Abstract
Exploring the mechanisms underlying the permeation of graphene quantum dots (GQDs) through different cell membranes is key for the practical application of GQDs in medicine. Here, the permeation process of GQDs through different lipid membranes was evaluated using molecular dynamics (MD) simulations. Our results showed that GQDs can easily permeate into 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) lipid membranes with low phospholipid molecule densities but cannot permeate into 1-palmitoyl-2-oleoyl phosphatidylethanolamine (POPE) lipid membranes with high phospholipid densities. Free energy calculation showed that a high-energy barrier exists on the surface of the POPE lipid membrane, which prevents GQDs from entering the cell membrane interior. Further analysis of the POPE membrane structure showed that sparsely arranged phospholipid molecules of the low-density lipid membrane facilitated the entry of GQDs into the interior of the membrane, compared to compactly arranged molecules in the high-density lipid membrane. Our simulation study provides new insights into the transmembrane transport of GQDs.
1. Introduction
As a new quasi-zero-dimensional nanomaterial, graphene quantum dots (GQDs) show broad applicability prospects in many fields owing to their ultra-small size, significant quantum size effects, and good biocompatibility [1,2,3,4,5,6]. GQDs are derived from graphene and graphene oxide (GO) and have excellent characteristics similar to graphene and GO, including mechanical and stable chemical properties [7,8,9,10]. In recent years, GQDs have been studied for application in the fields of biomedicine drug transmission and disease diagnosis [11,12,13]. Recently, GO has been used as an ideal drug-loading ligand to achieve targeted therapy, providing new prospects regarding the treatment of Parkinson’s disease [14]. Previous studies have shown that GQDs and related matrices can be used as platforms for cell adhesion, proliferation, and differentiation [15]. Similarly, GQDs and GO exhibit certain characteristics that make them suitable for anticancer therapy. For example, GO can be used to carry and effectively deliver doxorubicin to tumor cells [16].
The cytotoxicity of GQDs is lower than that of GO, so GQDs are suitable for biomedical applications [17]. Relevant studies showed that 3-nanometer GQDs had good membrane permeability, while 12-nanometer GQDs had weak permeability [18]. In addition, small-sized GQDs can be used for drug delivery because they can easily enter the intracellular space to attack tumor cells, eventually leading to tumor cell apoptosis [19]. Stable sulfur-doped GQDs can be used as efficient cell-imaging materials and can penetrate the cell membrane of HeLa cells [20]. As a crucial part of the cell, the cell membrane is not only a protective barrier to the cell but also plays an important role regarding compound exchange and metabolism. Some studies reported that GQDs have potential applications in medicine and can enter cells through direct infiltration, but the variation between organisms in terms of cell membrane structure should be considered [21,22]. The main lipid component of most bacteria is 1-palmitoyl-2-oleoyl phosphatidylethanolamine (POPE). For example, Escherichia coli (E. coli) contains 70–80% POPE, 20–25% POPG, and <5% cardiolipin [18,23]. In animal cells, the 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) phospholipid is the main component of the cell membrane, and the POPE content in blood cells is 6% [24,25]. Similarly, the neutral 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) cell membrane plays a significant role in gene transmission in biological cells [26]. However, the mechanisms underlying the interactions between GQDs and different cell membranes remain unclear. Therefore, understanding the permeation process of GQDs through different cell membranes is important for the advancement of GQD drug-loading therapy and biosensor research.
In addition to experimental approaches, molecular dynamics (MD) simulations are frequently used to explore the mechanisms of interaction between nanomaterials and biological macromolecules. MD can realize the visualization of atoms and visually observe the details of the interaction between nanoparticles and biomolecules (including proteins, DNA, and cell membranes) [22,27,28,29]. For example, Ji et al. found that GQDs and GO can help drug molecules to enter cell membranes [30]. Moreover, a complex of GO and anticancer drug molecules can be transported to specific cancer cells [31]. Similarly, MD simulations were used previously to explore the interaction mechanism between GO and 5-fluorouracil (5-FU) [32]. In this study, MD simulations were used to explore the penetration mechanism of GQDs through different cell membranes, including POPC, DOPE, and POPE. See the Supplementary Materials for simulation details and parameters of the system.
