Dynamin-Related Proteins Enhance Tomato Immunity by Mediating Pattern Recognition Receptor Trafficking
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Bioinformatic Analysis
2.3. Transient Expression
2.4. Cloning of SlDRP1B and Site-Directed Mutagenesis
2.5. Co-Immunoprecipitation
2.6. BiFluorescence Complementation
2.7. ROS Burst Assay
2.8. Ethylene Production Assay
2.9. Live-Cell Imaging
2.10. Confocal Image Analysis
3. Results
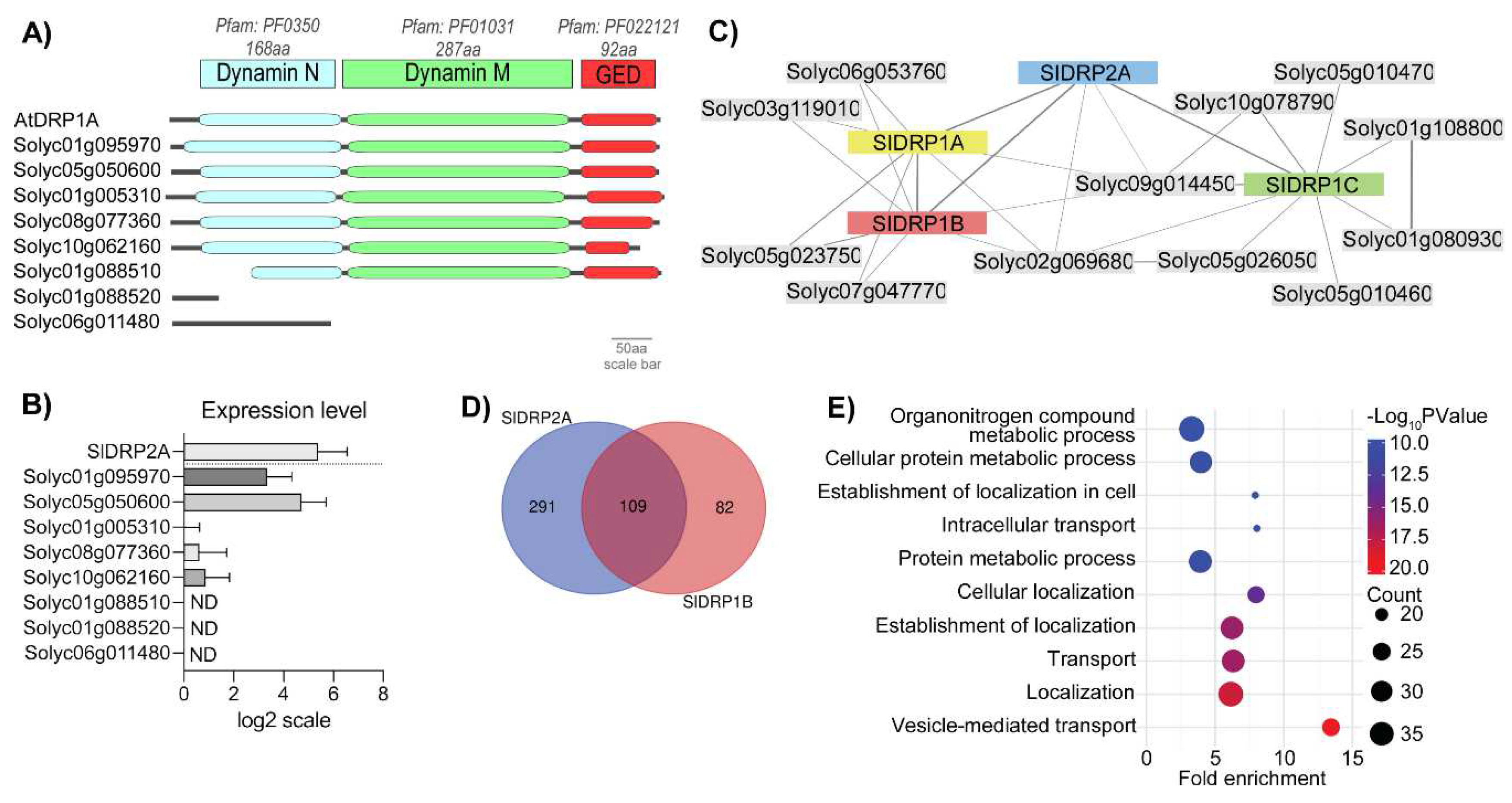
3.1. Searching for a Functional Tomato DRP1 Involved in PRR Endocytic Trafficking
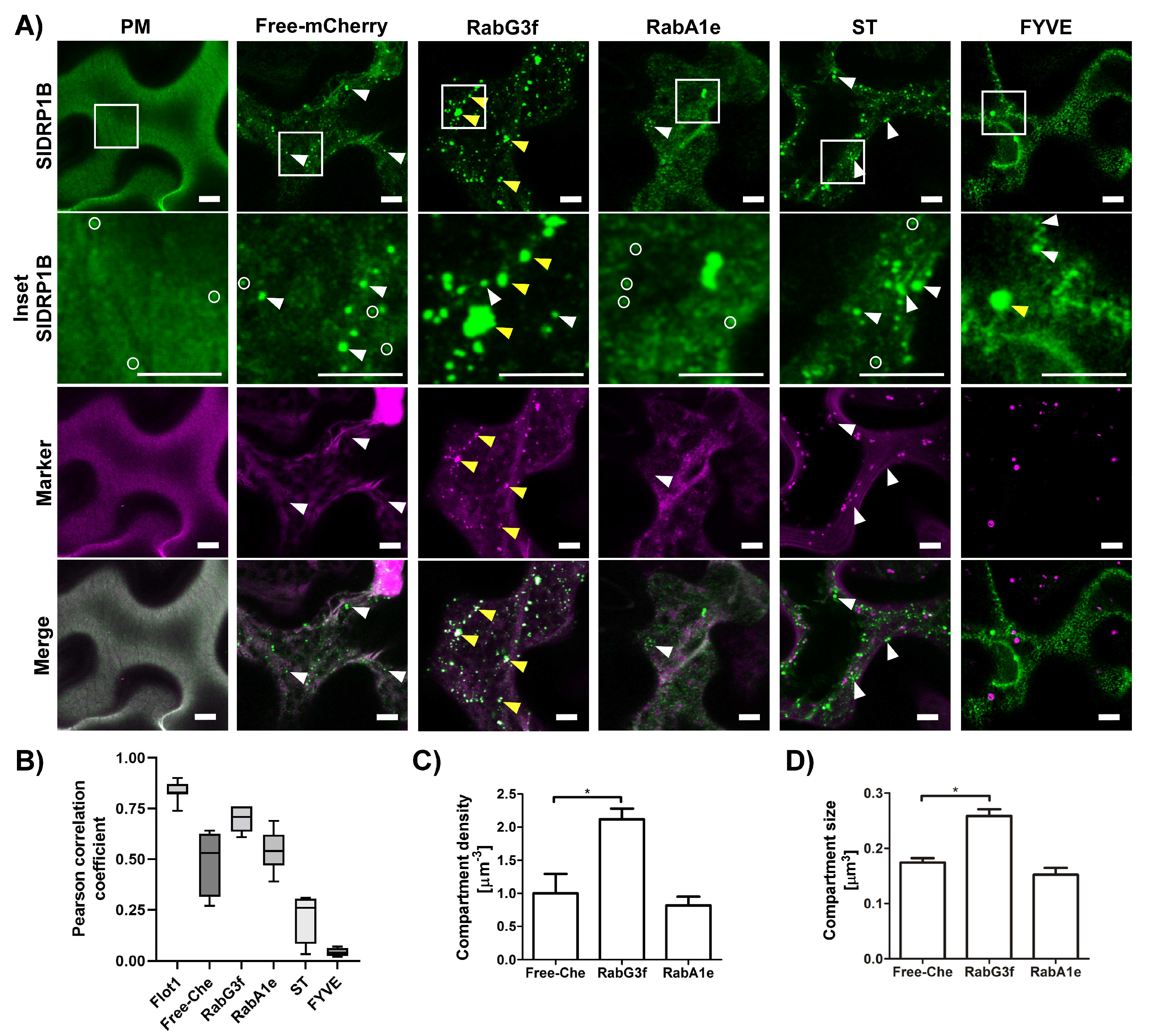
3.2. SlDRP1B Subcellular Localization
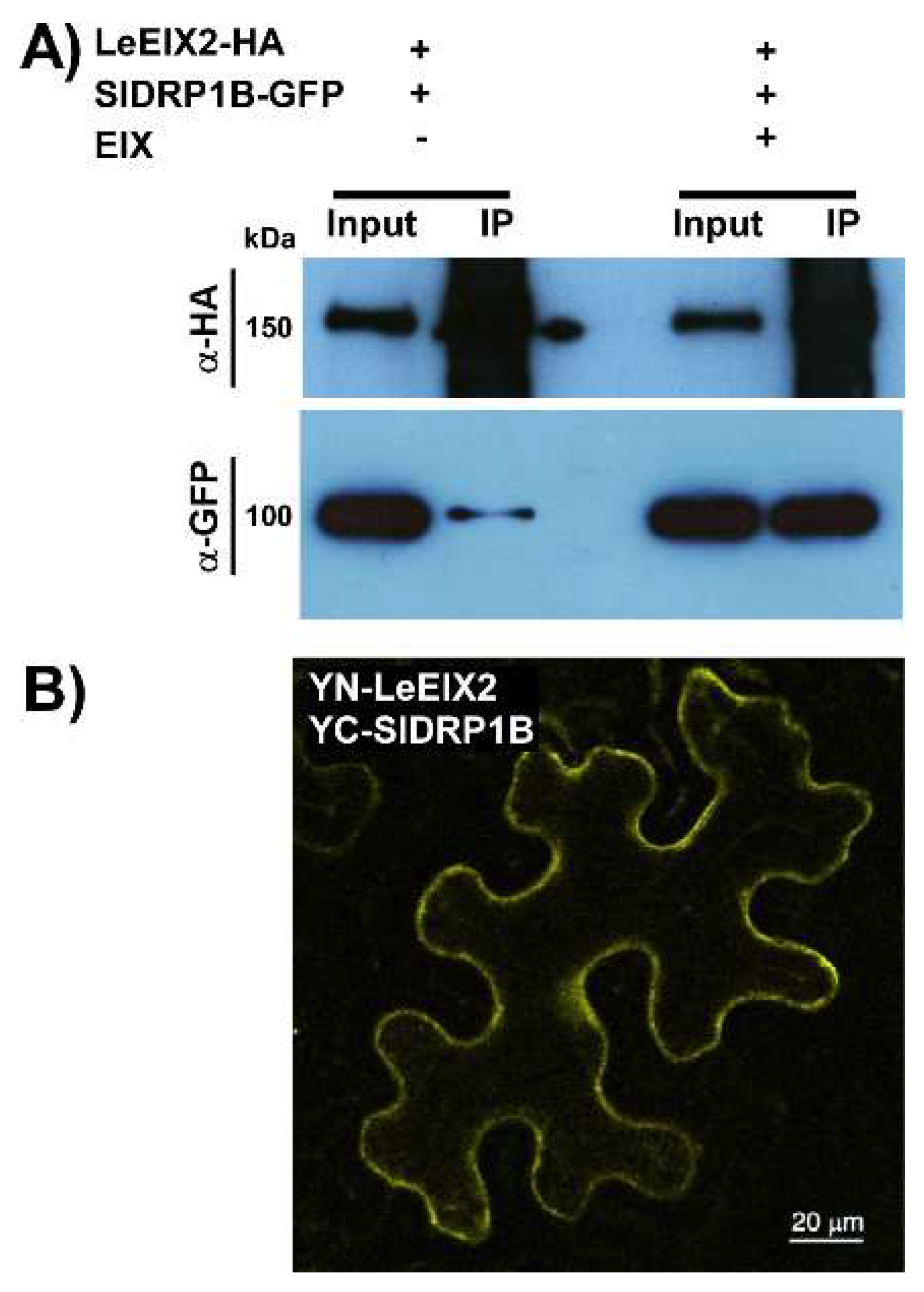
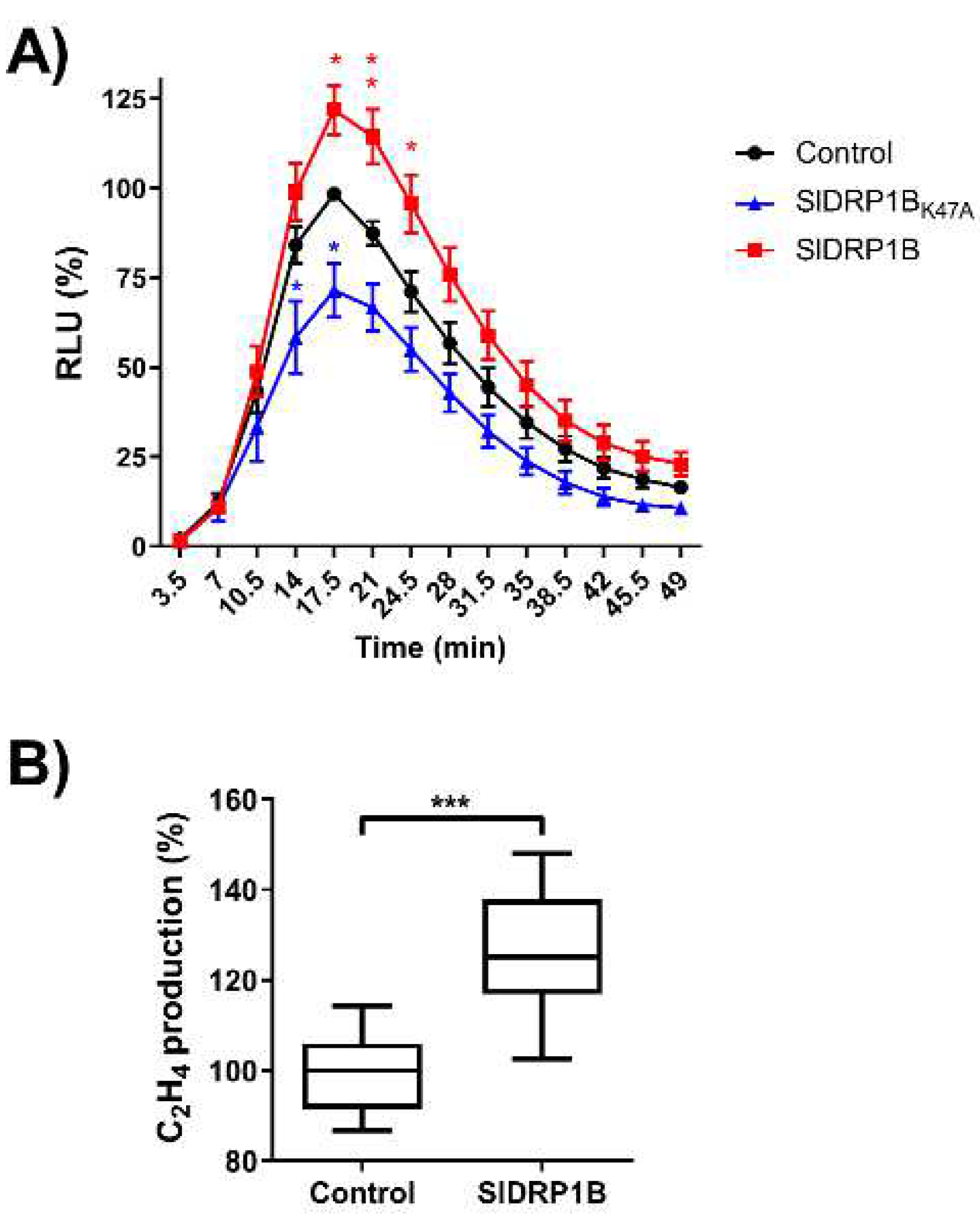
3.3. SlDRP1B Associates with LeEIX2 and Enhances Defense Responses Mediated by EIX
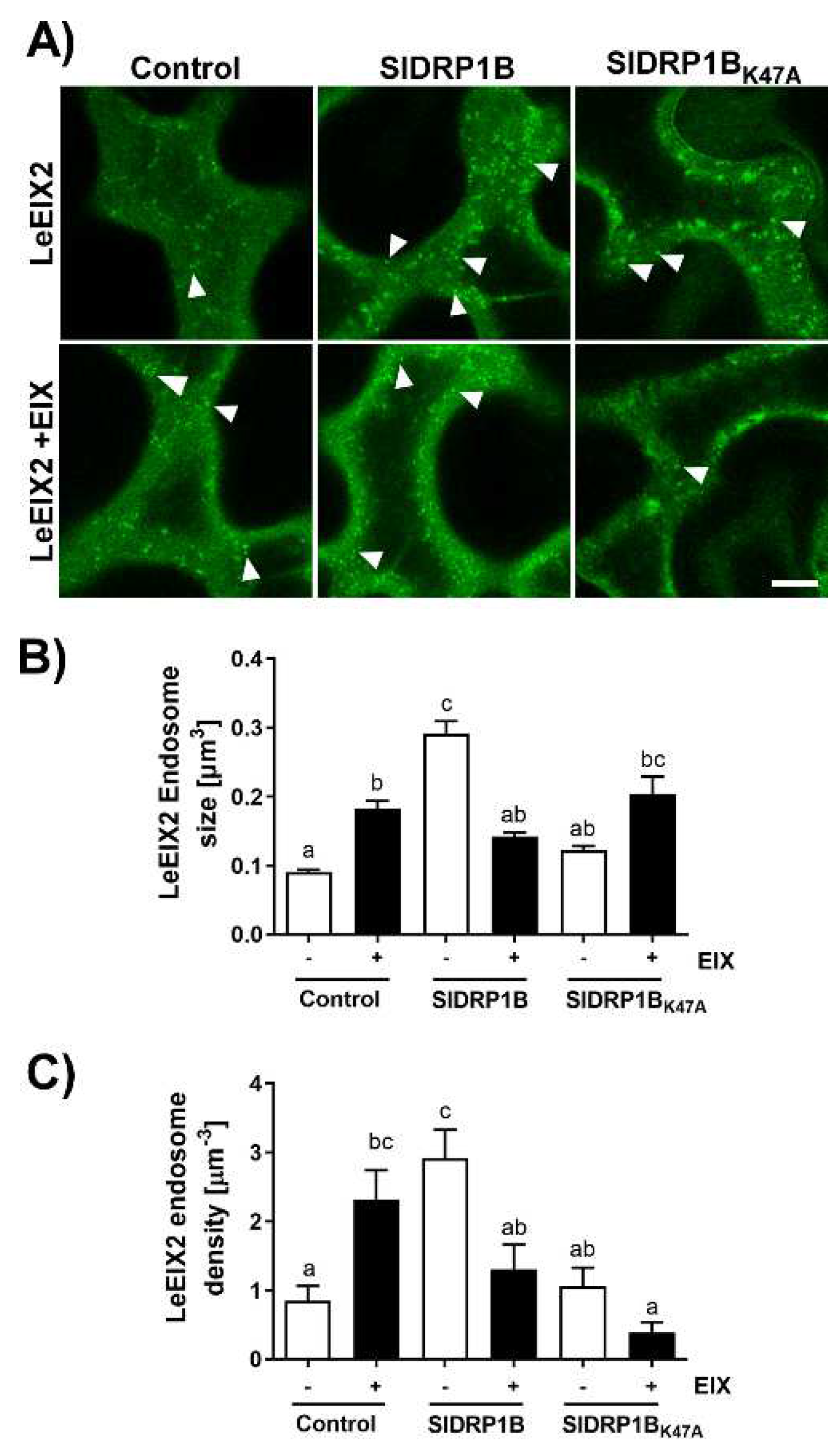
3.4. SlDRP1B Affects LeEIX2 Endosomal Distribution
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Boutrot, F.; Zipfel, C. Function, Discovery, and Exploitation of Plant Pattern Recognition Receptors for Broad-Spectrum Disease Resistance. Annu. Rev. Phytopathol. 2017, 55, 257–286. [Google Scholar] [CrossRef] [PubMed]
- Tor, M.; Lotze, M.T.; Holton, N. Receptor-mediated signalling in plants: Molecular patterns and programmes. J. Exp. Bot. 2009, 60, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Gust, A.A.; Felix, G. Receptor like proteins associate with SOBIR1-type of adaptors to form bimolecular receptor kinases. Curr. Opin. Plant Biol. 2014, 21, 104–111. [Google Scholar] [CrossRef]
- Liang, X.; Zhou, J.-M. Receptor-like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase–Mediated Signaling. Annu. Rev. Plant Biol. 2018, 69, 267–299. [Google Scholar] [CrossRef] [PubMed]
- Khaled, S.B.; Postma, J.; Robatzek, S. A Moving View: Subcellular Trafficking Processes in Pattern Recognition Receptor–Triggered Plant Immunity. Annu. Rev. Phytopathol. 2015, 53, 379–402. [Google Scholar] [CrossRef] [PubMed]
