, and Sensitization. Science 2004, 306, 1029–1032. [Google Scholar] [CrossRef]
- Thompson, A.; Lummis, S. Discriminating between 5-HT3A and 5-HT3AB receptors. Br. J. Pharmacol. 2013, 169, 736–747. [Google Scholar] [CrossRef]
- Thompson, A.J.; Lester, H.A.; Lummis, S.C.R. The structural basis of function in Cys-loop receptors. Q. Rev. Biophys. 2010, 43, 449–499. [Google Scholar] [CrossRef]
- Unwin, N.; Miyazawa, A.; Li, J.; Fujiyoshi, Y. Activation of the Nicotinic Acetylcholine Receptor Involves a Switch in Conformation of the α Subunits. J. Mol. Biol. 2002, 319, 1165–1176. [Google Scholar] [CrossRef]
- Unwin, N. Structure and action of the nicotinic acetylcholine receptor explored by electron microscopy. FEBS Lett. 2003, 555, 91–95. [Google Scholar] [CrossRef]
- Unwin, N. Refined Structure of the Nicotinic Acetylcholine Receptor at 4Å Resolution. J. Mol. Biol. 2005, 346, 967–989. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]

| Position | Mutation (α4/β2) | pEC50 (M) | EC50 (nM) | nH |
|---|---|---|---|---|
| WT | 7.60 ± 0.12 | 25.3 | 0.8 ± 0.2 | |
| WT+ | 7.94 ± 0.12 | 11.4 | 0.6 ± 0.1 | |
| IMA39A+ | I601A/I456A | 8.11 ± 0.12 | 8 | 1.3 ± 0.4 |
| VMA38A+ | V600A/V455A | NR | ||
| MMA37A+ | M599A/M454A | NR | ||
| VMA35A+ | V597A/V452A | NR | ||
| YMA34A+ | Y596A/Y451A | NR | ||
| KMA33A+ | K595A/K450A | NR | ||
| WMA32A+ | W594A/W449A | NR | ||
| DMA31A+ | D593A/D448A | NR | ||
| EMA30A+ | E592A/E447A | NR | ||
| K/RMA29A | K591A/R446A | 7.62 ± 0.06 | 24 | 1.1 ± 0.2 |
| VMA28A | V590A/V445A | 7.62 ± 0.06 | 24 | 1.5 ± 0.3 |
| SMA27A | S589A/S444A | 7.47 ± 0.06 | 34 | 1.2 ± 0.2 |
| F/QMA26A | F588A/Q443A | 7.83 ± 0.06 | 14.7 | 1.2 ± 0.2 |
| DMA25A | D587A/D442A | 7.79 ± 0.09 | 16.2 | 1.4 ± 0.4 |
| T/DMA24A+ | T586A/D441A | NR | ||
| DMA23A | D585A/D440A | 7.89 ± 0.08 | 12.8 | 1.1 ± 0.3 |
| EMA22A | E584A/E439A | 7.59 ± 0.05 | 25.3 | 2.1 ± 0.5 |
| WT/SMA21A | WT/S438A | 7.36 ± 0.03 | 43.2 | 1.6 ± 0.2 |
| K/RMA20A | K582A/R437A | 7.76 ± 0.08 | 17.2 | 1.3 ± 0.2 |
| L/MMA19A | L581A/M436A | 7.63 ± 0.09 | 23.5 | 1.5 ± 0.4 |
| HMA18A | H580A/H435A | 7.70 ± 0.19 | 20.1 | 1.9 ± 1.3 |
| DMA17A | D579A/D434A | 7.61 ± 0.13 | 24.7 | 2.6 ± 2.1 |
| IMA15A | I577A/I432A | 7.71 ± 0.10 | 19.6 | 1.3 ± 0.4 |
| Y/FMA14A | Y576A/F431A | 7.99 ± 0.05 | 10.2 | 1.2 ± 0.1 |
| Q/RMA13A | Q575A/R430A | 7.73 ± 0.13 | 18.6 | 1.1 ± 0.3 |
| VMA12A | V574A/V429A | 7.55 ± 0.14 | 28.0 | 1.2 ± 0.4 |
| GMA11A | G573A/G428A | 7.23 ± 0.27 | 58.0 | 0.8 ± 0.5 |
| E/DMA10A | E572A/D427A | 7.55 ± 0.11 | 28.5 | 1.3 ± 0.3 |
| VMA9A | V571A/V426A | 7.79 ± 0.10 | 16.4 | 1.2 ± 0.2 |
| R/EMA7A | R569A/E424A | 7.73 ± 0.16 | 18.5 | 1.6 ± 0.8 |
| T/RMA6A | T568A/R423A | 7.72 ± 0.24 | 19.3 | 0.9 ± 0.4 |
| LMA5A | L567A/L422A | 7.63 ± 0.10 | 23.2 | 0.9 ± 0.1 |
| WT/GMA4A | WT/G421A | 7.91 ± 0.10 | 12.1 | 1.7 ± 0.5 |
| P/CMA3A | P565A/C420A | 7.79 ± 0.06 | 16.1 | 1.1 ± 0.2 |
| SMA2A | S564A/S419A | 7.61 ± 0.08 | 24.3 | 1.2 ± 0.3 |