2. Computational Details
2.1. System Details
As described in our previous work [33,34,35], a graphene sheet was set on the xy plane, and a carbon atom was selected as the central atom (Cartesian coordinates 0, 0, 0). As shown in Figure 1, carbon atoms with x2 + y2 < R2 are part of the GQDs, where R is the radius of the GQDs [36]. In the MD simulation, the radius of the GQDs was set to 0.85 nm, the outermost carbon atoms of the GQDs were saturated with hydrogen atoms, and geometrical optimization based on density functional theory (DFT) was performed at the B3LYP/6-31G (d) level by Gaussian 03 [37]. Optimized GQDs were used for the initial MD configuration. Standardized lipid membranes were from CHARMM-GUI [38,39]. The varying areas of the different types of phospholipid molecules resulted in differences in the number of phospholipid molecules per unit area [25,40,41,42,43,44]. The detailed cell membrane information is presented in Table 1. Comparative studies of POPE and other phospholipids (POPC and DOPE) have shown that the area of POPE phospholipid molecules is the lowest [25]. The initialized GQDs were placed directly above the cell membrane with a distance of 4.5 nm from the centroid of the cell membrane.
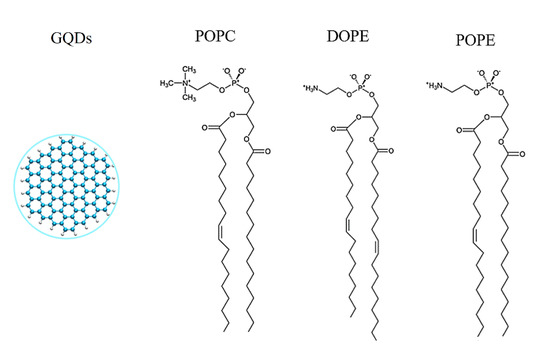
Figure 1.
Structural details of three phospholipids and GQDs.

Table 1.
Simulation details of the phospholipid molecules.
2.2. PMF Calculations
Based on the MD simulations, the free energy of GQDs’ translocation on different cell membranes was calculated using umbrella sampling. The zero coordinate was considered along the centroid of the cell membrane and 4.5 nm along the upper axis of the z-axis, and each equidistant window of 0.1 nm was extracted as the simulated reaction configuration. A harmonic force constant of 1000 kJmol−1nm −2 was used to position and suppress the ions in each window [45]. An independent 10-nanosecond simulation was performed for each window. The potential of the mean force (PMF) curve was produced using the g_wham tool of Gromacs 5.0, which implements a weighted histogram [46].
3. Results and Discussion
3.1. Translocation Phenomenon
The adsorption process of GQDs on different cell membranes was studied using MD simulations. The initial GQDs were placed 4.5 nm above the center of mass of the cell membrane. As shown in Figure 2a, the GQDs first reached the surface of the POPC cell membrane, and this process was completed in approximately 10 ns. Once the GQDs arrived at the surface of POPC, they rapidly permeated through the cell membrane. In the simulation of the DOPE cell membrane system, the GQDs were absorbed by the cell membrane within approximately 80 ns. Similar to the permeation of GQDs in POPC membranes, the GQDs were found to reach the surface of the membrane and rapidly penetrate it. However, unlike the POPC and DOPE lipid membranes, the GQDs were captured by the POPE membrane at 20 ns but remained on the upper surface of the membrane. Figure 2b shows the change in the number of phospholipid atoms adsorbed on the surface (d < 0.5 nm) of the GQDs. For the POPC cell membrane, the number of atomic contacts reached a maximum of 495 in 15 ns and then remained stable at 500. In this time, GQDs completely entered the POPC membrane. Similarly, GQDs completely entered the DOPE membrane at 110 ns, and then, the atomic contact number remained at 450. However, for the POPE lipid membrane, because the GQDs always remained on the surface of the cell membrane, only one side of the atom was attached to the lipid membrane, and the number of atoms in contact with the membrane was relatively low. In addition, some water molecules were also trapped on the surface of the GQDs and phospholipid molecular membrane head groups, which also led to low contact between the GQDs and POPE atoms.
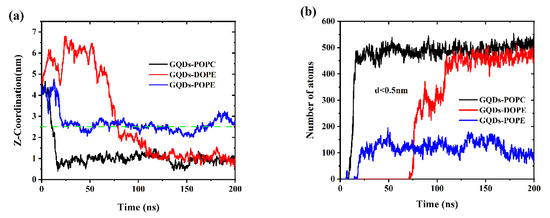
Figure 2.