- Ekanayake, G.; LaMontagne, E.D.; Heese, A. Never Walk Alone: Clathrin-Coated Vesicle (CCV) Components in Plant Immunity. Annu. Rev. Phytopathol. 2019, 57, 387–409. [Google Scholar] [CrossRef] [PubMed]
- Claus, L.A.N.; Savatin, D.V.; Russinova, E. The crossroads of receptor-mediated signaling and endocytosis in plants. J. Integr. Plant Biol. 2018, 60, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Zavaliev, R.; Dong, X. Membrane Trafficking in Plant Immunity. Mol. Plant 2017, 10, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, D.D.; Brandizzi, F. Plant endomembranes and cytoskeleton: Moving targets in immunity. Curr. Opin. Plant Biol. 2020, 58, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Agaoua, A.; Bendahmane, A.; Moquet, F.; Dogimont, C. Membrane Trafficking Proteins: A New Target to Identify Resistance to Viruses in Plants. Plants 2021, 10, 2139. [Google Scholar] [CrossRef] [PubMed]
- Matern, A.; Böttcher, C.; Eschen-Lippold, L.; Westermann, B.; Smolka, U.; Döll, S.; Trempel, F.; Aryal, B.; Scheel, D.; Geisler, M.; et al. A substrate of the ABC transporter PEN3 stimulates bacterial flagellin (flg22)-induced callose deposition in Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 6857–6870. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bao, H.; Wang, Z.; Wang, M.; Fan, B.; Zhu, C.; Chen, Z. Biogenesis and Function of Multivesicular Bodies in Plant Immunity. Front. Plant Sci. 2018, 9, 979. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.-J.; Schornack, S.; Spallek, T.; Geldner, N.; Chory, J.; Schellmann, S.; Schumacher, K.; Kamoun, S.; Robatzek, S. Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell. Microbiol. 2012, 14, 682–697. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, T.O.; Belhaj, K.; Dagdas, Y.F.; Chaparro-Garcia, A.; Wu, C.H.; Cano, L.M.; Kamoun, S. Rerouting of Plant Late Endocytic Trafficking Toward a Pathogen Interface. Traffic 2015, 16, 204–226. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nurnberger, T.; Jones, J.D.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Beck, M.; Zhou, J.; Faulkner, C.; MacLean, D.; Robatzek, S. Spatio-Temporal Cellular Dynamics of the Arabidopsis Flagellin Receptor Reveal Activation Status-Dependent Endosomal Sorting. Plant Cell 2012, 24, 4205–4219. [Google Scholar] [CrossRef]
- Choi, S.-W.; Tamaki, T.; Ebine, K.; Uemura, T.; Ueda, T.; Nakano, A. RABA Members Act in Distinct Steps of Subcellular Trafficking of the FLAGELLIN SENSING2 Receptor. Plant Cell 2013, 25, 1174–1187. [Google Scholar] [CrossRef]
- Mbengue, M.; Bourdais, G.; Gervasi, F.; Beck, M.; Zhou, J.; Spallek, T.; Bartels, S.; Boller, T.; Ueda, T.; Kuhn, H.; et al. Clathrin-dependent endocytosis is required for immunity mediated by pattern recognition receptor kinases. Proc. Natl. Acad. Sci. USA 2016, 113, 11034–11039. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Salamango, D.J.; Leslie, M.E.; Collins, C.A.; Heese, A. Sensitivity to Flg22 is modulated by ligand-induced degradation and de novo synthesis of the endogenous flagellin-receptor FLAGELLIN-SENSING2. Plant Physiol. 2014, 164, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Ron, M.; Avni, A. The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 2004, 16, 1604–1615. [Google Scholar] [CrossRef] [PubMed]
- Sharfman, M.; Bar, M.; Ehrlich, M.; Schuster, S.; Melech-Bonfil, S.; Ezer, R.; Sessa, G.; Avni, A. Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. Plant J. 2011, 68, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Leibman-Markus, M.; Pizarro, L.; Schuster, S.; Lin, Z.J.D.; Gershony, O.; Bar, M.; Coaker, G.; Avni, A. The intracellular nucleotide-binding leucine-rich repeat receptor (SlNRC4a) enhances immune signalling elicited by extracellular perception. Plant Cell Environ. 2018, 41, 2313–2327. [Google Scholar] [CrossRef] [PubMed]