| PMA0A | P562A/P417A | 7.67 ± 0.12 | 21.6 | 1.0 ± 0.2 |
| P558A/WT | 7.69 ± 0.14 | 20.3 | 1.4 ± 0.5 | |
| P557A/WT | 7.75 ± 0.18 | 17.6 | 1.1 ± 0.4 | |
| WT/P411A | 7.78 ± 0.07 | 16.6 | 1.1 ± 0.2 | |
| WT/P406A | 7.67 ± 0.08 | 21.4 | 1.0 ± 0.2 |
| Mutant α4 WT β2 | WT α4 Mutant β2 | |||||
|---|---|---|---|---|---|---|
| Position | pEC50 (M) | EC50 (nM) | nH | pEC50 (M) | EC50 (nM) | nH |
| WT | 7.60 ± 0.12 | 25.3 | 0.8 ± 0.2 | 7.60 ± 0.12 | 25.3 | 0.8 ± 0.2 |
| WT+ | 7.94 ± 0.12 | 11.4 | 0.6 ± 0.1 | 7.94 ± 0.12 | 11.4 | 0.6 ± 0.1 |
| IMA39A | 7.81 ± 0.06 | 15 | 1.9 ± 0.4 | 7.41 ± 0.08 | 39 | 1.0 ± 0.2 |
| VMA38A+ | 7.20 ± 0.24 | 63.3 | 0.8 ± 0.4 | 7.26 ± 0.11 | 55 | 1.2 ± 0.3 |
| MMA37A+ | 7.53 ± 0.06 | 30 | 1.0 ± 0.1 | NR | ||
| VMA35A+ | 7.32 ± 0.10 | 48.1 | 1.5 ± 0.4 | 7.67 ± 0.17 | 21 | 1.3 ± 0.7 |
| YMA34A+ | 7.46 ± 0.08 | 34.8 | 1.2 ± 0.2 | 7.46 ± 0.08 | 34 | 1.3 ± 0.3 |
| KMA33A+ | 7.63 ± 0.17 | 23.2 | 0.7 ± 0.2 | NR | ||
| WMA32A+ | NR | NR | ||||
| DMA31A+ | 7.53 ± 0.08 | 29.7 | 1.3 ± 0.3 | NR | ||
| EMA30A+ | 7.38 ± 0.08 | 41.9 | 0.8 ± 0.1 | 7.78 ± 0.07 | 17 | 1.5 ± 0.4 |
| T/DMA24A+ | NR | 7.59 ± 0.12 | 26.0 | 1.0 ± 0.2 | ||
| Position | pEC50 (M) | EC50 (nM) | nH |
|---|---|---|---|
| WT | 7.60 ± 0.12 | 25.3 | 0.8 ± 0.2 |
| WT+ | 7.94 ± 0.12 | 11.4 | 0.6 ± 0.1 |
| VMA38I+ | NR | ||
| VMA38T+ | NR | ||
| MMA37K+ | NR | ||
| VMA35I+ | NR | ||
| VMA35T+ | NR | ||
| YMA34F | 7.21 ± 0.16 | 61.2 | 0.88 ± 0.3 |
| YMA34S+ | NR | ||
| YMA34L+ | NR | ||
| YMA34Q+ | NR | ||
| KMA33E | 7.02 ± 0.06 | 95.4 | 1.1 ± 0.2 |
| KMA33Q+ | NR | ||
| KMA33M+ | NR | ||
| WMA32F+ | NR | ||
| WMA32Y+ | NR | ||
| DMA31E | 7.43 ± 0.09 | 37.4 | 1.2 ± 0.3 |
| DMA31K+ | NR | ||
| DMA31L+ | NR | ||
| DMA31N+ | NR | ||
| EMA30D | 7.48 ± 0.04 | 32.9 | 1.6 ± 0.2 |
| EMA30K+ | NR | ||
| EMA30L+ | NR | ||
| EMA30Q+ | NR | ||
| αTMA24D | 7.50 ± 0.05 | 31.4 | 1.6 ± 0.3 |
| αTMA24E | 7.14 ± 0.28 | 72.4 | 1.2 ± 0.8 |
| αTMA24K | 7.41 ± 0.10 | 39.4 | 1.4 ± 0.4 |
| αTMA24S | 7.80 ± 0.04 | 15.9 | 1.6 ± 0.2 |
| αTMA24V+ | NR | ||
| βDMA24T+ | 7.76 ± 0.10 | 17.3 | 1.1 ± 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fricska, D.I.; Mesoy, S.M.; Lummis, S.C.R. The MA Helix Is Important for Receptor Assembly and Function in the α4β2 nACh Receptor. Membranes 2023, 13, 891. https://doi.org/10.3390/membranes13120891
Fricska DI, Mesoy SM, Lummis SCR. The MA Helix Is Important for Receptor Assembly and Function in the α4β2 nACh Receptor. Membranes. 2023; 13(12):891. https://doi.org/10.3390/membranes13120891
Chicago/Turabian StyleFricska, Dorottya I., Susanne M. Mesoy, and Sarah C. R. Lummis. 2023. "The MA Helix Is Important for Receptor Assembly and Function in the α4β2 nACh Receptor" Membranes 13, no. 12: 891. https://doi.org/10.3390/membranes13120891
APA StyleFricska, D. I., Mesoy, S. M., & Lummis, S. C. R. (2023). The MA Helix Is Important for Receptor Assembly and Function in the α4β2 nACh Receptor. Membranes, 13(12), 891. https://doi.org/10.3390/membranes13120891