(a) Change in distance between the GQDs and the cell membrane in the z-direction. (b) Changes in contact numbers between GQDs and cell membrane atoms.
To visualize the translocation process of GQDs on lipid membranes, instantaneous screenshots of GQDs on membranes at different time points are shown in Figure 3. For the POPC and DOPE membranes, permeation into the cell membrane by GQDs from the surface was similar. Specifically, the GQDs were inclined at an angle to the POPC and DOPE membrane surfaces at 14 and 96 ns, respectively. The GQDs were first inserted into phospholipid molecules and then rapidly entered the cell membrane. Meanwhile, they maintained an orientation parallel to the conformation of the phospholipid molecule and stably adhered to the interior of the cell membrane. Figure 4 shows the change in the angle between the GQDs and the lipid membrane over time. The initialized GQDs moved freely in the liquid phase water, with large changes in the angle. When entering the cell membrane, the GQDs were maintained at an angle of 80° relative to the membrane surface. For the POPE membrane, the angle between the GQDs and the membrane (Figure 4b) showed a significant change at 130 ns, and the GQD angle was close to 50°. When the simulation reached 172 ns, the angle decreased to 10°, and the adsorption on the surface of the cell membrane was maintained until the end of the simulation.
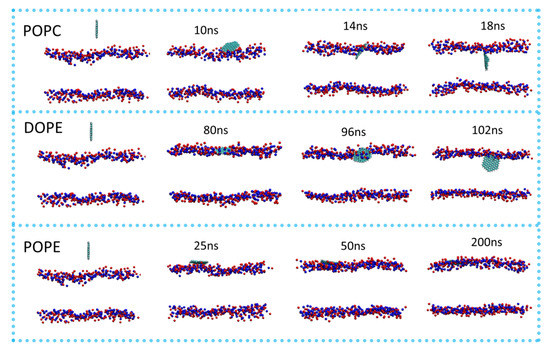
Figure 3.
Permeation of GQDs through different cell membranes. P atoms in the cell membrane are shown as blue spheres, and N atoms are shown as red spheres. All instantaneous screenshots were produced using VMD software.
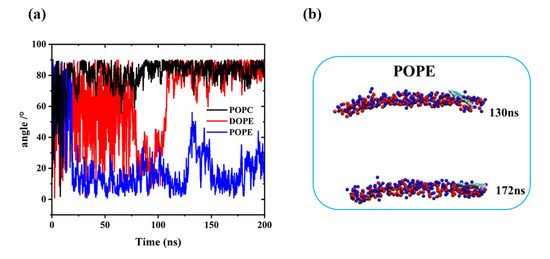
Figure 4.
(a) Change in the plane angle between GQDs and the cell membrane during simulation. (b) Angle of GQDs on POPE cell membrane at 130 and 172 ns.
3.2. Effect of the Phospholipid Molecule Concentration on Translocation
The MD simulation showed that the GQDs could not enter the POPE cell membrane. To explain this phenomenon, the cell membranes were analyzed based on the total number of phospholipid molecules. Due to the different types of phospholipid molecules, the density of phospholipid molecules per unit area was different. With the size of the initial box kept the same, the density of phospholipid molecules in POPE was the highest among the three cell membranes, with one box containing 342 molecules (Table 1). GQDs entering the cell membrane were inserted between the phospholipid molecules in the membrane. The size of the gap between adjacent phospholipid molecules decreased with the increasing density of phospholipid molecules. Therefore, it is more difficult for GQDs to cross this energy barrier and enter the cell membrane with a high phospholipid membrane density.
To explore whether the change in phospholipid molecular density in the membrane would affect the penetration of GQDs, we constructed a low-density POPE membrane (POPE-LD). Without changing the number of phospholipid molecules (i.e., 342), the density of phospholipid molecules in the system can be changed by expanding the size of the outer frame of the cell membrane. The size of the simulated box was expanded from 9.0 × 9.0 × 10 nm3 to 9.6 × 9.6 × 10 nm3, and the position of GQDs was kept unchanged. By adding water molecules, the cell membrane was completely balanced for 10 ns under NVT conditions. During the balancing process, the energy and temperature of the system tend to be stable (see Supplementary Materials for details). This shows that after 10 ns NVT simulation, the cell membrane tended to equilibrium.
The simulation results are shown in Figure 5. Similar to DOPE and POPC, the GQDs reached the surface of the membrane at 40 ns and had completely entered the membrane at 50 ns. Similarly, the fully stabilized GQDs remained in the upper center of the membrane.