- Pizarro, L.; Leibman-Markus, M.; Schuster, S.; Bar, M.; Avni, A. Tomato Dynamin Related Protein 2A Associates With LeEIX2 and Enhances PRR Mediated Defense by Modulating Receptor Trafficking. Front. Plant Sci. 2019, 10, 936. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Emonet, A.; Dénervaud Tendon, V.; Marhavy, P.; Wu, D.; Lahaye, T.; Geldner, N. Co-incidence of Damage and Microbial Patterns Controls Localized Immune Responses in Roots. Cell 2020, 180, 440–453.e18. [Google Scholar] [CrossRef] [PubMed]
- Pizarro, L.; Leibman-Markus, M.; Gupta, R.; Kovetz, N.; Shtein, I.; Bar, E.; Davidovich-Rikanati, R.; Zarivach, R.; Lewinsohn, E.; Avni, A.; et al. A gain of function mutation in SlNRC4a enhances basal immunity resulting in broad-spectrum disease resistance. Commun. Biol. 2020, 3, 404. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.M.; De Camilli, P. Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 2012, 13, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Antonny, B.; Burd, C.; De Camilli, P.; Chen, E.; Daumke, O.; Faelber, K.; Ford, M.; Frolov, V.A.; Frost, A.; Hinshaw, J.E.; et al. Membrane fission by dynamin: What we know and what we need to know. EMBO J. 2016, 35, 2270–2284. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Shaikh, A.; Mathur, N. Peroxisome Mitochondria Inter-relations in Plants. In Proteomics of Peroxisomes; Springer: Singapore, 2018; Volume 89, pp. 417–433. [Google Scholar] [CrossRef]
- Rose, R.J. Contribution of Massive Mitochondrial Fusion and Subsequent Fission in the Plant Life Cycle to the Integrity of the Mitochondrion and Its Genome. Int. J. Mol. Sci. 2021, 22, 5429. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Arimura, S.I.; Ueda, T.; Takanashi, H.; Hayashi, Y.; Nakano, A.; Tsutsumi, N. Arabidopsis dynamin-related proteins DRP2B and DRP1A participate together in clathrin-coated vesicle formation during endocytosis. Proc. Natl. Acad. Sci. USA 2010, 107, 6094–6099. [Google Scholar] [CrossRef] [PubMed]
- Ekanayake, G.; Smith, J.M.; Jones, K.B.; Stiers, H.M.; Robinson, S.J.; LaMontagne, E.D.; Kostos, P.H.; Cornish, P.V.; Bednarek, S.Y.; Heese, A. DYNAMIN-RELATED PROTEIN DRP1A functions with DRP2B in plant growth, flg22-immune responses, and endocytosis. Plant Physiol. 2021, 185, 1986–2002. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Leslie, M.E.; Robinson, S.J.; Korasick, D.A.; Zhang, T.; Backues, S.K.; Cornish, P.V.; Koo, A.J.; Bednarek, S.Y.; Heese, A. Loss of Arabidopsis thaliana Dynamin-Related Protein 2B Reveals Separation of Innate Immune Signaling Pathways. PLoS Pathog. 2014, 10, e1004578. [Google Scholar] [CrossRef] [PubMed]
- Chaparro-Garcia, A.; Schwizer, S.; Sklenar, J.; Yoshida, K.; Petre, B.; Bos, J.I.B.; Schornack, S.; Jones, A.M.E.; Bozkurt, T.O.; Kamoun, S. Phytophthora infestans RXLR-WY Effector AVR3a Associates with Dynamin-Related Protein 2 Required for Endocytosis of the Plant Pattern Recognition Receptor FLS2. PLoS ONE 2015, 10, e0137071. [Google Scholar] [CrossRef]
- Wu, G.; Cui, X.; Chen, H.; Renaud, J.B.; Yu, K.; Chen, X.; Wang, A. Dynamin-like proteins of endocytosis in plants are co-opted by potyviruses to enhance virus infection. J. Virol. 2018, 92, e01320-18. [Google Scholar] [CrossRef]
- Tang, D.; Ade, J.; Frye, C.A.; Innes, R.W. A mutation in the GTP hydrolysis site of Arabidopsis dynamin-related protein 1E confers enhanced cell death in response to powdery mildew infection. Plant J. 2006, 47, 75–84. [Google Scholar] [CrossRef]
- Leslie, M.E.; Rogers, S.W.; Heese, A. Increased callose deposition in plants lacking DYNAMIN-RELATED PROTEIN 2B is dependent upon POWDERY MILDEW RESISTANT 4. Plant Signal. Behav. 2016, 11, e1244594. [Google Scholar] [CrossRef]