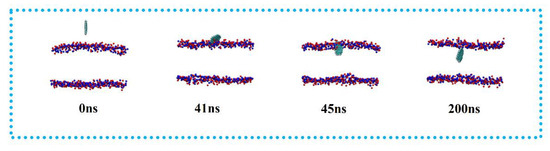
Figure 5.
Penetration of GQDs on POPE-LD membrane.
By expanding the size of the simulation box in the xy direction, the density of phospholipid molecules is reduced in the plane. In order to compare the density of phospholipid molecules more clearly, as shown in Figure 6, the density of the POPE membrane in a high-density state and a low-density state was calculated. There are large gaps between the phospholipid molecules with the lower density, which makes it easier for GQDs to enter the interior.
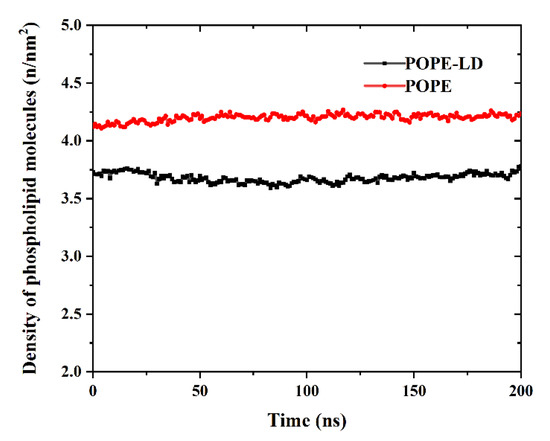
Figure 6.
Change in phospholipid molecular density of POPE membrane.
3.3. Free Energy Profile
To further explain the translocation mechanism of the GQDs on the cell membrane, PMF was used to calculate the shift in free energy of GQDs on POPE membranes, as shown in Figure 7. In the systems, the center of the cell membrane was defined as the zero point of free energy, and the surface free energy of GQDs from the liquid phase to the cell membrane decreased. It shows that GQDs are more inclined to adsorb on the surface of cell membranes than liquid phase water. However, compared with the POPE-LD membrane at low density, GQDs need to cross the higher energy barrier on the surface to enter the POPE membrane, which reduces the probability of GQDs penetrating the membrane surface of high-density phospholipid molecules. In addition, it is noted that the lowest points of free energy are located above the inner center of the cell membrane. In the PMF curve of POPC and DOPE cell membranes at low density (see Supplementary Materials for Figure S2), the free energy change of GQDs entering the cell membrane decreases, and the surface energy barrier is low. The lowest point of free energy is located inside the cell membrane, which is consistent with our MD results.
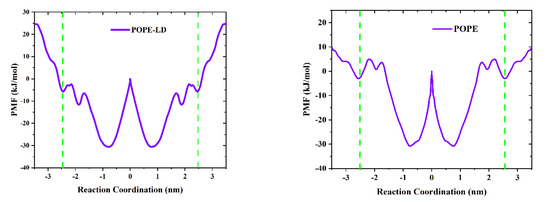
Figure 7.
The average force (PMF) potential of GQD transport through the membrane. The green dotted line indicates the two boundaries of the cell membrane.
GQDs can penetrate cell membranes, and interactions between them include electrostatic (Ele) and Van der Waals (VDW) forces. To explore the interactions between the GQDs and the cell membranes in all systems in the MD simulation, as shown in Figure 8, the changes in these two forces with time were calculated. Among them, VDW and Ele forces were zero, indicating that the GQDs were not yet in contact with the cell membrane. When touching the surface of the membrane, the GQDs were pulled into the membrane by the effects of VDW and Ele forces. From the overall calculation results, it can be observed that the effect of VDW force was significantly stronger than that of Ele force, which is the main driving force for GQDs to enter the cell membrane. However, in the high-density POPE system, the effects of VDW and Ele forces were weaker than those of the other systems, which is not sufficient to drive GQDs into the cell membrane.