- Fujimoto, M.; Arimura, S.I.; Nakazono, M.; Tsutsumi, N. Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep. 2008, 27, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef] [PubMed]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008, 2008, 420747. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Xu, W.; Ban, R.; Huang, S.; Miao, M.; Tang, X.; Liu, G.; Liu, Y. PTIR: Predicted Tomato Interactome Resource. Sci. Rep. 2016, 6, 25047. [Google Scholar] [CrossRef]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Leibman-Markus, M.; Marash, I.; Kovetz, N.; Rav-David, D.; Elad, Y.; Bar, M. Root zone warming represses foliar diseases in tomato by inducing systemic immunity. Plant Cell Environ. 2021, 44, 2277–2289. [Google Scholar] [CrossRef] [PubMed]
- Mehari, Z.H.; Elad, Y.; Rav-David, D.; Graber, E.R.; Meller Harel, Y. Induced systemic resistance in tomato (Solanum lycopersicum) against Botrytis cinerea by biochar amendment involves jasmonic acid signaling. Plant Soil 2015, 395, 31–44. [Google Scholar] [CrossRef]
- Leibman-Markus, M.; Schuster, S.; Avni, A. LeEIX2 Interactors’ Analysis and EIX-Mediated Responses Measurement. Methods Mol. Biol. 2017, 1578, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Fujimoto, M.; Fujiwara, M.; Fukao, Y.; Arimura, S.I.; Tsutsumi, N. Arabidopsis dynamin-related proteins, DRP2A and DRP2B, function coordinately in post-Golgi trafficking. Biochem. Biophys. Res. Commun. 2015, 456, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Konopka, C.A.; Backues, S.K.; Bednareka, S.Y. Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. Plant Cell 2008, 20, 1363–1380. [Google Scholar] [CrossRef]
- Li, R.; Liu, P.; Wan, Y.; Chen, T.; Wang, Q.; Mettbach, U.; Baluska, F.; Samaj, J.; Fang, X.; Lucas, W.J.; et al. A Membrane Microdomain-Associated Protein, Arabidopsis Flot1, Is Involved in a Clathrin-Independent Endocytic Pathway and Is Required for Seedling Development. Plant Cell 2012, 24, 2105–2122. [Google Scholar] [CrossRef]
- Zelazny, E.; Santambrogio, M.; Pourcher, M.; Chambrier, P.; Berne-Dedieu, A.; Fobis-Loisy, I.; Miege, C.; Jaillais, Y.; Gaude, T.; Miège, C.; et al. Mechanisms governing the endosomal membrane recruitment of the core retromer in Arabidopsis. J. Biol. Chem. 2013, 288, 8815–8825. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, Q.; Gao, C.; Ding, Y.; Zeng, Y.; Ueda, T.; Nakano, A.; Jiang, L. Activation of the Rab7 GTPase by the MON1-CCZ1 Complex Is Essential for PVC-to-Vacuole Trafficking and Plant Growth in Arabidopsis. Plant Cell 2014, 26, 2080–2097. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Dénervaud-Tendon, V.; Hyman, D.L.; Mayer, U.; Stierhof, Y.-D.D.; Chory, J.; Denervaud-Tendon, V.; Hyman, D.L.; Mayer, U.; Stierhof, Y.-D.D.; et al. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009, 59, 169–178. [Google Scholar] [CrossRef]
- Asaoka, R.; Uemura, T.; Ito, J.; Fujimoto, M.; Ito, E.; Ueda, T.; Nakano, A. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 2013, 73, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Saint-Jore, C.M.; Evins, J.; Batoko, H.; Brandizzi, F.; Moore, I.; Hawes, C. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 2002, 29, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Pizarro, L.; Leibman-Markus, M.; Schuster, S.; Bar, M.; Meltz, T.; Avni, A. Tomato prenylated RAB acceptor protein 1 modulates trafficking and degradation of the pattern recognition receptor LeEIX2, affecting the innate immune response. Front. Plant Sci. 2018, 9, 257. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Pizarro, L.; Leibman-Markus, M.; Marash, I.; Bar, M. Cytokinin response induces immunity and fungal pathogen resistance, and modulates trafficking of the PRR LeEIX2 in tomato. Mol. Plant Pathol. 2020, 21, 1287–1306. [Google Scholar] [CrossRef]