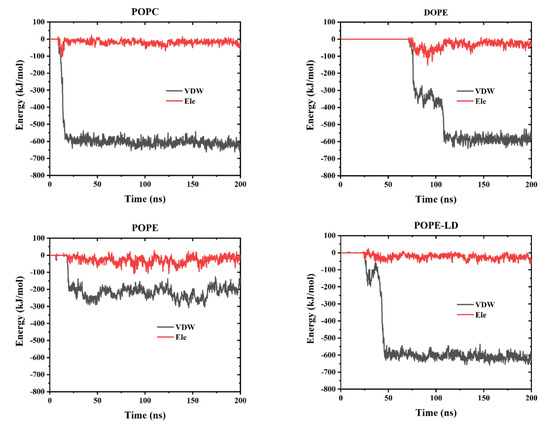
Figure 8.
The interaction energy between GQDs and cell membranes includes van der Waals interaction (VDW) and classical interaction (Ele).
4. Conclusions
MD simulations were used to explore the interactions between GQDs and different cell membranes, as well as to study the translocation process of GQDs in these settings. The simulation demonstrated that GQDs can easily permeate POPC and DOPE lipid membranes. For POPE lipid membranes, the GQDs can only be adsorbed onto the surface. This is consistent with the PMF calculation of the displacement of GQDs on this cell membrane. POPE has a high energy barrier, which makes it difficult for GQDs to cross. Analysis of the lipid membrane structure revealed that the density of phospholipid molecules is a key factor affecting the ability of GQDs to permeate the cell membrane. High-density POPE membranes are very difficult for GQDs to penetrate, whereas low-density POPE membranes allow GQDs to be transported into the intracellular space. The lipid density will have a great influence on the energy barrier affecting GQDs’ diffusion into the membrane. In real cell systems, the lipid densities are not uniform due to thermal fluctuation and cell movement. Some spots on the cell membrane will have a lower density, and these low-density spots will be the pathway for GQDs to diffuse into the cell membrane. This study provides a theoretical reference for further studies on the toxicity of GQDs and their transport across membranes.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/membranes12080753/s1, Figure S1: Changes in energy and temperature of the system, Figure S2: The average force (PMF) potential of GQDs transport through the membrane. The green dotted line indicates the two boundaries of the cell membrane. Table S1: Details of simulation calculations, Table S2: The LJ and charge parameters used in the MD. References [47,48,49,50,51,52] are cited in the supplementary materials.
Author Contributions
Formal analysis, F.J., L.W., W.H. and Y.Z.; Writing–review & editing, P.Z., Z.K. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant No. 21978060 and 11904300), the Key Research and Development Plan of Zhejiang Province (No. 2020C01008), and the Natural Science Foundation of Zhejiang Province (Grant No. LY21B060004). This research was also funded by the National Natural Science Foundation of China (NSFC) (No. 22073053), the Young Taishan Scholar Program of Shandong Province (No. tsqn201909139), and the Program for Scientific Research Innovation Team in Colleges and Universities of Jinan (No. 2021GXRC042).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare that there is no conflict of interest.
References
- Chung, S.; Revia, R.A.; Zhang, M. Graphene Quantum Dots and Their Applications in Bioimaging, Biosensing, and Therapy. Adv. Mater. 2021, 33, 1904362. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, G.F.; Zhu, L.; Li, G.X. Graphene quantum dots-based platform for the fabrication of electrochemical biosensors. Electrochem. Commun. 2011, 13, 31–33. [Google Scholar] [CrossRef]
- Kappen, J.; Ponkarpagam, S.; John, S.A. Study on the interactions between graphene quantum dots and Hg(II): Unraveling the origin of photoluminescence quenching of graphene quantum dots by Hg(II). Colloids Surf. A Physicochem. Eng. Asp. 2020, 591, 124551. [Google Scholar] [CrossRef]
- Mnasri, G.; Mansouri, S.; Yalcin, M.; ElMir, L. Characterization and study of CdS quantum dots solar cells based on Graphene-TiO2 nanocomposite photoanode. Results Phys. 2020, 18, 103253. [Google Scholar] [CrossRef]