- Spallek, T.; Beck, M.; Ben Khaled, S.; Salomon, S.; Bourdais, G.; Schellmann, S.; Robatzek, S. ESCRT-I Mediates FLS2 Endosomal Sorting and Plant Immunity. PLoS Genet. 2013, 9, e1004035. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.; Hu, J. The Arabidopsis peroxisome division mutant pdd2 is defective in the DYNAMIN-RELATED PROTEIN3A (DRP3A) gene. Plant Signal. Behav. 2009, 4, 542–544. [Google Scholar] [CrossRef][Green Version]
- Aung, K.; Hu, J. Differential roles of Arabidopsis Dynamin-Related Proteins DRP3A, DRP3B, and DRP5B in Organelle Division. J. Integr. Plant Biol. 2012, 54, 921–931. [Google Scholar] [CrossRef]
- Wang, F.; Liu, P.; Zhang, Q.; Zhu, J.; Chen, T.; Arimura, S.I.; Tsutsumi, N.; Lin, J. Phosphorylation and ubiquitination of dynamin-related proteins (AtDRP3A/3B) synergically regulate mitochondrial proliferation during mitosis. Plant J. 2012, 72, 43–56. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leibman-Markus, M.; Schuster, S.; Vasquez-Soto, B.; Bar, M.; Avni, A.; Pizarro, L. Dynamin-Related Proteins Enhance Tomato Immunity by Mediating Pattern Recognition Receptor Trafficking. Membranes 2022, 12, 760. https://doi.org/10.3390/membranes12080760
Leibman-Markus M, Schuster S, Vasquez-Soto B, Bar M, Avni A, Pizarro L. Dynamin-Related Proteins Enhance Tomato Immunity by Mediating Pattern Recognition Receptor Trafficking. Membranes. 2022; 12(8):760. https://doi.org/10.3390/membranes12080760
Chicago/Turabian StyleLeibman-Markus, Meirav, Silvia Schuster, Beatriz Vasquez-Soto, Maya Bar, Adi Avni, and Lorena Pizarro. 2022. "Dynamin-Related Proteins Enhance Tomato Immunity by Mediating Pattern Recognition Receptor Trafficking" Membranes 12, no. 8: 760. https://doi.org/10.3390/membranes12080760
APA StyleLeibman-Markus, M., Schuster, S., Vasquez-Soto, B., Bar, M., Avni, A., & Pizarro, L. (2022). Dynamin-Related Proteins Enhance Tomato Immunity by Mediating Pattern Recognition Receptor Trafficking. Membranes, 12(8), 760. https://doi.org/10.3390/membranes12080760