- Danial, W.H.; Mohamed, N.A.S.; Majid, Z.A.J.C.L. Recent advances on the preparation and application of graphene quantum dots for mercury detection: A systematic review. Carbon Lett. 2022, 32, 57–80. [Google Scholar] [CrossRef]
- Liu, Y.X.; Dong, X.C.; Chen, P. Biological and chemical sensors based on graphene materials. Chem. Soc. Rev. 2012, 41, 2283–2307. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Barnard, A.S.; Snook, I.K. Quantum mechanical properties of graphene nano-flakes and quantum dots. Nanoscale 2012, 4, 6761–6767. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.H.; Bu, H.; Zou, M.; Yi, H.; Bi, K.D.; Chen, Y.F. Anisotropic mechanical properties of graphene sheets from molecular dynamics. Phys. B Condens. Matter 2010, 405, 1301–1306. [Google Scholar] [CrossRef]
- Shen, J.L.; Chen, W.F.; Liu, X. Facile synthesis of graphene quantum dots from glucan and their application as a deoxidizer and in cell imaging. J. Chem. Res. 2020, 45, 242–247. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, Y.; Cheng, H.H.; Hu, Y.; Shi, G.Q.; Dai, L.M.; Qu, L.T. Nitrogen-doped graphene quantum dots with oxygen-rich functional groups. J. Am. Chem. Soc. 2012, 134, 15–18. [Google Scholar] [CrossRef]
- Wang, X.J.; Sun, X.W.; Lao, J.; He, H.; Cheng, T.T. Multifunctional graphene quantum dots for simultaneous targeted cellular imaging and drug delivery. Colloids Surf. B Biointerfaces 2014, 122, 638–644. [Google Scholar] [CrossRef]
- Ding, H.; Zhang, F.; Zhao, C.; Lv, Y.; Ma, G.; Wei, W.; Tian, Z. Beyond a Carrier: Graphene Quantum Dots as a Probe for Programmatically Monitoring Anti-Cancer Drug Delivery, Release, and Response. ACS Appl. Mater. Interfaces 2017, 9, 27396–27401. [Google Scholar] [CrossRef] [PubMed]
- Pasinszki, T.; Krebsz, M.; Tung, T.T.; Losic, D. Carbon Nanomaterial Based Biosensors for Non-Invasive Detection of Cancer and Disease Biomarkers for Clinical Diagnosis. Sensors 2017, 17, 1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, S.; Luo, J.S.; Wang, Q.; Li, Z.J.; Li, J.T.; Liu, Q.; Gao, L.Q.; Fang, S.H.; Li, Y.Y.; Pan, H.F. Targeted graphene oxide for drug delivery as a therapeutic nanoplatform against Parkinson’s disease. Biomater. Sci. 2021, 9, 1705–1715. [Google Scholar] [CrossRef]
- Bressan, E.; Ferroni, L.; Gardin, C.; Sbricoli, L.; Gobbato, L.; Ludovichetti, F.S.; Tocco, I.; Carraro, A.; Piattelli, A.; Zavan, B. Graphene based scaffolds effects on stem cells commitment. J. Transl. Med. 2014, 12, 296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, X.B.; Zhu, Z.; Gan, Z.; Chen, J.; Zhang, X.; Cheng, X.; Wan, Q.; Wang, J. PEGylated nano-graphene oxide as a nanocarrier for delivering mixed anticancer drugs to improve anticancer activity. Sci. Rep. 2020, 10, 2717. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.Y.; Wang, C.; Han, T.; Zhou, X.J. Insight into the cellular internalization and cytotoxicity of graphene quantum dots. Adv. Healthc. Mater. 2013, 2, 1613–1619. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Lei, R.; Huang, H.D.; Wang, N.; Yuan, L.; Xiao, R.Y.; Bai, L.D.; Li, X.; Li, L.M.; Yang, X.D. The permeability and transport mechanism of graphene quantum dots (GQDs) across the biological barrier. Nanoscale 2015, 7, 2034–2041. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.L.; Yu, T.W.; Chiang, W.H.; Chiu, H.C.; Chang, C.H.; Chiang, C.S.; Hu, S.H. Hierarchically Targeted and Penetrated Delivery of Drugs to Tumors by Size-Changeable Graphene Quantum Dot Nanoaircrafts for Photolytic Therapy. Adv. Funct. Mater. 2017, 27, 1700056. [Google Scholar] [CrossRef]
- Jin, K.X.; Gao, H.; Lai, L.H.; Pang, Y.Q.; Zheng, S.Y. Preparation of highly fluorescent sulfur doped graphene quantum dots for live cell imaging. J. Lumin. 2018, 197, 147–152. [Google Scholar] [CrossRef]
- Suwalsky, M.; Manrique, M.; Villena, F.; Sotomayor, C. Phospholipid Bilayers as Molecular Models for Drug-Cell Membrana Interactions. The Case of the Antiinflamatory Drug Diclofenac. Macromol. Symp. 2010, 296, 436–445. [Google Scholar] [CrossRef]
- Willems, N.; Urtizberea, A.; Verre, A.F.; Iliut, M.; Lelimousin, M.; Hirtz, M.; Vijayaraghavan, A.; Sansom, M.S. Biomimetic Phospholipid Membrane Organization on Graphene and Graphene Oxide Surfaces: A Molecular Dynamics Simulation Study. ACS Nano 2017, 11, 1613–1625. [Google Scholar] [CrossRef] [Green Version]
- Murzyn, K.; Rog, T.; Pasenkiewicz-Gierula, M. Phosphatidylethanolamine-phosphatidylglycerol bilayer as a model of the inner bacterial membrane. Biophys. J. 2005, 88, 1091–1103. [Google Scholar] [CrossRef] [Green Version]
- Urbina, J.A.; Moreno, B.; Arnold, W.; Taron, C.H.; Orlean, P.; Oldfield, E. A carbon-13 nuclear magnetic resonance spectroscopic study of inter-proton pair order parameters: A new approach to study order and dynamics in phospholipid membrane systems. Biophys. J. 1998, 75, 1372–1383. [Google Scholar] [CrossRef] [Green Version]
- Thurmond, R.; Dodd, S.; Brown, M. Molecular areas of phospholipids as determined by 2H NMR spectroscopy. Comparison of phosphatidylethanolamines and phosphatidylcholines. Biophys. J. 1991, 59, 108–113. [Google Scholar] [CrossRef] [Green Version]
- Galanis, E.; Hersh, E.M.; Stopeck, A.T.; Gonzalez, R.; Rubin, J. Immunotherapy of advanced malignancy by direct gene transfer of an interleukin-2 DNA/DMRIE/DOPE lipid complex: Phase I/II experience. J. Clin. Oncol. 1999, 17, 3313–3323. [Google Scholar] [CrossRef] [PubMed]
- Lindorff-Larsen, K.; Trbovic, N.; Maragakis, P.; Piana, S.; Shaw, D.E. Structure and dynamics of an unfolded protein examined by molecular dynamics simulation. J. Am. Chem. Soc. 2012, 134, 3787–3791. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhou, G.; Chen, L.; Wang, Y.; Wang, X.; Zeng, S. Interaction of Graphene and its Oxide with Lipid Membrane: A Molecular Dynamics Simulation Study. J. Phys. Chem. C 2016, 120, 6225–6231. [Google Scholar] [CrossRef]
- Cheng, C.L.; Zhao, G.J. Steered molecular dynamics simulation study on dynamic self-assembly of single-stranded DNA with double-walled carbon nanotube and graphene. Nanoscale 2012, 4, 2301–2305. [Google Scholar] [CrossRef]
- Ji, Y.; Zhu, R.Y.; Shen, Y.; Tan, Q.; Chen, J.L. Comparison of loading and unloading of different small drugs on graphene and its oxide. J. Mol. Liq. 2021, 341, 117454. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, S.; Li, Y.; Wang, M.; Shi, P.; Huang, X. Covalent functionalization of graphene oxide with biocompatible poly(ethylene glycol) for delivery of paclitaxel. ACS Appl. Mater. Interfaces 2014, 6, 17268–17276. [Google Scholar] [CrossRef] [PubMed]
- Safdari, F.; Raissi, H.; Shahabi, M.; Zaboli, M. DFT Calculations and Molecular Dynamics Simulation Study on the Adsorption of 5-Fluorouracil Anticancer Drug on Graphene Oxide Nanosheet as a Drug Delivery Vehicle. J. Inorg. Organomet. Polym. Mater. 2017, 27, 805–817. [Google Scholar] [CrossRef]
- Liang, L.J.; Kong, Z.; Kang, Z.S.; Wang, H.B.; Zhang, L.; Shen, J.W. Theoretical Evaluation on Potential Cytotoxicity of Graphene Quantum Dots. ACS Biomater. Sci. Eng. 2016, 2, 1983–1991. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Hu, W.; Jiao, F.F.; Zhang, P.Z.; Shen, J.W.; Cui, B.; Wang, H.B.; Liang, L.L. Theoretical Evaluation of DNA Genotoxicity of Graphene Quantum Dots: A Combination of Density Functional Theory and Molecular Dynamics Simulations. J. Phys. Chem. B 2020, 124, 9335–9342. [Google Scholar] [CrossRef]
- Xue, Z.; Sun, Q.; Zhang, L.; Kang, Z.; Liang, L.; Wang, Q.; Shen, J.W. Graphene quantum dot assisted translocation of drugs into a cell membrane. Nanoscale 2019, 11, 4503–4514. [Google Scholar] [CrossRef]
- Kang, Y.; Zhang, Z.S.; Shi, H.; Zhang, J.Q.; Liang, L.J.; Wang, Q.; Agren, H.; Tu, Y.Q. Na(+) and K(+) ion selectivity by size-controlled biomimetic graphene nanopores. Nanoscale 2014, 6, 10666–10672. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Trucks, G.; Schlegel, H.e.a.; Scuseria, G.; Robb, M.; Cheeseman, J.; Montgomery Jr., J.; Vreven, T.; Kudin, K.; Burant, J. Gaussian 03, revision C. 02; Gaussian, Inc.: Wallingford, CT, USA, 2004. [Google Scholar]
- Wu, E.L.; Cheng, X.; Jo, S.; Rui, H.; Song, K.C.; Davila-Contreras, E.M.; Qi, Y.; Lee, J.; Monje-Galvan, V.; Venable, R.M.; et al. CHARMM-GUI Membrane Builder toward realistic biological membrane simulations. J. Comput. Chem. 2014, 35, 1997–2004. [Google Scholar] [CrossRef] [Green Version]
- Witzke, S.; Petersen, M.; Carpenter, T.S.; Khalid, S. Molecular Dynamics Simulations Reveal the Conformational Flexibility of Lipid II and Its Loose Association with the Defensin Plectasin in the Staphylococcus aureus Membrane. Biochemistry 2016, 55, 3303–3314. [Google Scholar] [CrossRef] [Green Version]
- Inui, N. Equilibrium shape of a suspended graphene sheet under electrostatic and van der Waals forces. J. Phys. D Appl. Phys. 2018, 51, 115303. [Google Scholar] [CrossRef]
- Klauda, J.B.; Venable, R.M.; Freites, J.A.; O’Connor, J.; Tobias, D.J.; Mondragon-Ramirez, C.; Vorobyov, I.; Mackerell, A.D.; Pastor, R.W. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J. Phys. Chem. B 2010, 114, 7830–7843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucerka, N.; Tristram-Nagle, S.; Nagle, J.F. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J. Membr. Biol. 2005, 208, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Janosi, L.; Gorfe, A.A. Simulating POPC and POPC/POPG Bilayers: Conserved Packing and Altered Surface Reactivity. J. Chem. Theory Comput. 2010, 6, 3267. [Google Scholar] [CrossRef] [PubMed]
- Smaby, J.M.; Momsen, M.M.; Brockman, H.L.; Brown, R.E. Phosphatidylcholine acyl unsaturation modulates the decrease in interfacial elasticity induced by cholesterol. Biophys. J. 1997, 73, 1492–1505. [Google Scholar] [CrossRef] [Green Version]
- Torrie, G.M.; Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: Umbrella sampling. J. Comput. Phys. 1977, 23, 187–199. [Google Scholar] [CrossRef]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Mja, A.; Tm, D.; Rsb, C.; Sp, A.; Jcsb, C.; Bh, A.; Ela, D. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers - ScienceDirect. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Cohen-Tanugi, D.; Grossman, J.C. Water Desalination across Nanoporous Graphene. Nano Lett. 2012, 12, 3602–3608. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Y.; Yang, J.; Bi, K.; Ni, Z.; Li, D.; Chen, Y. Molecular dynamics study of DNA translocation through graphene nanopores. Phys. Rev. E Stat. Nonlin Soft Matter. Phys. 2013, 87, 062707. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, B.; Wang, X. Cholesterol Extraction from Cell Membrane by Graphene Nanosheets: A Computational Study. J. Phys. Chem. B 2016, 120, 957–964. [Google Scholar] [CrossRef]
- Beu, T.A. Molecular dynamics simulations of ion transport through carbon nanotubes. I. Influence of geometry, ion specificity, and many-body interactions. J. Chem. Phys. 2010, 132, 164513. [Google Scholar] [CrossRef] [Green Version]
- Mooney, D.A.; Müller-Plathe, F.; Kremer, K.J.C.P.L. Simulation studies for liquid phenol: Properties evaluated and tested over a range of temperatures. Chem. Phys. Lett. 1998, 294, 135–142. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).